A Concerted Action of UBA5 C-Terminal Unstructured Regions Is Important for Transfer of Activated UFM1 to UFC1
Abstract
:1. Introduction
2. Results
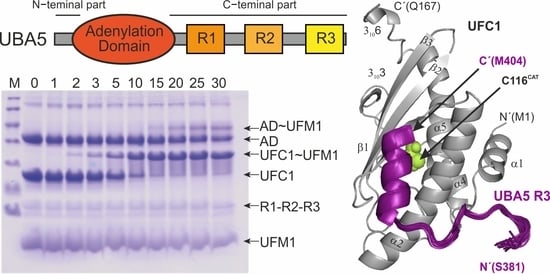
2.1. The UBA5 C-Terminal Part Is a Regulatory Platform for the Ufmylation Cascade
2.2. Interactions between Different UBA5 C-Terminal Regions and UFC1, UFM1 and LC3/GABRAP Proteins
2.3. Structure of UFC1 in Complex with the UBA5 R3 Peptide
2.4. Interactions within the Ufmylation Cascade
3. Discussion
3.1. The UFC1:UBA5 Interaction
3.2. Interaction between GABARAPL2 and UBA5 C-Terminal Part
3.3. The Role of the A371T Mutation in the Ufmylation Cascade
4. Materials and Methods
4.1. DNA Constructs Used in This Study
4.2. Expression, Isolation and Purification of the Peptides and Proteins
4.3. In Vitro Thioester Formation Assay
4.4. Isothermal Titration Calorimetry
4.5. NMR Spectroscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | UBA5 adenylation domain |
| ASC1 | activating signal co-integrator 1 |
| ATP | adenosine triphosphate |
| BEST | band-selective excitation short-transient |
| CSP | chemical shift perturbation |
| FL | full length |
| GABARAP | GABAA-receptor-associated protein |
| HMQC | heteronuclear multiple quantum coherence |
| HSQC | heteronuclear single quantum coherence |
| ITC | isothermal titration calorimetry |
| LC3 | microtubule-associated protein 1 light chain 3 |
| LIR | LC3-interacting region |
| NEDD8 | neural precursor cell expressed developmentally downregulated protein 8 |
| NMR | nuclear magnetic resonance |
| NOESY | nuclear Overhauser and exchange spectroscopy |
| R1, R2, R3 | UBA5 C-terminal regions R1, R2 and R3 |
| SD | standard deviation |
| SOFAST | band-selective optimized-flip-angle short-transient |
| SUMO | small ubiquitin related modifier |
| TOCSY | total correlation spectroscopy |
| TROSY | transverse relaxation optimized spectroscopy |
| UBA5 | UFM1-activating enzyme 5 |
| UBL | ubiquitin-like |
| UFBP1 | UFM1-binding protein 1 |
| UFC1 | UFM1-conjugating enzyme 1 |
| UFIM | UFM1-interacting motive |
| UFL1 | UFM1 ligase 1 |
| UFM1 | Ubiquitin fold modifier 1 |
| UfSP1/2 | UFM1-specific proteases 1 and 2 |
References
- Komatsu, M.; Chiba, T.; Tatsumi, K.; Iemura, S.i.; Tanida, I.; Okazaki, N.; Ueno, T.; Kominami, E.; Natsume, T.; Tanaka, K. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 2004, 23, 1977–1986. [Google Scholar] [CrossRef] [Green Version]
- Sasakawa, H.; Sakata, E.; Yamaguchi, Y.; Komatsu, M.; Tatsumi, K.; Kominami, E.; Tanaka, K.; Kato, K. Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1. Biochem. Biophys. Res. Commun. 2006, 343, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Kumar, M.; Wiener, R. Decrypting UFMylation: How Proteins Are Modified with UFM1. Biomolecules 2020, 10, 1442. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.; Liebau, E. The ufm1 cascade. Cells 2014, 3, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Ha, B.H.; Ahn, H.-C.; Kang, S.H.; Tanaka, K.; Chung, C.H.; Kim, E.E. Structural basis for Ufm1 processing by UfSP1. J. Biol. Chem. 2008, 283, 14893–14900. [Google Scholar] [CrossRef] [Green Version]
- Ha, B.H.; Jeon, Y.J.; Shin, S.C.; Tatsumi, K.; Komatsu, M.; Tanaka, K.; Watson, C.M.; Wallis, G.; Chung, C.H.; Kim, E.E. Structure of ubiquitin-fold modifier 1-specific protease UfSP2. J. Biol. Chem. 2011, 286, 10248–10257. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.H.; Kim, G.R.; Seong, M.; Baek, S.H.; Seol, J.H.; Bang, O.S.; Ovaa, H.; Tatsumi, K.; Komatsu, M.; Tanaka, K. Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2. J. Biol. Chem. 2007, 282, 5256–5262. [Google Scholar] [CrossRef] [Green Version]
- Bacik, J.-P.; Walker, J.R.; Ali, M.; Schimmer, A.D.; Dhe-Paganon, S. Crystal Structure of the Human Ubiquitin-activating Enzyme 5 (UBA5) Bound to ATP Mechanistic Insights into a Minimalistic E1 Enzyme. J. Biol. Chem. 2010, 285, 20273–20280. [Google Scholar] [CrossRef] [Green Version]
- Gavin, J.M.; Hoar, K.; Xu, Q.; Ma, J.; Lin, Y.; Chen, J.; Chen, W.; Bruzzese, F.J.; Harrison, S.; Mallender, W.D. Mechanistic study of Uba5 enzyme and the Ufm1 conjugation pathway. J. Biol. Chem. 2014, 289, 22648–22658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padala, P.; Oweis, W.; Mashahreh, B.; Soudah, N.; Cohen-Kfir, E.; Todd, E.A.; Berndsen, C.E.; Wiener, R. Novel insights into the interaction of UBA5 with UFM1 via a UFM1-interacting sequence. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, T.; Tatsumi, K.; Ozaki, Y.; Kawakami, T.; Suzuki, A.; Ogasahara, K.; Komatsu, M.; Kominami, E.; Tanaka, K.; Yamane, T. Crystal structure of Ufc1, the Ufm1-conjugating enzyme. Biochem. Biophys. Res. Commun. 2007, 362, 1079–1084. [Google Scholar] [CrossRef]
- Tatsumi, K.; Sou, Y.-S.; Tada, N.; Nakamura, E.; Iemura, S.-I.; Natsume, T.; Kang, S.H.; Chung, C.H.; Kasahara, M.; Kominami, E. A novel type of E3 ligase for the Ufm1 conjugation system. J. Biol. Chem. 2010, 285, 5417–5427. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.R.; Lingeman, E.; Luong, T.; Ahmed, S.; Muhar, M.; Nguyen, T.; Olzmann, J.A.; Corn, J.E. A Genome-wide ER-phagy Screen Highlights Key Roles of Mitochondrial Metabolism and ER-Resident UFMylation. Cell 2020, 180, 1160–1177.e1120. [Google Scholar] [CrossRef]
- Walczak, C.P.; Leto, D.E.; Zhang, L.; Riepe, C.; Muller, R.Y.; DaRosa, P.A.; Ingolia, N.T.; Elias, J.E.; Kopito, R.R. Ribosomal protein RPL26 is the principal target of UFMylation. Proc. Natl. Acad. Sci. USA 2019, 116, 1299–1308. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Wang, H.; Kang, B.; Chen, B.; Shi, Y.; Yang, S.; Sun, L.; Liu, Y.; Xiao, W.; Zhang, T. CDK5RAP3, a UFL1 substrate adaptor, is crucial for liver development. Development 2019, 146, dev169235. [Google Scholar] [CrossRef] [Green Version]
- Yoo, H.M.; Kang, S.H.; Kim, J.Y.; Lee, J.E.; Seong, M.W.; Lee, S.W.; Ka, S.H.; Sou, Y.-S.; Komatsu, M.; Tanaka, K. Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol. Cell 2014, 56, 261–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Guan, D.; Dong, M.; Yang, J.; Wei, H.; Liang, Q.; Song, L.; Xu, L.; Bai, J.; Liu, C. UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nat. Cell Biol. 2020, 22, 1056–1063. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Yu, J.; Nowsheen, S.; Wang, M.; Tu, X.; Liu, T.; Li, H.; Wang, L.; Lou, Z. UFL1 promotes histone H4 ufmylation and ATM activation. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gong, Y.; Peng, B.; Shi, R.; Fan, D.; Zhao, H.; Zhu, M.; Zhang, H.; Lou, Z.; Zhou, J. MRE11 UFMylation promotes ATM activation. Nucleic Acids Res. 2019, 47, 4124–4135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gak, I.A.; Vasiljevic, D.; Zerjatke, T.; Yu, L.; Brosch, M.; Roumeliotis, T.I.; Horenburg, C.; Klemm, N.; Bakos, G.; Herrmann, A. UFMylation regulates translational homeostasis and cell cycle progression. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Pi, W.; Sivaprakasam, S.; Zhu, X.; Zhang, M.; Chen, J.; Makala, L.; Lu, C.; Wu, J.; Teng, Y. UFBP1, a key component of the Ufm1 conjugation system, is essential for ufmylation-mediated regulation of erythroid development. PLoS Genet. 2015, 11, e1005643. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhang, H.; Song, Y.; Zhuang, L.; Yang, Q.; Pan, M.; Chen, F. Ubiquitin fold modifier 1 activates NF-κB pathway by down-regulating LZAP expression in the macrophage of diabetic mouse model. Biosci. Rep. 2020, 40, BSR20191672. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, K.; Moura, R.F.; Granvik, M.; Igoillo-Esteve, M.; Hohmeier, H.E.; Hendrickx, N.; Newgard, C.B.; Waelkens, E.; Cnop, M.; Schuit, F. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS ONE 2011, 6, e18517. [Google Scholar] [CrossRef] [Green Version]
- Roberts, A.M.; Miyamoto, D.K.; Huffman, T.R.; Bateman, L.A.; Ives, A.N.; Akopian, D.; Heslin, M.J.; Contreras, C.M.; Rape, M.; Skibola, C.F. Chemoproteomic screening of covalent ligands reveals UBA5 as a novel pancreatic cancer target. ACS Chem. Biol. 2017, 12, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Muona, M.; Ishimura, R.; Laari, A.; Ichimura, Y.; Linnankivi, T.; Keski-Filppula, R.; Herva, R.; Rantala, H.; Paetau, A.; Pöyhönen, M. Biallelic variants in UBA5 link dysfunctional UFM1 ubiquitin-like modifier pathway to severe infantile-onset encephalopathy. Am. J. Hum. Genet. 2016, 99, 683–694. [Google Scholar] [CrossRef] [Green Version]
- Nahorski, M.S.; Maddirevula, S.; Ishimura, R.; Alsahli, S.; Brady, A.F.; Begemann, A.; Mizushima, T.; Guzmán-Vega, F.J.; Obata, M.; Ichimura, Y. Biallelic UFM1 and UFC1 mutations expand the essential role of ufmylation in brain development. Brain 2018, 141, 1934–1945. [Google Scholar] [CrossRef] [Green Version]
- Tatsumi, K.; Yamamoto-Mukai, H.; Shimizu, R.; Waguri, S.; Sou, Y.-S.; Sakamoto, A.; Taya, C.; Shitara, H.; Hara, T.; Chung, C.H. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2011, 2, 1–7. [Google Scholar] [CrossRef]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like protein conjugation: Structures, chemistry, and mechanism. Chem. Rev. 2018, 118, 889–918. [Google Scholar] [CrossRef] [PubMed]
- Oweis, W.; Padala, P.; Hassouna, F.; Cohen-Kfir, E.; Gibbs, D.R.; Todd, E.A.; Berndsen, C.E.; Wiener, R. Trans-binding mechanism of ubiquitin-like protein activation revealed by a UBA5-UFM1 complex. Cell Rep. 2016, 16, 3113–3120. [Google Scholar] [CrossRef] [Green Version]
- Soudah, N.; Padala, P.; Hassouna, F.; Kumar, M.; Mashahreh, B.; Lebedev, A.A.; Isupov, M.N.; Cohen-Kfir, E.; Wiener, R. An N-terminal extension to Uba5 adenylation domain boosts Ufm1 activation: Isoform-specific differences in ubiquitin-like protein activation. J. Mol. Biol. 2019, 431, 463–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habisov, S.; Huber, J.; Ichimura, Y.; Akutsu, M.; Rogova, N.; Loehr, F.; McEwan, D.G.; Johansen, T.; Dikic, I.; Doetsch, V. Structural and functional analysis of a novel interaction motif within UFM1-activating enzyme 5 (UBA5) required for binding to ubiquitin-like proteins and ufmylation. J. Biol. Chem. 2016, 291, 9025–9041. [Google Scholar] [CrossRef] [Green Version]
- Xie, S. Characterization, crystallization and preliminary X-ray crystallographic analysis of the human Uba5 C-terminus–Ufc1 complex. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 1093–1097. [Google Scholar] [CrossRef] [Green Version]
- Huber, J.; Obata, M.; Gruber, J.; Akutsu, M.; Löhr, F.; Rogova, N.; Güntert, P.; Dikic, I.; Kirkin, V.; Komatsu, M. An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5. Autophagy 2020, 16, 256–270. [Google Scholar] [CrossRef] [PubMed]
- Colin, E.; Daniel, J.; Ziegler, A.; Wakim, J.; Scrivo, A.; Haack, T.B.; Khiati, S.; Denommé, A.-S.; Amati-Bonneau, P.; Charif, M. Biallelic variants in UBA5 reveal that disruption of the UFM1 cascade can result in early-onset encephalopathy. Am. J. Hum. Genet. 2016, 99, 695–703. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Forouhar, F.; Eletsky, A.; Atreya, H.S.; Aramini, J.M.; Xiao, R.; Huang, Y.J.; Abashidze, M.; Seetharaman, J.; Liu, J. NMR and X-ray structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway. J. Struct. Funct. Genom. 2009, 10, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Rogov, V.V.; Rozenknop, A.; Rogova, N.Y.; Löhr, F.; Tikole, S.; Jaravine, V.; Güntert, P.; Dikic, I.; Dötsch, V. A universal expression tag for structural and functional studies of proteins. ChemBioChem 2012, 13, 959–963. [Google Scholar] [CrossRef]
- von Delbrück, M.; Kniss, A.; Rogov, V.V.; Pluska, L.; Bagola, K.; Löhr, F.; Güntert, P.; Sommer, T.; Dötsch, V. The CUE domain of Cue1 aligns growing ubiquitin chains with Ubc7 for rapid elongation. Mol. Cell 2016, 62, 918–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buetow, L.; Gabrielsen, M.; Anthony, N.G.; Dou, H.; Patel, A.; Aitkenhead, H.; Sibbet, G.J.; Smith, B.O.; Huang, D.T. Activation of a primed RING E3-E2–ubiquitin complex by non-covalent ubiquitin. Mol. Cell 2015, 58, 297–310. [Google Scholar] [CrossRef] [Green Version]
- Dou, H.; Buetow, L.; Sibbet, G.J.; Cameron, K.; Huang, D.T. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat. Struct. Mol. Biol. 2013, 20, 982–986. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Magala, P.; Geiger-Schuller, K.R.; Majumdar, A.; Tolman, J.R.; Wolberger, C. Role of a non-canonical surface of Rad6 in ubiquitin conjugating activity. Nucleic Acids Res. 2015, 43, 9039–9050. [Google Scholar] [CrossRef] [Green Version]
- Eck, F.; Phuyal, S.; Smith, M.D.; Kaulich, M.; Wilkinson, S.; Farhan, H.; Behrends, C. ACSL3 is a novel GABARAPL2 interactor that links ufmylation and lipid droplet biogenesis. J. Cell Sci. 2020, 133, jcs243477. [Google Scholar] [CrossRef] [PubMed]
- Farjon, J.; Boisbouvier, J.; Schanda, P.; Pardi, A.; Simorre, J.-P.; Brutscher, B. Longitudinal-relaxation-enhanced NMR experiments for the study of nucleic acids in solution. J. Am. Chem. Soc. 2009, 131, 8571–8577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solyom, Z.; Schwarten, M.; Geist, L.; Konrat, R.; Willbold, D.; Brutscher, B. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J. Biomol. NMR 2013, 55, 311–321. [Google Scholar] [CrossRef]
- Logan, T.M.; Olejniczak, E.T.; Xu, R.X.; Fesik, S.W. A general method for assigning NMR spectra of denatured proteins using 3D HC (CO) NH-TOCSY triple resonance experiments. J. Biomol. NMR 1993, 3, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Montelione, G.T.; Lyons, B.A.; Emerson, S.D.; Tashiro, M. An efficient triple resonance experiment using carbon-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically-enriched proteins. J. Am. Chem. Soc. 1992, 114, 10974–10975. [Google Scholar] [CrossRef]
- Löhr, F.; Hänsel, R.; Rogov, V.V.; Dötsch, V. Improved pulse sequences for sequence specific assignment of aromatic proton resonances in proteins. J. Biomol. NMR 2007, 37, 205–224. [Google Scholar] [CrossRef]
- Brutscher, B.; Boisbouvier, J.; Pardi, A.; Marion, D.; Simorre, J.-P. Improved sensitivity and resolution in 1H− 13C NMR experiments of RNA. J. Am. Chem. Soc. 1998, 120, 11845–11851. [Google Scholar] [CrossRef]
- Pervushin, K.; Riek, R.; Wider, G.; Wüthrich, K. Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in 13C-labeled proteins. J. Am. Chem. Soc. 1998, 120, 6394–6400. [Google Scholar] [CrossRef]
- Zwahlen, C.; Legault, P.; Vincent, S.J.; Greenblatt, J.; Konrat, R.; Kay, L.E. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage λ N-peptide/boxB RNA complex. J. Am. Chem. Soc. 1997, 119, 6711–6721. [Google Scholar] [CrossRef]
- Güntert, P. Automated structure determination from NMR spectra. Eur. Biophys. J. 2009, 38, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Berjanskii, M.V.; Neal, S.; Wishart, D.S. PREDITOR: A web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006, 34, W63–W69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koradi, R.; Billeter, M.; Güntert, P. Point-centered domain decomposition for parallel molecular dynamics simulation. Comput. Phys. Commun. 2000, 124, 139–147. [Google Scholar] [CrossRef]




| DNA Construct | Expressed Protein/Peptide | Short Description | References |
|---|---|---|---|
| pET39_Ub19_UBA51–404 | FL1–404 | Full length UBA5, residues 1–404 | [31] |
| pET39_Ub19_UBA5325–404 | R1-R2-R3325–404 | UBA5 C-terminal part, residues 325–404 | [31] |
| pET39_Ub19_UFM1 | UFM1 | Full length UFM1, residues 2–83 | [31] |
| pETm60_Ub3_LC3A | LC3A | LC3A, residues 4–120 | [36] |
| pETm60_Ub3_LC3B | LC3B | LC3B, residues 5–120 | [36] |
| pET39_Ub19_LC3C | LC3C | LC3C, residues 5–126 | [36] |
| pET39_Ub19_GABARAP | GABARAP | GABARAP, residues 3–116 | [36] |
| pETm60_Ub3_GABARAPL1 | GABARAPL1 | GABARAPL1, residues 2–116 | [36] |
| pET39_Ub19_GABARAPL2 | GABARAPL2 | GABARAPL2, residues 3–116 | [36] |
| pETm60_Ub | Ubiquitin | Ubiquitin, residues 1–76 | [37] |
| pET39_Ub19_UFC1 | UFC1 | Full length UFC1, residues 1–167 | This work |
| pET39_Ub19_UBA51–330 | AD1–330 | UBA5 adenylation domain, residues 1–330 | This work |
| pET39_Ub19_UBA5325–376 | R1-R2325–376 | UBA5 C-terminal regions R1 and R2, residues 325–376 | This work |
| pET39_Ub19_UBA5359–404 | R2-R3359–404 | UBA5 C-terminal regions R2 and R3, residues 359–404 | This work |
| pET39_Ub19_UBA5325–357 | R1325–357 | UBA5 C-terminal region R1, residues 325–357 | This work |
| pET39_Ub19_UBA5359–376 | R2359–376 | UBA5 C-terminal region R2, residues 359–376 | This work |
| pET39_Ub19_UBA5388–404 | R3388–404 | UBA5 C-terminal region R3, residues 388–404 | This work |
| pET39_Ub19_UBA5381–404W | R3381–404W | Optimized R3, residues 381–404 with C-terminal W | This work |
| pET39_Ub19_UBA5325–404 A371T | R1-R2-R3325–404 A371T | UBA5 C-terminal part with A371T mutant (res. 325–404) | This work |
| pET39_Ub19_UBA5325–404 A371E | R1-R2-R3325–404 A371E | UBA5 C-terminal part with A371E mutant (res. 325–404) | This work |
| pET39_Ub19_UBA51–380 | ΔR31–380 | UBA5 with deleted R3 region, residues 1–380 | This work |
| pNiC-CTH0_UBA51–404 C250K | FL1–404 C250K | Full length UBA5 (res. 1–404) with C250K mutant | This work |
| pET39_Ub19_UBA51–330 C250K 0 | AD1–330 C250K | UBA5 adenylation domain with C250K mutant | This work |
| pNiC-CTH0_UFC1 | UFC1_His6 | Full length UFC1 with C-terminal hexahistidine-tag | This work |
| Proteins | UBA5 Regions | ΔH (kcal mol−1) | ΔS (cal mol−1 K−1) | −T*ΔS (kcal mol−1) | ΔG (kcal mol−1) | KA*10−6 (M−1) | KD (µM) | N |
|---|---|---|---|---|---|---|---|---|
| UFM1 | R1-R2-R3325–404 | −5.83 ± 0.20 * | 4.41 | −1.31 | −7.15 | 0.17 ± 0.02 | 5.8 | 1.04 ± 0.03 |
| R1-R2325–376 | −5.61 ± 0.21 | 4.99 | −1.49 | −7.10 | 0.16 ± 0.01 | 6.2 | 0.96 ± 0.03 | |
| R2359–376 | ND | ND | >100 ** | ND | ||||
| R1-R2-R3325–404 A371T | −10.99 ± 0.25 | −13.3 | 3.96 | −7.02 | 0.14 ± 0.01 | 7.1 | 1.12 ± 0.02 | |
| R1-R2-R3325–404 A371E | −11.42 ± 0.42 | −15.3 | 4.56 | −6.86 | 0.11 ± 0.01 | 9.2 | 1.01 ± 0.01 | |
| UFC1 | R1-R2-R3325–404 | −7.04 ± 0.07 | 2.09 | −0.62 | −7.66 | 0.41 ± 0.02 | 2.4 | 1.03 ± 0.01 |
| R2-R3359–404 | −8.08 ± 0.10 | 1.59 | −0.47 | −7.60 | 0.37 ± 0.01 | 2.7 | 0.97 ± 0.01 | |
| R3388–404 | −4.99 ± 0.22 | 6.91 | −2.06 | −7.05 | 0.03 ± 0.003 | 10 | 0.95 ± 0.03 | |
| R2359–376 | ND | ND | >50 ** | ND | ||||
| R1-R2-R3325–404 A371T | −7.88 ± 0.08 | −0.19 | 0.06 | −7.82 | 0.54 ± 0.03 | 1.8 | 1.03 ± 0.008 | |
| R1-R2-R3325–404 A371E | −7.78 ± 0.05 | 0.36 | −0.11 | −7.88 | 0.60 ± 0.02 | 1.6 | 1.03 ± 0.005 | |
| FL1–404 | −7.62 ± 0.01 | 1.26 | −0.38 | −8.00 | 0.72 ± 0.04 | 1.4 | 0.97 ± 0.009 | |
| FL1–404 C250K~Ufm1 | −8.21 ± 0.01 | −0.34 | 0.10 | −8.10 | 0.87 ± 0.05 | 1.2 | 1.03 ± 0.009 | |
| GABARAPL2 | R1-R2-R3325–404 | −8.64 ± 0.06 | 2.04 | −0.61 | −9.25 | 5.99 ± 0.49 | 0.17 | 0.97 ± 0.004 |
| R1-R2325–376 | −8.08 ± 0.05 | 4.44 | −1.32 | −9.41 | 7.87 ± 0.74 | 0.13 | 0.91 ± 0.003 | |
| R2359–376 | ND | ND | >100 ** | ND | ||||
| R1-R2-R3325–404 A371T | −7.58 ± 0.07 | 7.76 | −2.31 | −9.89 | 17.90 ± 3.75 | 0.06 | 0.937 ± 0.005 | |
| R1-R2-R3325–404 A371E | −7.79 ± 0.05 | 7.23 | −2.16 | −9.95 | 19.60 ± 2.57 | 0.06 | 1.01 ± 0.003 | |
| GABARAP | R1-R2-R3325–404 | −0.93 ± 0.04 | 24.2 | −7.22 | −8.15 | 0.96 ± 0.16 | 1.1 | 0.99 ± 0.03 |
| R1-R2325–376 | −1.1 ± 0.02 | 23.1 | −6.89 | −7.99 | 0.72 ± 0.05 | 1.4 | 0.94 ± 0.01 | |
| LC3B | R1-R2-R3325–404 | −4.47 ± 0.10 | 8.86 | −2.64 | −7.33 | 0.24 ± 0.09 | 4.2 | 0.92 ± 0.02 |
| R1-R2325–376 | −4.23 ± 0.09 | 10.7 | −3.19 | −7.42 | 0.27 ± 0.14 | 3.7 | 0.98 ± 0.02 | |
| R2359–376 | ND | ND | >100 * | ND | ||||
| R1-R2-R3325–404 A371T | −3.76 ± 0.05 | 14.3 | −4.26 | −8.02 | 0.76 ± 0.04 | 1.3 | 0.937 ± 0.009 | |
| R1-R2-R3325–404 A371E | −3.93 ± 0.05 | 15.3 | −4.56 | −8.49 | 1.66 ± 0.12 | 0.6 | 0.944 ± 0.009 | |
| LC3A | R1-R2-R3325–404 | 4.25 ± 10.5 | 10.5 | −3.13 | −7.38 | 0.26 ± 0.03 | 3.8 | 0.91 ± 0.04 |
| R1-R2325–376 | −3.81 ± 0.18 | 11.6 | −3.46 | −7.26 | 0.21 ± 0.02 | 4.7 | 0.94 ± 0.03 | |
| UBA5 AD1–330 | R1-R2-R3325–404 | ND | ND | - | ND | |||
| Ub | R1-R2-R3325–404 | ND | ND | - | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wesch, N.; Löhr, F.; Rogova, N.; Dötsch, V.; Rogov, V.V. A Concerted Action of UBA5 C-Terminal Unstructured Regions Is Important for Transfer of Activated UFM1 to UFC1. Int. J. Mol. Sci. 2021, 22, 7390. https://doi.org/10.3390/ijms22147390
Wesch N, Löhr F, Rogova N, Dötsch V, Rogov VV. A Concerted Action of UBA5 C-Terminal Unstructured Regions Is Important for Transfer of Activated UFM1 to UFC1. International Journal of Molecular Sciences. 2021; 22(14):7390. https://doi.org/10.3390/ijms22147390
Chicago/Turabian StyleWesch, Nicole, Frank Löhr, Natalia Rogova, Volker Dötsch, and Vladimir V. Rogov. 2021. "A Concerted Action of UBA5 C-Terminal Unstructured Regions Is Important for Transfer of Activated UFM1 to UFC1" International Journal of Molecular Sciences 22, no. 14: 7390. https://doi.org/10.3390/ijms22147390
APA StyleWesch, N., Löhr, F., Rogova, N., Dötsch, V., & Rogov, V. V. (2021). A Concerted Action of UBA5 C-Terminal Unstructured Regions Is Important for Transfer of Activated UFM1 to UFC1. International Journal of Molecular Sciences, 22(14), 7390. https://doi.org/10.3390/ijms22147390