Integrating Omics and Gene Editing Tools for Rapid Improvement of Traditional Food Plants for Diversified and Sustainable Food Security
Abstract
:1. Introduction
2. Importance of Traditional Food Plants
2.1. Diversity of Traditional Food Plants across the Globe
2.2. Traditional Food Plants Possess Important Nutritional Traits
2.3. Traditional Food Plants Show Varying Degrees of Tolerance to Stresses
2.4. Traditional Food Plants Ensure Stable and Sustainable Food Security
2.5. Traditional Food Plants Provide Alternative Sources of Income to the Farmers and Unorganized Workers
3. Multi-Omics Tools to Dissect Nutritional and Stress-Related Traits for Ensuring Sustainable Global Food Security
4. Examples of Application of Multi-Omics Tools to Traditional Food Plants
4.1. Lysine Biosynthesis in Amaranthus
4.2. Transcriptional Regulation of Anti-Nutritional Saponins in Chenopodium quinoa
4.3. Genetic Mechanism of Stress Tolerance in Manihot esculenta
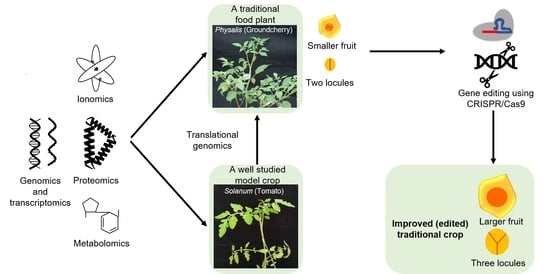
4.4. Genetic Dissection of Pathogen Resistance and the Early Fruit Development and Evolution in Physalis
4.5. Detection of Genes Regulating Uptake and Storage of Micronutrients in Traditional Food Plants
4.6. Unraveling the Mechanism behind High Amount of α-Linolenic Acid and Salinity Tolerance in Portulaca oleracea
4.7. Higher Accumulation of Lycopene in Elaeagnus
4.8. Nutritional Composition of Dioscorea, a Neglected Staple Tuber Crop of the Indigenous Communities
4.9. Transcriptional Basis of Lipid Biosynthesis in Salvia, a Wonder Seed for the 21st Century
4.10. The Adansonia digitata Contains More Vitamin C Than Oranges
5. Integrating Omics and Gene Editing Tools for Improvement/Domestication of Traditional Food Plants
6. Recent Successful Examples of Gene Editing and Translational Genomics in Traditional Food Plants
7. Challenges to Translational Genomics Using Gene Editing Technology/Tools
8. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AOCC | African Orphan Crops Consortium |
| Cas9 | CRISPR-Associated Protein 9 |
| CFF | Crops For the Future |
| CRISPR-Cas9 | Clustered Regularly Interspaced Short Palindromic Repeat-Associated Protein 9 |
| DGE | Differential Gene Expression |
| DNA | Deoxyribonucleic Acid |
| FAO | Food and Agricultural Organization |
| GC | Gas Chromatography |
| gRNA | Guide ribonucleic Acid |
| HDR | Homology Directed Recombination |
| HPLC | High Performance Liquid Chromatography |
| ICP-MS | Inductively Coupled Plasma Mass Spectroscopy |
| ICRAF | International Council for Research in Agroforestry |
| mRNA | Messenger Ribonucleic Acid |
| NCBI | National Center for Biotechnology Information |
| NHEJ | Non-Homologous End Joining |
| PEG | Poly Ethylene Glycol |
| sgRNA | Single Guide Ribonucleic Acid |
| RNA | Ribonucleic Acid |
| RT-PCR | Real-Time Polymerase Chain Reaction |
| QTLs | Quantitative Trait Locus |
| TALENs | Transcriptional Activator-Like Effector Nucleases |
| TFPs | Traditional Food Plants |
| Trex2 | Three prime Repair Exonuclease 2 |
| ZFNs | Zinc Finger Nucleases |
References
- FAO. Proceedings of the Expert Meeting on How to Feed the World in 2050; Food and Agriculture Organization: Rome, Italy, 2009. [Google Scholar]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [Green Version]
- Beddington, J.R.; Asaduzzaman, M.; Fernández, A.; Clark, M.E.; Guillou, M.; Jahn, M.M.; Erda, L.; Mamo, T.; Van, B.N.; Nobre, C.A.; et al. Achieving Food Security in the Face of Climate Change: Summary for Policy Makers from the Commission on Sustainable Agriculture and Climate Change; CGIAR Research Program on Climate Change, Agriculture and Food Security: Wageningen, The Netherlands, 2011. [Google Scholar]
- Dhyani, A. Plants of the World—Diverse, Fascinating and Threatened; Science Reporter, NISCAIR-CSIR India: Delhi, India, 2020; Volume 57, p. 3. Available online: http://nopr.niscair.res.in/handle/123456789/54100 (accessed on 7 April 2021).
- Willis, K.J. State of the World’s Plants. Available online: https://stateoftheworldsplants.org/ (accessed on 7 June 2021).
- Meyer, R.S.; DuVal, A.E.; Jensen, H.R. Patterns and Processes in Crop Domestication: An Historical Review and Quantitative Analysis of 203 Global Food Crops. New Phytol. 2012, 196, 29–48. [Google Scholar] [CrossRef]
- Ross-Ibarra, J.; Morrell, P.L.; Gaut, B.S. Plant Domestication, a Unique Opportunity to Identify the Genetic Basis of Adaptation. Proc. Natl. Acad. Sci. USA 2007, 104, 8641–8648. [Google Scholar] [CrossRef] [Green Version]
- Pimentel, D.; Jackson, W.; Bender, M.; Pickett, W. Perennial Grains—An Ecology of New Crops. Interdiscip. Sci. Rev. 1986, 11, 42–49. [Google Scholar] [CrossRef]
- Hawtin, G.; Collins, W. Conserving and Using Crop Plant Biodiversity in Agroecosystems. In Biodiversity in Agroecosystems; Collin, W.W., Quaslet, C.O., Eds.; CRC Press: Boca Raton, FL, USA, 1998; pp. 267–281. [Google Scholar]
- Hambrey, J. The 2030 Agenda and the Sustainable Development Goals: The Challenge for Aquaculture Development and Management, FAO Fisheries and Aquaculture Circular. Available online: http://www.fao.org/policy-support/tools-and-publications/resources-details/en/c/1153661/ (accessed on 24 June 2020).
- Thrupp, L. Linking Agricultural Biodiversity and Food Security: The Valuable Role of Agrobiodiversity for Sustainable Agriculture. Int. Aff. 2000, 76, 283–297. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature Increase Reduces Global Yields of Major Crops in Four Independent Estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tigchelaar, M.; Battisti, D.; Naylor, R.; Ray, D. Future Warming Increases Probability of Globally Synchronized Maize Production Shocks. Proc. Natl. Acad. Sci. USA 2018, 115, 6644–6649. [Google Scholar] [CrossRef] [Green Version]
- Byg, A.; Salick, J. Local Perspectives on a Global Phenomenon—Climate Change in Eastern Tibetan Villages. Tradit. Peoples Clim. Change 2009, 19, 156–166. [Google Scholar] [CrossRef]
- Kotir, J. Climate Change and Variability in Sub-Saharan Africa: A Review of Current and Future Trends and Impacts on Agriculture and Food Security. Environ. Dev. Sustain. 2011, 13, 587–605. [Google Scholar] [CrossRef]
- Speranza, C.I. Resilient Adaptation to Climate Change in African Agriculture, 54th ed.; German Development Institute: Bonn, Germany, 2010. [Google Scholar]
- Padulosi, S.; Bhag, M.; Bala, R.S.; Shanthakumar, G.; Yenagi, N.; Dutta, M. Food Security and Climate Change: Role of Plant Genetic Resources of Minor Millets. Indian J. Plant Genet. Resour. 2009, 22, 1–16. [Google Scholar]
- Hughes, J. Just Famine Foods? What Contributions Can Underutilized Plants Make to Food Security? Acta Hortic. 2009, 806, 39–48. [Google Scholar] [CrossRef]
- Jarvis, D.I.; Hodgkin, T.; Sthapit, B.R.; Fadda, C.; Lopez-Noriega, I. An Heuristic Framework for Identifying Multiple Ways of Supporting the Conservation and Use of Traditional Crop Varieties within the Agricultural Production System. Crit. Rev. Plant Sci. 2011, 30, 125–176. [Google Scholar] [CrossRef] [Green Version]
- Wolter, F.; Schindele, P.; Puchta, H. Plant Breeding at the Speed of Light: The Power of CRISPR/Cas to Generate Directed Genetic Diversity at Multiple Sites. BMC Plant Biol. 2019, 19, 176. [Google Scholar] [CrossRef] [Green Version]
- Maundu, P.M. The Status of Traditional Vegetable Utilization in Kenya. In Proceedings of the IPGRI International Workshop on Genetic Resources of Traditional Vegetables in Africa: Conservation and Use 29–31 August 1995, Guarino, Nairobi, Kenya. Institute of Plant Genetics and Crop Plant Research, Gatersleben/International Plant Genetic Resources Institute, Rome, Italy, 1998, L. Ed.: ICRAF-HQ; Volume 16, pp. 66–75.
- Muthoni, J.; Nyamongo, D. Traditional Food Crops and Their Role in Food and Nutritional Security in Kenya. J. Agric. Food Inf. 2010, 11, 36–50. [Google Scholar] [CrossRef]
- Adhikari, L.; Hussain, A.; Rasul, G. Tapping the Potential of Neglected and Underutilized Food Crops for Sustainable Nutrition Security in the Mountains of Pakistan and Nepal. Sustainability 2017, 9, 291. [Google Scholar] [CrossRef] [Green Version]
- Longin, C.F.H.; Würschum, T. Back to the Future—Tapping into Ancient Grains for Food Diversity. Trends Plant Sci. 2016, 21, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.L.; Lammerts van Bueren, E.T.; Ceccarelli, S.; Grando, S.; Upadhyaya, H.D.; Ortiz, R. Diversifying Food Systems in the Pursuit of Sustainable Food Production and Healthy Diets. Trends Plant Sci. 2017, 22, 842–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthamilarasan, M.; Singh, N.; Prasad, M. Multi-omics Approaches for Strategic Improvement of Stress Tolerance in Underutilized Crop Species: A Climate Change Perspective. Adv. Genet. 2019, 103, 1–38. [Google Scholar] [CrossRef]
- Adhikari, L.; Tuladhar, S.; Hussain, A.; Aryal, K. Are Traditional Food Crops Really ‘Future Smart Foods? ’ A Sustainability Perspective. Sustainability 2019, 11, 5236. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, R.; Kumar, G.V.; Kumar, S.P.J. OMICS–Based Approaches in Plant Biotechnology; John Wiley & Sons: Hoboken, NJ, USA, 2019. [Google Scholar]
- Esquinas-Alcázar, J. Science and Society: Protecting Crop Genetic Diversity for Food Security: Political, Ethical and Technical Challenges. Nat. Rev. Genet. 2005, 6, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Milner, S.; Jost, M.; Taketa, S.; Mazón, E.; Himmelbach, A.; Oppermann, M.; Weise, S.; Knüpffer, H.; Basterrechea Salido, M.; König, P.; et al. Genebank Genomics Highlights the Diversity of a Global Barley Collection. Nat. Genet. 2019, 51, 319–326. [Google Scholar] [CrossRef] [Green Version]
- Langridge, P.; Waugh, R. Harnessing the Potential of Germplasm Collections. Nat. Genet. 2019, 51, 200–201. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Zavaglia, A.; Mejuto, J.C.; Simal-Gandara, J. Mitigation of Emerging Implications of Climate Change on Food Production Systems. Food Res. Int. 2020, 134, 1–12. [Google Scholar] [CrossRef]
- Gráda, C.Ó. Black ’47 and Beyond; The Great Irish Famine in History, Economy, and Memory, Ed.; Princeton University Press: Princeton, NJ, USA, 1999. [Google Scholar]
- Bruns, H. Southern Corn Leaf Blight: A Story Worth Retelling. Agron. J. 2017, 109, 1218–1224. [Google Scholar] [CrossRef] [Green Version]
- Risch, S.; Andow, D.; Altieri, M. Agroecosystem Diversity and Pest Control: Data, Tentative Conclusions, and New Research Directions. Environ. Entomol. 1983, 12, 625–629. [Google Scholar] [CrossRef]
- Altieri, M. Altieri, M. A Monocultures and their impacts on biodiversity. In Red Sugar, Green Deserts: Latin American Report on Monocultures and Violations of the Human Rights to Adequate Food and Housing, to Water, to Land and to Territory; FIAN International: Heidelberg, Germany, 2009; pp. 67–76. [Google Scholar]
- Turner, M.; Calder, W.; Cumming, G.; Hughes, T.; Jentsch, A.; LaDeau, S.; Lenton, T.; Shuman, B.; Turetsky, M.; Ratajczak, Z.; et al. Climate Change, Ecosystems and Abrupt Change: Science Priorities. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomiyama, J.-M.; Takagi, D.; Kantar, M. The Effect of Acute and Chronic Food Shortage on Human Population Equilibrium in a Subsistence Setting. Agric. Food Secur. 2020, 9, 1–12. [Google Scholar] [CrossRef]
- Ciaccia, C.; Testani, E.; Roccuzzo, G.; Stefano, C. The Role of Agrobiodiversity in Sustainable Food Systems Design and Management. In Genetic Diversity in Horticultural Plants. Sustainable Development and Biodiversity; Springer: Berlin/Heidelberg, Germany, 2019; Volume 22, pp. 245–271. [Google Scholar]
- Chaudhary, P.; Bhatta, S.; Aryal, K.; Joshi, B.; Gauchan, D. Threats, Drivers and Conservation Imperative of Agrobiodiversity. J. Agric. Environ. 2020, 21, 44–61. [Google Scholar]
- Choi, H.-K. Translational Genomics and Multi-Omics Integrated Approaches as a Useful Strategy for Crop Breeding. Genes Genom. 2019, 41, 133–146. [Google Scholar] [CrossRef] [Green Version]
- El Bilali, H.; Callenius, C.; Strassner, C.; Probst, L. Food and Nutrition Security and Sustainability Transitions in Food Systems. Food Energy Secur. 2018, 8, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Negi, G.C.S.; Samal, P.; Kuniyal, J.C.; Sharma, R.; Dhyani, P.P. Impacts of Climate Change on Western Himalayan Mountain Ecosystems: An Overview. Trop. Ecol. 2012, 53, 345–356. [Google Scholar]
- Akinola, R.; Pereira, L.M.; Mabhaudhi, T.; de Bruin, F.-M.; Rusch, L. A Review of Indigenous Food Crops in Africa and the Implications for More Sustainable and Healthy Food Systems. Sustainability 2020, 12, 3493. [Google Scholar] [CrossRef] [Green Version]
- Gregory, P.; Mayes, S.; Chai, H.H.; Jahanshiri, E.; Julkifle, A.; Kuppusamy, G.; Kuan, H.; Lin, T.; Massawe, F.; Syaheerah, T.; et al. Crops For the Future (CFF): An Overview of Research Efforts in the Adoption of Underutilised Species. Planta 2019, 250, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hanafiah, N.M.; Mispan, M.S.; Lim, P.E.; Baisakh, N.; Cheng, A. The 21st Century Agriculture: When Rice Research Draws Attention to Climate Variability and How Weedy Rice and Underutilized Grains Come in Handy. Plants 2020, 9, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agulanna, F.T. The Role of Indigenous and Underutilized Crops in The Enhancement of Health and Food Security in Nigeria. Afr. J. Biomed. Res. 2020, 23, 305–312. [Google Scholar]
- Borelli, T.; Hunter, D.; Padulosi, S.; Amaya, N.; Meldrum, G.; de Oliveira Beltrame, D.M.; Samarasinghe, G.; Wasike, V.W.; Güner, B.; Tan, A.; et al. Local Solutions for Sustainable Food Systems: The Contribution of Orphan Crops and Wild Edible Species. Agronomy 2020, 10, 231. [Google Scholar] [CrossRef] [Green Version]
- Conti, M.V.; Campanaro, A.; Coccetti, P.; De Giuseppe, R.; Galimberti, A.; Labra, M.; Cena, H. Potential Role of Neglected and Underutilized Plant Species in Improving Women’s Empowerment and Nutrition in Areas of Sub-Saharan Africa. Nutr. Rev. 2019, 77, 817–828. [Google Scholar] [CrossRef]
- FAO and the 17 Sustainable Development Goals. Sustainable Development Knowledge Platform. Available online: https://sustainabledevelopment.un.org/index.php?page=view&type=400&nr=2205&menu=1515 (accessed on 7 June 2021).
- Dawson, I.K.; Hendre, P.; Powell, W.; Sila, D.; McMullin, S.; Simons, T.; Revoredo-Giha, C.; Odeny, D.A.; Barnes, A.P.; Graudal, L.; Working Paper, No.; et al. 276 ed.; World Agroforestry United Nations; World Agroforestry Centre: Niarobi, Kenya, 2018. [Google Scholar] [CrossRef]
- Jamnadass, R.; Mumm, R.H.; Hale, I.; Hendre, P.; Muchugi, A.; Dawson, I.K.; Powell, W.; Graudal, L.; Yana-Shapiro, H.; Simons, A.J.; et al. Enhancing African Orphan Crops with Genomics. Nat. Genet. 2020, 52, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Ramdwar, M.; Siew, N. Strategic Approaches to Food Security in Developing Countries. In Agricultural Development and Food Security in Developing Nations; Ganpat, W.G., Ed.; IGI Global: Hershey, PA, USA, 2017; pp. 197–221. [Google Scholar]
- Tadele, Z. Orphan Crops: Their Importance and the Urgency of Improvement. Planta 2019, 250, 677–694. [Google Scholar] [CrossRef] [Green Version]
- Hendre, P.; Muchugi, A.; Chang, Y.; Fu, Y.; Song, Y.; Liu, M.; Liao, X.; Liu, H.; Song, B.; Xu, X.; et al. Generation of Open-Source Genomics Resources for African Orphan Tree Crops by African Orphan Crops Consortium (AOCC), a Public-Private Partnership for Promoting Food and Nutritional Security in Africa. Acta Hortic. 2020, 615–622. [Google Scholar] [CrossRef]
- Yssel, A.; Kao, S.-M.; Peer, V. Sterck ORCAE-AOCC: A Centralized Portal for the Annotation of African Orphan Crop Genomes. Genes 2019, 10, 950. [Google Scholar] [CrossRef]
- Department for International Development. Crops for the Future Strategic Plan 2009–2013. Available online: https://www.gov.uk/research-for-development-outputs/crops-for-the-future-strategic-plan-2009-2013 (accessed on 7 June 2021).
- FAO. Promoting Neglected and Underutilized Crop Species. Available online: http://www.fao.org/news/story/en/item/1032516/icode/ (accessed on 7 June 2021).
- Maundu, M.P.; Ngugi, W.G.; Kabuye, H.S.C. Traditional Food Plants of Kenya; National Museums of Kenya: Nairobi, Kenya, 1999. [Google Scholar]
- Campbell, J. Development, Global Change and Traditional Food Security in Pacific Island Countries. Reg. Environ. Change 2014, 15, 1–12. [Google Scholar] [CrossRef]
- Shelef, O.; Weisberg, P.; Provenza, F. The Value of Native Plants and Local Production in an Era of Global Agriculture in: Mirás-Avalos & Baveye 2018 Agroecosystems Facing Global Climate Change The Search for Sustainability. Front. Plant Sci. 2019, 8, 2069. [Google Scholar] [CrossRef] [Green Version]
- Rajapaksha, U. Traditional Food Plants in Sri Lanka; Hector Kobbekaduwa Agrarian Research and Training Institute: Colombo, Sri Lanka, 1998. [Google Scholar]
- Kristbergsson, K.; Oliveira, J. Traditional foods: General and consumer aspects. In Integrating Food Science and Engineering Knowledge into the Food Chain; Kristbergsson, K., Ed.; Springer: New York, NY, USA, 2016; pp. 85–86. [Google Scholar]
- Molina, M.; Tardío, J.; Aceituno-Mata, L.; Morales, R.; Reyes-García, V.; Pardo-de-Santayana, M. Weeds and Food Diversity: Natural Yield Assessment and Future Alternatives for Traditionally Consumed Wild Vegetables. J. Ethnobiol. 2014, 34, 44–67. [Google Scholar] [CrossRef]
- Gamba, G.; Donno, D.; Mellano, M.G.; Riondato, I.; De Biaggi, M.; Randriamampionona, D.; Beccaro, G.L. Phytochemical Characterization and Bioactivity Evaluation of Autumn Olive (Elaeagnus umbellata Thunb.) Pseudo drupes as Potential Sources of Health-Promoting Compounds. Appl. Sci. 2020, 10, 4354. [Google Scholar] [CrossRef]
- Kozioł, M.J. Chemical Composition and Nutritional Evaluation of Quinoa (Chenopodium quinoa Willd.). J. Food Compos. Anal. 1992, 5, 35–68. [Google Scholar] [CrossRef]
- Mwanri, W.A.; Mamboleo, F.T.; Msuya, M.J.; Gowele, F.V. Oxalate, Phytate and Nitrate Content in African Nightshade, Spider Plant and Amaranths at Different Stages of Maturity. Afr. J. Food Sci. 2018, 12, 316–322. [Google Scholar] [CrossRef]
- Sedbrook, J.C.; Phippen, W.B.; Marks, M.D. New Approaches to Facilitate Rapid Domestication of a Wild Plant to an Oilseed Crop: Example Pennycress (Thlaspi arvense L.). Plant Sci. Int. J. Exp. Plant Biol. 2014, 227, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.; Fritz, G.; Patton, P.; Carmody, S.; Horton, E. Growing the Lost Crops of Eastern North America’s Original Agricultural System. Nat. Plants 2017, 3, 1–5. [Google Scholar] [CrossRef]
- Peng, W.; Berry, E. The Concept of Food Security. In Encyclopedia of Food Security and Sustainability, 1st ed.; Ferranti, P., Berry, E., Jock, A., Eds.; Elsevier Science & Technology: Amsterdam, The Netherlands, 2018; pp. 1–7. [Google Scholar]
- Schmidhuber, J.; Tubiello, F.N. Global Food Security under Climate Change. Proc. Natl. Acad. Sci. USA 2007, 104, 19703–19708. [Google Scholar] [CrossRef] [Green Version]
- Van Berkum, S.; Ruben, R. The Food System Approach: Sustainable Solutions for a Sufficient Supply of Healthy Food; Wageningen Economic Research: Memorandum 2018-064; Wageningen University & Research: Wageningen, The Netherlands, 2018. [Google Scholar]
- Ashby, S.; Kleve, S.; McKechnie, R.; Palermo, C. Measurement of the Dimensions of Food Insecurity in Developed Countries: A Systematic Literature Review. Public Health Nutr. 2016, 19, 2887–2896. [Google Scholar] [CrossRef] [PubMed]
- Abdulkadyrova, M.A.; Dikinov, A.H.; Tajmashanov, H.È.; Shidaev, L.A.; Shidaeva, E.A. Global Food Security Problems in the Modern World Economy. Int. J. Environ. Sci. Educ. 2016, 11, 5320–5330. [Google Scholar]
- Dresselhaus, T.; Hückelhoven, R. Biotic and Abiotic Stress Responses in Crop Plants. Agronomy 2018, 8, 267. [Google Scholar] [CrossRef] [Green Version]
- Porter, J.; Semenov, M. Crop Response to Climatic Variation. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2005, 360, 2021–2035. [Google Scholar] [CrossRef] [PubMed]
- Dhankher, O.P.; Foyer, C.H. Climate Resilient Crops for Improving Global Food Security and Safety. Plant Cell Environ. 2018, 41, 877–884. [Google Scholar] [CrossRef]
- Scholes, R.; Biggs, R. Ecosystem Services in Southern Africa: A Regional Assessment; Council for Scientific and Industrial Research: Pretoria, South Africa, 2004. [Google Scholar]
- Chen, Y.; Michalak, M.; Agellon, L.B. Importance of Nutrients and Nutrient Metabolism on Human Health. Yale J. Biol. Med. 2018, 91, 95–103. [Google Scholar] [PubMed]
- Houghton, J.E.T.; Ding, Y.; Griggs, D.; Noguer, M.; van der Linden, P.; Dai, X.; Maskell, M.; Johnson, C. Climate Change 2001: The Scientific Basis. In Climate Change 2001: The Scientific Basis. Contribution of Working Group I to the Third Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2001; Volume 881. [Google Scholar]
- Ebert, A.W. Potential of Underutilized Traditional Vegetables and Legume Crops to Contribute to Food and Nutritional Security, Income and More Sustainable Production Systems. Sustainability 2014, 6, 319–335. [Google Scholar] [CrossRef] [Green Version]
- Berkelaar, D.; The Importance of Indegenous Food Plants. Echo Community. Available online: https://www.echocommunity.org/en/resources/a118dadf-50d6-492c-a19f-948a23c93e83 (accessed on 12 October 2020).
- Zhang, F.; Batley, J. Exploring the Application of Wild Species for Crop Improvement in a Changing Climate. Curr. Opin. Plant Biol. 2020, 56, 218–222. [Google Scholar] [CrossRef]
- Schnyder, H.; Seo, S.; Rademacher, I.F.; Kühbauch, W. Spatial Distribution of Growth Rates and of Epidermal Cell Lengths in the Elongation Zone during Leaf Development in Lolium perenne L. Planta 1990, 181, 423–431. [Google Scholar] [CrossRef]
- Mishra, S.; Mohanara, S.K. Dash. Review on Cleome gynandra. Int. J. Res. Pharm. Chem. 2011, 1, 681–689. [Google Scholar]
- Rao, A.P.; Rajendrudu, G. Net Photosynthetic Rate in Relation to Leaf Anatomical Characteristics of C3, C3-C4 and C4 Dicotyledons. Proc. Indian Acad. Sci. Plant Sci. 1989, 99, 529–538. [Google Scholar]
- Kumar, U.D.J.; Saraswathy, R.; Rama Das, V.S. Differential Performance of Cleome gynandra L. (C4) and Cleome speciosa L. (C3) under Water Stress and Recovery. Environ. Exp. Bot. 1984, 24, 305–310. [Google Scholar] [CrossRef]
- Bamidele, O.; Akinnuga, A.M.; Olorunfemi, J.O.; Tony, O.A.; Oparaji, C.K.; Ezeigbo, N. Effects of Aqueous Extract of Basella alba Leaves on Haematological and Biochemical Parameters in Albinorats. Afr. J. Biotechnol. 2010, 9, 6952–6955. [Google Scholar] [CrossRef]
- Adhikari, R.; Kumar, H.N.N; Shruthi, S.D. A Review on Medicinal Importance of Basella alba L. Int. J. Pharm. Sci. Drug Res. 2012, 4, 110–114. [Google Scholar]
- Deshmukh, S.; Gaikwad, D. A Review of the Taxonomy, Ethnobotany, Phytochemistry and Pharmacology of Basella alba (Basellaceae). J. Appl. Pharm. Sci. 2014, 4, 153–165. [Google Scholar] [CrossRef]
- Murevanhema, Y.Y.; Jideani, V.A. Potential of Bambara Groundnut (Vigna subterranea (L.) Verdc) Milk as a Probiotic Beverage-a Review. Crit. Rev. Food Sci. Nutr. 2013, 53, 954–967. [Google Scholar] [CrossRef] [PubMed]
- Adebowale, K.O.; Adeniyi Afolabi, T.; Lawal, O.S. Isolation, Chemical Modification and Physicochemical Characterisation of Bambarra Groundnut (Voandzeia subterranean) Starch and Flour. Food Chem. 2002, 78, 305–311. [Google Scholar] [CrossRef]
- Omoikhoje, S.O. Assessment of the Nutritive Value of Bambara Groundnut as Influenced by Cooking Time. Livest. Res. Rural Dev. 2008, 20, 55–60. [Google Scholar]
- Aberoumand, A.; Deokule, S.S. Chemical Analysis and Nutritional Value of Chlorophytum comosum: A Plant Food in Iran. J. Med. Food Plants 2009, 1, 87–91. [Google Scholar]
- Schippers, R.R. African Indigenous Vegetables: An Overview of the Cultivated Species; Natural Resources Institute/ACP-EU Technical Centre for Agricultural and Rural Cooperation: Chatham, UK, 2002. [Google Scholar]
- Lee, C.-F.; Fan, C.-W.; Chiang, N.-N.; Chang, H.-C.; Chen, C.; Huang, Y.-S.; Wang, H.-Y.; Lin, W.-C.; Chen, F.-A. Protective Effect of Corchorus capsularis L. Leaves on Ethanol-Induced Acute Gastric Mucosal Lesion in Rats. J. Vet. Med. Sci. 2019, 81, 1636–1642. [Google Scholar] [CrossRef] [Green Version]
- Bhartiya, A.; Aditya, J.; Kant, L. Nutritional and Remedial Potential of an Underutilized Food Legume Horsegram (Macrotyloma uniflorum): A Review. J. Anim. Plant Sci. 2015, 25, 908–920. [Google Scholar]
- Campbell, C.G.; Heller, J.; Engels, J. Buckwheat. Fagopyrum Esculentum Moench, 19th ed.; International Plant Genetic Resources Institute: Rome, Italy, 1997. [Google Scholar]
- Cartea, M.E.; Francisco, M.; Soengas, P.; Velasco, P. Phenolic Compounds in Brassica Vegetables. Molecules 2011, 16, 251–280. [Google Scholar] [CrossRef] [PubMed]
- Grubben, G.J.H.; Denton, O.A. Plant Resources of Tropical Africa; Netherlands/Backhuys Publishers: Wageningen, The Netherlands, 2004. [Google Scholar]
- Temesgen, M.; Retta, N. Nutritional Potential, Health and Food Security Benefits of Taro Colocasia esculenta (L.): A Review. Open Food Sci. J. 2015, 36, 23–30.
- Kaushal, P.; Kumar, V.; Sharma, H. Utilization of Taro (Colocasia esculenta): A Review. J. Food Sci. Technol. 2013, 52, 27–40. [Google Scholar] [CrossRef]
- Khalafalla, M.M.; Daffalla, H.M.; Abdellatef, E.; Agabna, E.; El-Shemy, H.A. Establishment of an in Vitro Micropropagation Protocol for Boscia senegalensis (Pers.) Lam. Ex Poir. J. Zhejiang Univ. Sci. 2011; 12. [Google Scholar] [CrossRef]
- Kim, T.R.; Pastuszyn, A.; Vanderjagt, D.J.; Glew, R.S.; Millson, M.; Glew, R.H. The Nutritional Composition of Seeds From Boscia senegalensis (Dilo) from the Republic of Niger. J. Food Compos. Anal. 1997, 10, 73–81. [Google Scholar] [CrossRef]
- FAO Food and Nutrition Paper (FAO). Traditional Food Plants. A Resource Book for Promoting the Exploitation and Consumption of Food Plants in Arid, Semi-Arid and Sub-Humid Lands of Eastern Africa; FAO: Rome, Italy, 1998. [Google Scholar]
- Adewale, D.; Odoh, N. A Review on Genetic Resources, Diversity and Agronomy of African Yam Bean (Sphenostylis stenocarpa (Hochst. Ex A. Rich.) Harms): A Potential Future Food Crop. Sustain. Agric. Res. 2012; 2. [Google Scholar] [CrossRef]
- Adegboyega, T.T.; Abberton, M.T.; AbdelGadir, A.H.; Dianda, M.; Maziya-Dixon, B.; Oyatomi, O.A.; Ofodile, S.; Babalola, O.O. Evaluation of Nutritional and Antinutritional Properties of African Yam Bean (Sphenostylis stenocarpa (Hochst Ex. A. Rich.) Harms.) Seeds. J. Food Qual. 2020; 11. [Google Scholar] [CrossRef]
- Okoli, B.E.; Mgbeogu, C.M. Fluted Pumpkin, Telfairia occidentalis: West African Vegetable Crop. Econ. Bot. 1983, 37, 145–149. [Google Scholar] [CrossRef]
- Glew, R.H.; Laabes, E.P.; Presley, J.M.; Schulze, J.; Andrews, R.; Wang, Y.-C.; Chang, Y.-C.; Chuang, L.-T. Fatty Acid, Amino Acid, Mineral and Antioxidant Contents of Acha (Digitaria exilis) Grown on the Jos Plateau, Nigeria. Int. J. Nutr. Metab. 2013, 5, 1–8. [Google Scholar] [CrossRef]
- Istifanus, M.F.; Agbo, E.B. Nutritional and Health Benefits of Acha (Digitaria exilis) in the Human Diet—A Review. Niger. Food J. 2016, 34, 72–78. [Google Scholar] [CrossRef]
- Jideani, I.A. Traditional and Possible Technological Uses of Digitaria exilis (Acha) and Digitaria iburua (Iburu): A Review. Plant Foods Hum. Nutr. Dordr. Neth. 1999, 54, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Abukutsa-Onyango, M. Response of Slenderleaf (Crotalaria brevidens Benth) to Inorganic Nitrogen Application. Afr. J. Food Agric. Nutr. Dev. 2007, 7, 1–10. [Google Scholar]
- Ajibesin, K. Dacryodes edulis (G. Don) H.J. Lam: A Review on Its Medicinal, Phytochemical and Economical Properties. Res. J. Med. 2011; 5. [Google Scholar] [CrossRef]
- Stadlmayr, B.; Charrondiere, U.; Eisenwagen, S.; Jamnadass, R.; Kehlenbeck, K. Review: Nutrient Composition of Selected Indigenous Fruits from Sub-Saharan Africa. J. Sci. Food Agric. 2013, 93, 2627–2636. [Google Scholar] [CrossRef] [PubMed]
- Ene-Obong, H.; Igile, G.; Ekpo, A.; Egbung, E.; Agbo, M. Variations in the Nutrients and Bioactive Compounds of Different Accessions of the West African Pear (Dacryodes edulis): Implications for Dietary Intake Assessment and Health. J. Food Compos. Anal. 2019, 79, 80–86. [Google Scholar] [CrossRef]
- Nuga, O.O.; Ofodile, E.A.U. Potentials of Treculia africana Decne—An Endangered Species of Southern Nigeria. J. Agric. Soc. Res. 2010, 10, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Okafor, J.C.; Okolo, H.C. Potentials and Some Indigenous Fruit Trees of Nigeria. In Proceedings of the 5th Annual Conference on Forestry Association of Nigeria, Jos, Nygeria, 1–6 December 1974. [Google Scholar]
- Thakur, G.S.; Bag, M.; Sanodiya, B.S.; Bhadouriya, P.; Debnath, M.; Prasad, G.B.K.S.; Bisen, P.S. Momordica balsamina: A Medicinal and Neutraceutical Plant for Health Care Management. Curr. Pharm. Biotechnol. 2009, 10, 667–682. [Google Scholar] [CrossRef]
- Flyman, M.V.; Afolayan, A.J. Proximate and Mineral Composition of the Leaves of Momordica balsamina L.: An under-Utilized Wild Vegetable in Botswana. Int. J. Food Sci. Nutr. 2007; 58. [Google Scholar] [CrossRef]
- Gebauer, J.; El-Siddig, K.; Ebert, G. Baobab (Adansonia digitata L.): A Review on a Multipurpose Tree with Promising Future in the Sudan. Gartenbauwissenschaft 2002, 67, 155–160.
- Yazzie, D.; VanderJagt, D.J.; Pastuszyn, A.; Okolo, A.; Glew, R.H. The Amino Acid and Mineral Content of Baobab (Adansonia digitata L.) Leaves. J. Food Compos. Anal. 1994; 7. [Google Scholar] [CrossRef]
- Lusepani, N.E. Reproductive Biology and Utilisation of Berchemia discolor (Klotzsch) Hemsley (Rhamnaceae). Ph.D. Dissertation, Stellenbosch University, Stellenbosch, South Africa, 1999. [Google Scholar]
- Udosen, E.O.; Udok, U.E.; Unuigbe, O.S. The Comparison of the Nutrient Compositions of Lasianthera africana and Hejnsia crinita. J. Food Biochem. 1999, 23, 571–576. [Google Scholar] [CrossRef]
- Lepcha, P.; Egan, A.; Doyle, J.; Narayana, N.S. A Review on Current Status and Future Prospects of Winged Bean (Psophocarpus tetragonolobus) in Tropical Agriculture. Plant Foods Hum. Nutr. 2017, 72, 1–11. [Google Scholar] [CrossRef]
- Amoo, I.A.; Adebayo, O.; Oyeleye, A. Chemical Evaluation of Winged Beans (Psophocarpus tetragonolobus), Pitanga Cherries (Eugenia uniflora) and Orchid Fruit (Orchid Fruit Myristica). Afr. J. Food Agric. Nutr. Dev. 2011, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Misra, P.S.; Misra, G.; Prakash, D.; Tripathi, R.D.; Chaudhary, A.R.; Mishra, P.N. Assay of Some Nutritional and Antinutritional Factors in Different Cultivars of Winged Bean (Psophocarpus tetragonolobus (L.) DC) Seeds. Plant Foods Hum. Nutr. 1987; 36. [Google Scholar] [CrossRef] [Green Version]
- Jaffe, W.G.; Korte, R. Nutritional Characteristics of the Winged Bean (Psophocarpus tetragonolobus) in Rats (A Little Known Crop Presently Cultivated in Parts of South East Asia, Some Parts of Africa, and Mostly in Papua New Guinea). Nutr. Rep. Int. USA 1976, 14, 449–455. [Google Scholar]
- Ticona, L.N.A.; Pérez, V.T.; Benito, P.B. Local/Traditional Uses, Secondary Metabolites and Biological Activities of Mashua (Tropaeolum tuberosum Ruíz & Pavón). J. Ethnopharmacol. 2020, 247, 112–152. [Google Scholar] [CrossRef]
- María Elena, J.H.; Yamilet Irene, G.G.; Iván, Y.G.; Migdalia, M.M. Chemical Study and Determination of the Antioxidant Activity of Three Varieties Tropaeolum tuberosum (Mashua). Am. J. Plant Sci. 2019, 10, 2279–2297. [Google Scholar] [CrossRef] [Green Version]
- Campos, D.; Chirinos, R.; Gálvez Ranilla, L.; Pedreschi, R. Bioactive Potential of Andean Fruits, Seeds, and Tubers. In Advances in Food and Nutrition Research; Michael Eskin N., A., Ed.; Elsevier, 2018; Vol. 84, pp. 287–343 ISBN 9780128149904.
- Flores, H.; Walker, T.; Guimarães, R.; Bais, H.; Vivanco, J. Andean Root and Tuber Crops: Underground Rainbows. HortScience 2003, 38, 161–167. [Google Scholar] [CrossRef] [Green Version]
- Ojansivu, I.; Ferreira, C.L.; Salminen, S. Yacon, A New Source of Prebiotic Oligosaccharides with a History of Safe Use. Trends Food Sci. Technol. 2011, 22, 40–46. [Google Scholar] [CrossRef]
- Lachman, J.; Fernández, E.; Orsák, M. Yacon [Smallanthus sonchifolia (Poepp. et Endl.) H. Robinson] Chemical Composition and Use—A Review. Plant Soil Environ. 2003; 49. [Google Scholar] [CrossRef] [Green Version]
- Repo-Carrasco-Valencia, R.; Acevedo de La Cruz, A.; Icochea Alvarez, J.C.; Kallio, H. Chemical and Functional Characterization of Kañiwa (Chenopodium pallidicaule) Grain, Extrudate and Bran. Plant Foods Hum. Nutr. Dordr. Neth. 2009, 64, 94–101. [Google Scholar] [CrossRef]
- White, P.L.; Alvistur, E.; Dias, C.; Visas, E.; White, H.S.; Collazos, C. Nutrient Content and Protein Quality of Quinua and Caflihua, Edible Seed Products of the Andes Mountains. J. Agric. Food Chem. 1955, 3, 531–534. [Google Scholar] [CrossRef]
- Gross, R.; Koch, F.; Malaga, I.; de Miranda, A.F.; Schoeneberger, H.; Trugo, L.C. Chemical Composition and Protein Quality of Some Local Andean Food Sources. Food Chem. 1989, 34, 25–34. [Google Scholar] [CrossRef]
- Maass, B.; Knox, M.; Chinnegowda, V.; Angessa, T.T.; Ramme, S.; Pengelly, B.C. Lablab purpureus-A Crop Lost for Africa? Trop. Plant Biol. 2010, 3, 123–135. [Google Scholar] [CrossRef] [Green Version]
- Engle, L.M.; Altoveros, N.C. Collection, Conservation and Utilization of Indigenous Vegetables. Proceedings of World Vegetable Center, a Workshop on Collection, Conservation and Utilization of Indigenous Vegetables, Shanhua, Taiwan, 16–18 August 1999; Mecozzi, M., Ed.; AVRDC Publication: Tainan, Taiwan, 2000. [Google Scholar]
- Naeem, M.; Aftab, T.; Khan, M.M. Hyacinth Bean (Lablab purpureus L.)—An Underutilised Crop with Future Potential. Sci. Hortic. 2020; 12. [Google Scholar] [CrossRef]
- Mariod, A.A.; Abdelwahab, S.I. Sclerocarya birrea (Marula), An African Tree of Nutritional and Medicinal Uses: A Review. Food Rev. Int. 2012, 28, 375–388. [Google Scholar] [CrossRef]
- Behera, A.; Kumar, S.; Jena, P.K. A Review on Amorphophallus Species: Important Medicinal Wild Food Crops of Odisha. Int. J. Pharm. Life Sci. 2014, 5, 3512–3516. [Google Scholar]
- Tripathi, A.; Chitra, V.; Sheikh, D.; Mohale, D.; Dewani, A. Immunomodulatory Activity of the Methanol Extract of Amorphophallus campanulatus (Araceae) Tuber. Trop. J. Pharm. Res. 2010, 9, 451–454. [Google Scholar] [CrossRef] [Green Version]
- Acosta, O.; Pérez, A.M.; Vaillant, F. Chemical Characterization, Antioxidant Properties, and Volatile Constituents of Naranjilla (Solanum quitoense Lam.) Cultivated in Costa Rica. Arch. Latinoam. Nutr. 2009, 59, 88–94.
- Kubmarawa, D.; Magomya, A.M.; Yebpella, G.G.; Adedayo, S.A. Nutrient Content and Amino Acid Composition of the Leaves of Cassia tora and Celtis integrifolia. Int. Res. J. Biochem. Bioinforma. 2011, 1, 222–225. [Google Scholar]
- Shukla, S.; Kumar, A.; Terrence, M.; Yusuf, J.; Singh, V.; Mishra, M. The Probable Medicinal Usage of Cassia tora: An Overview. OnLine J. Biol. Sci. 2013, 13, 109–125. [Google Scholar] [CrossRef] [Green Version]
- Li, J.-W.; Fan, L.-P.; Ding, S.-D.; Ding, X.-L. Nutritional Composition of Five Cultivars of Chinese Jujube. Food Chem. 2007, 103, 454–460. [Google Scholar] [CrossRef]
- Guil-Guerrero, J.; Delgado, A.; González, M.; Isasa, M.E. Fatty Acids and Carotenes in Some Ber (Ziziphus jujuba Mill) Varieties. Plant Foods Hum. Nutr. Dordr. Neth. 2004, 59, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.T. Edible Medicinal and Non-Medicinal Plants: Volume 1, Fruits; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Marcone, M.F.; Jahaniaval, F.; Aliee, H.; Kakuda, Y. Chemical Characterization of Achyranthes bidentata Seed. Food Chem. 2003, 81, 7–12. [Google Scholar] [CrossRef]
- Shen, R.; Yang, S.; Zhao, G.; Shen, Q.; Diao, X. Identification of Carotenoids in Foxtail Millet (Setaria italica) and the Effects of Cooking Methods on Carotenoid Content. J. Cereal Sci. 2015, 61, 86–93. [Google Scholar] [CrossRef]
- Yadav, A.K. Phalsa: A Potential New Small Fruit for Georgia. In Perspectives on New Crops and New Uses; Janik, J., Ed.; ASHS Press: Alexandria, Egypt, 1999; pp. 348–352. [Google Scholar]
- Khan, R.; Asghar, W.; Khalid, N.; Nazir, W.; Farooq, M.; Ahmed, I.; Syed, Q.A. Phalsa (Grewia asiatica L) Fruit Berry a Promising Functional Food Ingredient: A Comprehensive Review. J. Berry Res. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Venthodika, A.; Chhikara, N.; Mann, S.; Garg, M.K.; Sofi, S.A.; Panghal, A. Bioactive Compounds of Aegle marmelos L., Medicinal Values and Its Food applications: A Critical Review. Phytother. Res. 2021; 35. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, P.C.; Kumar, A.; Meena, M.D.; Sharma, D.K. Genotypic Differences for Salt Tolerance in Bael (Aegle marmelos) Cultivars. Indian J. Agric. Sci. 2018, 88, 435–441. [Google Scholar]
- Jayakumar, K.; Muthuraman, B. Traditional Uses and Nutrient Status of Indian Native Plant Fruit (Carissa carandas Linn.). World Sci. News 2018, 96, 217–224.
- Dalal, R.P.S. ; Navjot; Thakur, A.; Singh, A. Nutritional Value of Karonda (Carissa carandas Linn.)—A Non-Conventional Fruit under Semi-Arid Condition of Punjab. Indian J. Agrofor. 2020, 12, 102–104.
- Rodrigues, B.; Souza, B.; Nogueira, R.; Mauro, E.; Santos, M. Tolerance to Water Deficit in Young Trees of Jackfruit and Sugar Apple. Rev. Cienc. Agron. 2010, 41, 245–252. [Google Scholar] [CrossRef]
- Ranasinghe, R.A.S.N.; Maduwanthi, S.D.T.; Marapana, R.A.U.J. Nutritional and Health Benefits of Jackfruit (Artocarpus heterophyllus Lam.): A Review. Int. J. Food Sci. 2019; 12. [Google Scholar] [CrossRef] [Green Version]
- Busch, J.M.; Sangketkit, C.; Savage, G.P.; Martin, R.J.; Halloy, S.; Deo, B. Nutritional Analysis and Sensory Evaluation of Ulluco (Ullucus tuberosus Loz) Grown in New Zealand. J. Sci. Food Agric. 2000, 80, 2232–2240. [Google Scholar] [CrossRef]
- Lim, T. Arracacia xanthorrhiza. In Edible Medicinal and Non Medicinal Plants; Lim, T.K., Ed.; Springer: Cham, Switzerland, 2015; pp. 361–366. [Google Scholar]
- Manner, H.; Buker, R.; Smith, V.; Ward, D.; Elevitch, C. Species Profiles for Pacific Island Agroforestry; Permanent Agriculture Resources: New York, NY, USA, 2006. [Google Scholar]
- Chan-Blanco, Y.; Vaillant, F.; Mercedes Perez, A.; Reynes, M.; Brillouet, J.-M.; Brat, P. The Noni Fruit (Morinda citrifolia L.): A Review of Agricultural Research, Nutritional and Therapeutic Properties. Biodivers. Nutr. 2006; 19. [Google Scholar] [CrossRef]
- Ekanayake, S.; Jansz, E.; Nair, B. Literature Review of an Underutilized Legume: Canavalia gladiata L. Plant Foods Hum. Nutr. Dordr. Neth. 2000, 55, 305–321. [Google Scholar] [CrossRef]
- Popoola, J.; Ojuederie, O.; Omonhinmin, C.; Adegbite, A. Neglected and Underutilized Legume Crops: Improvement and Future Prospects. In Recent Advances in Grain Crops Research; IntechOpen: London, UK; 2019; ISBN 978-1-78985-449-7. [Google Scholar]
- Mohan, V.R.; Janardhanan, K. The Biochemical Composition and Nutrient Assessment of Less Known Pulses of the Genus Canavalia. Int. J. Food Sci. Nutr. 1994, 45, 255–262. [Google Scholar] [CrossRef]
- Eastwood, R.J.; Hughes, C.E. Lupinus mutabilis. Curtis’s Bot. Mag. 2018, 35, 134–148. [CrossRef]
- Carvajal-Larenas, F.E.; Linnemann, A.R.; Nout, M.J.R.; Koziol, M.; van Boekel, M.A.J.S. Lupinus mutabilis: Composition, Uses, Toxicology, and Debittering. Crit. Rev. Food Sci. Nutr. 2016, 56, 1454–1487. [Google Scholar] [CrossRef] [Green Version]
- Gulisano, A.; Alves, S.; Martins, J.N.; Trindade, L.M. Genetics and Breeding of Lupinus mutabilis: An Emerging Protein Crop. Front. Plant Sci. 2019, 10, 1385–1385. [Google Scholar] [CrossRef]
- Vijayvargia, P.; Vijayvergia, R. A Review on Limonia acidissima l.: Multipotential Medicinal Plant. Int. J. Pharm. Sci. Rev. Res. 2014, 28, 191–195.
- Ratnayake, S.S.; Kumar, L.; Kariyawasam, C.S. Neglected and Underutilized Fruit Species in Sri Lanka: Prioritisation and Understanding the Potential Distribution under Climate Change. Agronomy 2020, 10, 34. [Google Scholar] [CrossRef] [Green Version]
- Meghwal, P.; Singh, A. Lasoda or Gonda (Cordia myxa L.). In Lasoda or Gonda (Cordia myxa L.), Ghos, S.,N., Ed.; Jaya Publishing House: New Delhi, India, 2015; pp. 247–253. [Google Scholar]
- Singh, A.; Uppal, G. A Review on Carissa carandas Phytochemistry, Ethnopharmacology, and Micropropagation as Conservation Strategy. Asian J. Pharm. Clin. Res. 2015, 8, 26–30. [Google Scholar]
- Arif, M.; Kamal, M.; Jawaid, T. Carissa carandas Linn. (Karonda): An Exotic Minor Plant Fruit with Immense Value in Nutraceutical and Pharmaceutical Industries. Asian J. Biomed. Pharm. Sci. 2016, 6, 14–19.
- Muhammad, I.; Zhao, J.; Dunbar, D.C.; Khan, I.A. Constituents of Lepidium meyenii ‘Maca. 2002; 59. [Google Scholar] [CrossRef]
- Peres, N.d.S.L.; Bortoluzzi, L.C.P.; Marques, L.L.M.; Formigoni, M.; Fuchs, R.H.B.; Droval, A.A.; Cardoso, F.A.R. Medicinal Effects of Peruvian Maca (Lepidium meyenii): A Review. Food Funct. 2020, 11, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Lim, T.K. Pastinaca sativa. In Edible Medicinal and Non Medicinal Plants; Lim, T.K., Ed.; Springer: Dordrecht, The Netherlands, 2015; pp. 417–428. [Google Scholar]
- Tutin, T.G. Umbellifers of the British Isles; Botanical Society of the British Isles: London, UK, 1980. [Google Scholar]
- Boakye, A.A.; Wireko-Manu, F.D.; Oduro, I.; Ellis, W.O.; Gudjónsdóttir, M.; Chronakis, I.S. Utilizing Cocoyam (Xanthosoma sagittifolium) for Food and Nutrition Security: A Review. Food Sci. Nutr. 2018, 6, 703–713. [Google Scholar] [CrossRef]
- Miller, C.D. Food Values of Poi, Taro, and Limu, Periodicals Service Co: Hudson, NY, USA, 1971; p. 25.
- Nyman, L.P.; Gonzales, C.J.; Arditti, J. In-Vitro Selection for Salt Tolerance of Taro (Colocasia esculenta var antiquorum). Ann. Bot. 1983, 51, 229–236. [Google Scholar] [CrossRef]
- Rai, S.; Wahile, A.; Mukherjee, K.; Saha, B.P.; Mukherjee, P.K. Antioxidant Activity of Nelumbo nucifera (Sacred Lotus) Seeds. J. Ethnopharmacol. 2006, 104, 322–327. [Google Scholar] [CrossRef]
- Shad, M.; Nawaz, H.; Siddique, F.; Zahra, J.; Mushtaq, A. Nutritional and Functional Characterization of Seed Kernel of Lotus (Nelumbo nucifera): Application of Response Surface Methodology. Food Sci. Technol. Res. 2013, 19, 163–172. [Google Scholar] [CrossRef] [Green Version]
- Nohara, S.; Kimura, M. Growth Characteristics of Nelumbo nucifera Gaertn. in Response to Water Depth and Flooding. Ecol. Res. 1997, 12, 11–20. [Google Scholar] [CrossRef]
- Manikandan, S.; Lakshmanan, G.M.; Chandran, C. Phytochemical Screening and Evaluation of Tuber Extract of Plectranthus rotundifolius Spreng. By GC-MS and FTIR Spectrum Analysis. Eur. J. Herb. Med. 2016, 4, 36–40.
- Sethuraman, G.; Nizar, M.; Nadia, F.; Syaheerah, T.; Jahanshiri, E.; Gregory, P.; Azam-Ali, S. Nutritional Composition of Black Potato (Plectranthus rotundifolius (Poir.) Spreng.) (Synonym: Solenostemon rotundifolius). Int. J. Sci. Eng. Res. 2020, 11, 1145–1150.
- Priya, M.H.; Anbuselvi, S. Physico Chemical Analysis of Plectranthus rotundifolius. J. Chem. Pharm. Res. 2013, 5, 12–14. [Google Scholar]
- Hidalgo, A.; Brandolini, A. Nutritional Properties of Einkorn Wheat (Triticum monococcum L.). J. Sci. Food Agric. 2014, 94, 601–612.
- Prażak, R. Salt Tolerance of Triticum monococcum L., T. dicoccum (Schrank) Schubl., T. durum Desf. and T. aestivum L. Seedlings. J. Appl. Genet. 2001, 42, 289–292.
- Dhanavath, S.; Prasada Rao, U.J.S. Nutritional and Nutraceutical Properties of Triticum dicoccum Wheat and Its Health Benefits: An Overview. J. Food Sci. 2017, 82, 2243–2250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaharieva, M.; Ayana, N.G.; Hakimi, A.A; Misra, S.C.; Monneveux, P. Cultivated Emmer Wheat (Triticum dicoccon Schrank), An Old Crop with Promising Future: A Review. Genet. Resour. Crop Evol. 2010, 57, 937–962. [Google Scholar] [CrossRef]
- Zlatica, K.; Jolana, K. Nutritional Value and Baking Application of Spelt Wheat. Acta Sci. Pol. Technol. Aliment. 2008, 7, 5–14. [Google Scholar]
- Ruibal-Mendieta, N.L.; Delacroix, D.L.; Mignolet, E.; Pycke, J.-M.; Marques, C.; Rozenberg, R.; Petitjean, G.; Habib-Jiwan, J.-L.; Meurens, M.; Quetin-Leclercq, J.; et al. Spelt (Triticum aestivum Ssp. spelta) as a Source of Breadmaking Flours and Bran Naturally Enriched in Oleic Acid and Minerals but Not Phytic Acid. J. Agric. Food Chem. 2005; 53. [Google Scholar] [CrossRef]
- Burgos, M.; Messmer, M.; Stamp, P.; Schmid, J.E. Flooding Tolerance of Spelt (Triticum spelta L.) Compared to Wheat (Triticum aestivum L.)—A Physiological and Genetic Approach. 2001. [Google Scholar] [CrossRef]
- Chandra, D.; Chandra, S. ; Pallavi; Sharma, A.K. Review of Finger Millet (Eleusine coracana (L.) Gaertn): A Power House of Health Benefiting Nutrients. Food Sci. Hum. 2016; 5. [Google Scholar] [CrossRef] [Green Version]
- Hittalmani, S.; Mahesh, H.B.; Shirke, M.D.; Biradar, H.; Uday, G.; Aruna, Y.R.; Lohithaswa, H.C.; Mohanrao, A. Genome and Transcriptome Sequence of Finger Millet (Eleusine coracana (L.) Gaertn.) Provides Insights into Drought Tolerance and Nutraceutical Properties. BMC Genom. 2017; 18. [Google Scholar] [CrossRef]
- Devi, P.B.; Vijayabharathi, R.; Sathyabama, S.; Malleshi, N.G.; Priyadarisini, V.B. Health Benefits of Finger Millet (Eleusine coracana L.) Polyphenols and Dietary Fiber: A Review. J. Food Sci. Technol. 2011; 51. [Google Scholar] [CrossRef] [Green Version]
- Kalinova, J.; Moudry, J. Content and Quality of Protein in Proso Millet (Panicum miliaceum L.) Varieties. Plant Foods Hum. Nutr. Dordr. Neth. 2006; 61. [Google Scholar] [CrossRef]
- Zhang, Y. Comparative Analysis of Proso Millet (Panicum miliaceum L.) Leaf Transcriptomes for Insight into Drought Tolerance Mechanisms. BMC Plant Biol. 2019, 19, 397.
- Jukanti, A.K.; Gowda, C.L.L.; Rai, K.N.; Manga, V.K.; Bhatt, R.K. Crops That Feed the World 11. Pearl Millet (Pennisetum glaucum L.): An Important Source of Food Security, Nutrition and Health in the Arid and Semi-Arid Tropics. Food Secur. 2016; 8. [Google Scholar] [CrossRef]
- Sade, F.Q. Proximate, Antinutritional Factors and Functional Properties of Processed Pearl Millet (Pennisetum glaucum). J. Food Technol. 2009, 7, 92–97. [Google Scholar]
- Onyango, C.; Ochanda, S.; Mwasaru, M.; Ochieng, J.; Mathooko, F.; Kinyuru, J. Effects of Malting and Fermentation on Anti-Nutrient Reduction and Protein Digestibility of Red Sorghum, White Sorghum and Pearl Millet. J. Food Res. 2013, 2, 41–49. [Google Scholar] [CrossRef]
- Newman, Y.; Jennings, E.D.; Vendramini, J.; Blount, A. Pearl Millet (Pennisetum glaucum): Overview and Management; Institute of Food and Agricultural Sciences, University of Florida: Gainesville, FL, USA, 2020; Available online: https://edis.ifas.ufl.edu/publication/AG347 (accessed on 21 March 2021).
- Rzedowski, J. The Northern Limit of Tropical Rain Forests in Continental North America. Vegetatio 1963, 11, 173–198. [Google Scholar] [CrossRef]
- Subiria-Cueto, R.; Larqué-Saavedra, A.; Reyes-Vega, M.L.; de la Rosa, L.A.; Santana-Contreras, L.E.; Gaytán-Martínez, M.; Vázquez-Flores, A.A.; Rodrigo-García, J.; Corral-Avitia, A.Y.; Núñez-Gastélum, J.A.; et al. Brosimum alicastrum Sw. (Ramón): An Alternative to Improve the Nutritional Properties and Functional Potential of the Wheat Flour Tortilla. 2019; 8. [Google Scholar] [CrossRef] [Green Version]
- Taylor, M.B.; Tuia, V.S. Breadfruit in the Pacific Region. Acta Hortic. 2007, 757, 43–50. [Google Scholar] [CrossRef]
- Tukura, B.W.; Obliva, O. Proximate and Nutritional Compositions of Breadfruit (Artocarpus altilis) Seeds. IOSR J. Environ. Sci. 2015, 9, 68–73. [Google Scholar] [CrossRef]
- Encalada, S.V.; Campos, M.R.S. Mucuna pruriens Fiber: Nutritional, Functional And Biological Properties. Food Sci. Tech. 2020, 41, 120–126. [Google Scholar] [CrossRef]
- Lampariello, L.R.; Cortelazzo, A.; Guerranti, R.; Sticozzi, C.; Valacchi, G. The Magic Velvet Bean of Mucuna pruriens. J. Tradit. Complement. Med. 2012, 2, 331–339. [Google Scholar] [CrossRef]
- Rodrigues, A.P.; Pereira, G.A.; Tomé, P.H.F.; Arruda, H.S.; Eberlin, M.N.; Pastore, G.M. Chemical Composition and Antioxidant Activity of Monguba (Pachira aquatica) Seeds. Food Res. Int. 2019, 121, 880–887. [Google Scholar] [CrossRef]
- Oliveira, J.T.A.; Vasconcelos, I.; Bezerra, L.C.N.M.; Silveira, S.B.; Monteiro-Moreira, A.; Moreira, R. Composition and Nutritional Properties of Seeds from Pachira aquatica Aubl, Sterculia striata St Hil et Naud and Terminalia catappa Linn. Food Chem. 2000, 70, 185–191. [Google Scholar] [CrossRef]
- Orwa, C.; Mutua, C.; Kindt, R.; Jamnadass, R.; Anthony, S. Strychnos cocculoides. Available online: http://apps.worldagroforestry.org/treedb/AFTPDFS/Strychnos_cocculoides.PDF (accessed on 2 March 2021).
- Ngadze, R.T.; Linnemann, A.R.; Nyanga, L.K.; Fogliano, V.; Verkerk, R. Local Processing and Nutritional Composition of Indigenous Fruits: The Case of Monkey Orange (Strychnos Spp.) from Southern Africa. Food Rev. Int. 2017; 33. [Google Scholar] [CrossRef] [Green Version]
- Maikhuri, R.K.; Semwal, R.L.; Rao, K.S.; Nautiyal, S.; Saxena, K.G. Eroding Traditional Crop Diversity Imperils the Sustainability of Agricultural Systems in Central Himalaya. Curr. Sci. 1997, 73, 777–782. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Prasad, M. Small Millets for Enduring Food Security Amidst Pandemics. Trends Plant Sci. 2020, 26, 1–8. [Google Scholar] [CrossRef]
- Zsögön, A.; Cermak, T.; Voytas, D.; Peres, L. Genome Editing as a Tool to Achieve the Crop Ideotype and de novo Domestication of Wild Relatives: Case Study in Tomato. Plant Sci. 2016, 256, 120–130. [Google Scholar] [CrossRef]
- Hammer, K.; Arrowsmith, N.; Gladis, T. Agrobiodiversity with Emphasis on Plant Genetic Resources. Sci. Nat. 2003, 90, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Ogwu, M.; Osawaru, M.; Ahana, C. Challenges in Conserving and Utilizing Plant Genetic Resources (PGR). Int. J. Genet. Mol. Biol. 2014, 6, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Mabhaudhi, T.; Chibarabada, T.P.; Chimonyo, V.G.P.; Murugani, V.G.; Pereira, L.M.; Sobratee, N.; Govender, L.; Slotow, R.; Modi, A.T. Mainstreaming Underutilized Indigenous and Traditional Crops into Food Systems: A South African Perspective. Sustainability 2019, 11, 172. [Google Scholar] [CrossRef] [Green Version]
- Moose, S.; Mumm, R. Molecular Plant Breeding as the Foundation for 21st Century Crop Improvement. Plant Physiol. 2008, 147, 969–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukushima, A.; Kusano, M.; Redestig, H.; Arita, M.; Saito, K. Integrated Omics Approaches in Plant Systems Biology. Curr. Opin. Chem. Biol. 2009, 13, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Friel, S.; Hattersley, L.; Snowdon, W.; Thow, A.-M.; Lobstein, T.; Sanders, D.; Barquera, S.; Mohan, S.; Hawkes, C.; Kelly, B.; et al. Monitoring the Impacts of Trade Agreements on Food Environments. Obes. Rev. Off. J. Int. Assoc. Study Obes. 2013, 14, 120–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grote, U. Can We Improve Global Food Security? A Socio-Economic and Political Perspective. Food Secur. 2014, 6, 187–200. [Google Scholar] [CrossRef]
- Glauber, J.; Laborde Debucquet, D.; Martin, W.; Vos, R. COVID-19: Trade Restrictions Are Worst Possible Response to Safeguard Food Security. Available online: https://ebrary.ifpri.org/digital/collection/p15738coll2/id/133833/ (accessed on 7 June 2021).
- Laborde, D.; Martin, W.; Swinnen, J.; Vos, R. COVID-19 Risks to Global Food Security. Science 2020, 369, 500–502. [Google Scholar] [CrossRef]
- Espitia, A.; Rocha, N.; Ruta, M. COVID-19 and Food Protectionism: The Impact of the Pandemic and Export Restrictions on World Food Markets; Policy Research Working Paper; No. 9253; Social Science Research Network: Rochester, NY, USA; World Bank, Washington, DC, USA, 2020; Available online: https://openknowledge.worldbank.org/handle/10986/33800 (accessed on 9 March 2021).
- Béné, C. Resilience of Local Food Systems and Links to Food Security—A Review of Some Important Concepts in the Context of COVID-19 and Other Shocks. Food Secur. 2020, 12, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Paudyal, A.; Regmi, B. Climate Change and Agrobiodiversity in Nepal: Opportunities to Include Agrobiodiversity Maintenance to Support Nepal’s National Adaptation Programme of Action; A Report Prepared by LI-BIRD for the Platform for Agrobiodiversity Research in Collaboration with FAO and Bioversity International; FAO: Rome, Italy, 2009. [Google Scholar]
- Coelho, F.C.; Coelho, E.M.; Egerer, M. Local Food: Benefits and Failings Due to Modern Agriculture. Sci. Agric. 2018, 75, 84–94. [Google Scholar] [CrossRef] [Green Version]
- Sheil, D.; Wunder, S. The Value of Tropical Forest to Local Communities: Complications, Caveats, and Cautions. Ecol. Soc. 2002, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Legwaila, G.M.; Mojeremane, W.; Madisa, M.; Mmolotsi, R.; Rampart, M. Potential of Traditional Food Plants in Rural Household Food Security in Botswana. J. Hortic. For. 2011, 3, 171–177. [Google Scholar] [CrossRef]
- Nesamvuni, C.; Steyn, N.; Potgieter, M. Nutritional Value of Wild, Leafy Plants Consumed by the Vhavenda. South Afr. J. Sci. 2001, 97, 51–54. [Google Scholar]
- Kadu, C.A.C.; Imbuga, M.; Jamnadass, R.; Dawson, I.K. Genetic Management of Indigenous Fruit Trees in Southern Africa: A Case Study of Sclerocarya birrea Based on Nuclear and Chloroplast Variation. South Afr. J. Bot. 2006, 72, 421–427. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Garcia, G.S.; Price, L.L. Gathering of Wild Food Plants in Anthropogenic Environments across the Seasons: Implications for Poor and Vulnerable Farm Households. Ecol. Food Nutr. 2014, 53, 363–389. [Google Scholar] [CrossRef]
- Bourgeois, R. Secondary crops, rural poverty and policy bias. In Proceedings of the Regional Workshop on Rural Prosperity and Secondary Crops: Towards Applied Pro-Poor Research and Policies in Asia and the Pacific, Bogor, Indonesia; 6–9 December, 2005, Bourgeois, R., Svensson, L., Burrows, M., Eds.; UNESCAP-CAPSA. Publisher: Bogor, Indonesia, 2005. [Google Scholar]
- Hart, T. The Significance of African Vegetables in Ensuring Food Security for South Africa’s Rural Poor. Agric. Hum. Values 2011, 28, 321–333. [Google Scholar] [CrossRef]
- Lockett, C.T.; Calvert, C.C.; Grivetti, L.E. Energy and Micronutrient Composition of Dietary and Medicinal Wild Plants Consumed during Drought. Study of Rural Fulani, Northeastern Nigeria. Int. J. Food Sci. Nutr. 2000; 51. [Google Scholar] [CrossRef]
- Amadou, I.; Gbadamosi, O.; Le, G. Millet-Based Traditional Processed Foods and Beverages: A Review. Cereal Foods World 2011, 56, 115–121. [Google Scholar] [CrossRef]
- Yenagi, N.; Handigol, J.; Ravi, S.; Mal, B.; Padulosi, S. Nutritional and Technological Advancements in the Promotion of Ethnic and Novel Foods Using the Genetic Diversity of Minor Millets in India. Indian J. Plant Genet. Resour. 2010, 23, 82–86. [Google Scholar]
- Islam, M.; Das, P.R.; Salehin, M.F.; Mahmud, B.; Hasan, M.; Jahan, I.; Seraj, S.; Islam, F.; Khatun, Z.; Chowdhury, A.; et al. A Survey of Non-Conventional Plant Items Consumed During Food Scarcity in Two Randomly Selected Villages of Kurigram District, Bangladesh. Am. Eurasian J. Sustain. Agric. 2011, 5, 233–239. [Google Scholar]
- Bhattacharjee, R. Harnessing Biotechnology for Conservation and Increased Utilization of Orphan Crops. Afr. Technol. Dev. Forum J. 2009, 6, 24–82. [Google Scholar]
- Hu, H.; Scheben, A.; Edwards, D. Advances in Integrating Genomics and Bioinformatics in the Plant Breeding Pipeline. Agriculture 2018, 8, 75. [Google Scholar] [CrossRef] [Green Version]
- Mosa, K.A.; Ismail, A.; Helmy, M. Plant Stress Tolerance: An Integrated Omics Approach; SpringerBriefs in Systems Biology; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Yokoyama, S.; Yura, K. Special Issue: Big Data Analyses in Structural and Functional Genomics. J. Struct. Funct. Genom. 2017, 17, 67–70. [Google Scholar] [CrossRef] [Green Version]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics Technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [Green Version]
- Appleby, N.; Edwards, D.; Batley, J. New Technologies for Ultra-High Throughput Genotyping in Plants. In Plant Genomics: Methods and Protocols; Gustafson, J.P., Langridge, P., Somers, D.J., Eds.; Humana Press: Totowa, NJ, USA, 2009; pp. 19–39. [Google Scholar]
- Vu, L.D.; Gevaert, K.; De Smet, I. Protein Language: Post-Translational Modifications Talking to Each Other. Trends Plant Sci. 2018, 23, 1068–1080. [Google Scholar] [CrossRef] [PubMed]
- Eldakak, M.; Milad, S.I.M.; Nawar, A.I.; Rohila, J.S. Proteomics: A Biotechnology Tool for Crop Improvement. Front. Plant Sci. 2013, 4, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahner, B.; Gong, J.; Mahmoudian, M.; Smith, E.L.; Abid, K.B.; Rogers, E.E.; Guerinot, M.L.; Harper, J.F.; Ward, J.M.; McIntyre, L.; et al. Genomic Scale Profiling of Nutrient and Trace Elements in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 1215–1221. [Google Scholar] [CrossRef]
- Van Emon, J. Omics Revolution in Agricultural Research. J. Agric. Food Chem. 2015, 64, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Benkeblia, N. Omics Technologies and Crop Improvement; Taylor & Francis, Milten Park, Australia, 2014.
- Xia, E.-H.; Tong, W.; Wu, Q.; Wei, S.; Zhao, J.; Zhang, Z.-Z.; Wei, C.-L.; Wan, X.-C. Tea Plant Genomics: Achievements, Challenges and Perspectives. Hortic. Res. 2020, 7, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ausubel, F.M. Arabidopsis Genome. A Milestone in Plant Biology. Plant Physiol. 2000, 124, 1451–1454. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, J.P.; Buell, C.R. Advances in Plant Genome Sequencing. Plant J. Cell Mol. Biol. 2012, 70, 177–190. [Google Scholar] [CrossRef]
- Michael, T.P.; Jackson, S. The First 50 Plant Genomes. Plant Genome 2013, 6, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Heck, M.; Neely, B.A. Proteomics in Non-Model Organisms: A New Analytical Frontier. J. Proteome Res. 2020, 19, 3595–3606. [Google Scholar] [CrossRef]
- Fabres, P.J. A Multiple “Omics” Approach to Study the Interaction between the Vitis vinifera Transcriptome and Epigenome and the Barossa Valley Terroir. Ph.D. Dissertation, University of Adelaide, Australia, 2020. [Google Scholar]
- Kersey, P. Plant Genome Sequences: Past, Present, Future. Curr. Opin. Plant Biol. 2019, 48, 1–8. [Google Scholar] [CrossRef]
- Pareek, C.S.; Smoczynski, R.; Tretyn, A. Sequencing Technologies and Genome Sequencing. J. Appl. Genet. 2011, 52, 413–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pryer, K.M.; Schneider, H.; Zimmer, E.A.; Ann Banks, J. Deciding among Green Plants for Whole Genome Studies. Trends Plant Sci. 2002, 7, 550–554. [Google Scholar] [CrossRef]
- Steinwand, M.; Ronald, P. Crop Biotechnology and the Future of Food. Nat. Food 2020, 1, 273–283. [Google Scholar] [CrossRef]
- Mochida, K.; Shinozaki, K. Advances in Omics and Bioinformatics Tools for Systems Analyses of Plant Functions. Plant Cell Physiol. 2011, 52, 2017–2038. [Google Scholar] [CrossRef]
- Lepcha, P.; Kumar, P.R.; Sathyanarayana, N. Exploring Genomics Research in the Context of Some Underutilized Legumes—A Review. In OMICS-Based Approaches in Plant Biotechnology; Banerjee, R., Kumar, G.V., Kumar, S.P.J., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019; pp. 1–18. [Google Scholar]
- Khound, R.; Santra, D. Omics for Proso Millet Genetic Improvement. Nucl. India 2020, 63, 241–247. [Google Scholar] [CrossRef]
- Moe, K.T.; Kwon, S.-W.; Park, Y. Trends in Genomics and Molecular Marker Systems for the Development of Some Underutilized Crops. Genes Genom. 2012, 34, 451–456. [Google Scholar] [CrossRef]
- Chang, Y.; Liu, H.; Liu, M.; Liao, X.; Sahu, S.K.; Fu, Y.; Song, B.; Cheng, S.; Kariba, R.; Muthemba, S.; et al. The Draft Genomes of Five Agriculturally Important African Orphan Crops. GigaScience 2019, 8, 1–16. [Google Scholar] [CrossRef]
- Li, Q.; Yan, J. Sustainable Agriculture in the Era of Omics: Knowledge-Driven Crop Breeding. Genome Biol. 2020, 21, 1–5. [Google Scholar] [CrossRef]
- Singh, N.; Rai, V.; Singh, N. Multi-omics Strategies and Prospects to Enhance Seed Quality and Nutritional Traits in Pigeonpea. Nucleus 2020, 21, 1–8. [Google Scholar] [CrossRef]
- Verma, V.; Patel, S. Value Added Products from Nutri-Cereals: Finger Millet (Eleusine coracana). Emir. J. Food Agric. 2012, 25, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Nirgude, M.; Babu, B.; Shambhavi, Y.; Singh, U.; Upadhyaya, H.; Kumar, A. Development and Molecular Characterization of Genic Molecular Markers for Grain Protein and Calcium Content in Finger Millet (Eleusine coracana (L.) Gaertn.). Mol. Biol. Rep. 2014; 41. [Google Scholar] [CrossRef]
- Kumar, A.; Babu, B.; Yadav, S.; Agrawal, P. Allele Mining for Resistance Gene Analogs (RGAs) in Crop Plants: A Special Emphasis on Blast Resistance in Finger Millet (Eleusine coracana L.). Indian J. Genet. Plant Breed. 2016; 76. [Google Scholar] [CrossRef]
- Kumar, A.; Gaur, V.; Goel, A.; Gupta, A. De Novo Assembly and Characterization of Developing Spikes Transcriptome of Finger Millet (Eleusine coracana): A Minor Crop Having Nutraceutical Properties. Plant Mol. Biol. Report. 2014, 33, 905–922. [Google Scholar] [CrossRef]
- Singh, M.; Metwal, M.; Kumar, V.; Kumar, A. Identification and Molecular Characterization of 48 KDa Calcium Binding Protein as Calreticulin from Finger Millet (Eleusine coracana) Using Peptide Mass Fingerprinting and Transcript Profiling. J. Sci. Food Agric. 2015, 96, 672–679. [Google Scholar] [CrossRef]
- Anatala, T.; Gajera, H.; Mandavia, M.; Dave, R.; Vallabhbhai, K.; Golakiya, B.A. Leaf Proteome Alterations in Tolerant Pearl Millet (Pennisetum glaucum L.) Genotype under Water Stress. Int. J. Agric. Environ. Biotechnol. 2015; 8. [Google Scholar] [CrossRef]
- Bandyopadhyay, T.; Muthamilarasan, M.; Prasad, M. Millets for Next Generation Climate-Smart Agriculture. Front. Plant Sci. 2017, 8, 1266. [Google Scholar] [CrossRef] [Green Version]
- Lata, C.; Sahu, P.P.; Prasad, M. Comparative Transcriptome Analysis of Differentially Expressed Genes in Foxtail Millet (Setaria italica L.) during Dehydration Stress. Biochem. Biophys. Res. Commun. 2010. [Google Scholar] [CrossRef]
- Shi, W.; Cheng, J.; Wen, X.; Wang, J.; Shi, G.; Yao, J.; Liyuan, H.; Sun, Q.; Xiang, P.; Yuan, X.; et al. Transcriptomic Studies Reveal a Key Metabolic Pathway Contributing to a Well-Maintained Photosynthetic System under Drought Stress in Foxtail Millet (Setaria italica L.). 2018; 6. [Google Scholar] [CrossRef] [Green Version]
- Sreenivasulu, N.; Miranda, M.; Prakash, H.S.; Wobus, U.; Weschke, W. Transcriptome Changes in Foxtail Millet Genotypes at High Salinity: Identification and Characterization of a PHGPX Gene Specifically up-Regulated by NaCl in a Salt-Tolerant Line. J. Plant Physiol. 2004, 161, 467–477. [Google Scholar] [CrossRef]
- Leone, A.; Fiorillo, G.; Criscuoli, F.; Ravasenghi, S.; Santagostini, L.; Fico, G.; Spadafranca, A.; Battezzati, A.; Schiraldi, A.; Pozzi, F.; et al. Nutritional Characterization and Phenolic Profiling of Moringa oleifera Leaves Grown in Chad, Sahrawi Refugee Camps, and Haiti. Int. J. Mol. Sci. 2015, 16, 18923–18937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosch, C.H. Moringa oleifera Lam. In Plant Resources of Tropical Africa Vegetables; Grubben, G.J.H., Denton, O.A., Eds.; Backhuys Publishers: Kerkwerve, The Netherlands, 2004. [Google Scholar]
- Tian, Y.; Zeng, Y.; Zhang, J.; Yang, C.; Yan, L.; Wang, X.; Shi, C.; Xie, J.; Dai, T.; Peng, L.; et al. High Quality Reference Genome of Drumstick Tree (Moringa oleifera Lam.), a Potential Perennial Crop. Sci. China Life Sci. 2015; 58. [Google Scholar] [CrossRef] [Green Version]
- Pirrò, S.; Matic, I.; Guidi, A.; Zanella, A.; Gisondi, A.; Cicconi, A.; Bernardini, R.; Colizzi, V.; Canini, A.; Mattei, M.; et al. Identification of microRNAs and Relative Target Genes in Moringa oleifera leaf and callus. Sci. Rep. 2019, 9, 15145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasha, S.N.; Shafi, K.M.; Joshi, A.G.; Meenakshi, I.; Harini, K.; Mahita, J.; Sajeevan, R.S.; Karpe, S.D.; Ghosh, P.; Nitish, S.; et al. The Transcriptome Enables the Identification of Candidate Genes behind Medicinal Value of Drumstick Tree (Moringa oleifera). Genomics 2020, 112, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Makita, C.S. Metabolomic Exploration of Pharmacologically Relevant Metabolites in Moringa oleifera and Moringa ovalifolia through the Use of UPLC-QTOF-MS and Multivariate Models. Ph.D. Thesis, Faculty of Science, University of the Witwatersrand, Johannesburg, South Africa, 2017. [Google Scholar]
- Fuentes, F.; Martínez, E.; Hinrichsen, P.; Jellen, R.; Maughan, J. Assessment of Genetic Diversity Patterns in Chilean Quinoa (Chenopodium quinoa Willd.) Germplasm Using Multiplex Fluorescent Microsatellite Markers. Conserv. Genet. 2009; 10. [Google Scholar] [CrossRef]
- Nowak, V.; Du, J.; Charrondière, U.R. Assessment of the Nutritional Composition of Quinoa (Chenopodium quinoa Willd.). Food Chem. 2016; 54. [Google Scholar] [CrossRef]
- Aranda, M.; Vega-Galvez, A.; Quispe, I.; Rodriguez, M.; Martínez, E. Nutritional Aspects of Six Quinoa (Chenopodium quinoa Willd.) Ecotypes from the Geographical Areas of Chile. Chil. J. Agric. Res. 2012, 72, 175–181.
- Yasui, Y.; Hirakawa, H.; Oikawa, T.; Toyoshima, M.; Matsuzaki, C.; Ueno, M.; Mizuno, N.; Nagatoshi, Y.; Imamura, T.; Miyago, M.; et al. Draft Genome Sequence of an Inbred Line of Chenopodium quinoa, an Allotetraploid Crop with Great Environmental Adaptability and Outstanding Nutritional Properties. DNA Res. 2016, 23, 215–224. [Google Scholar] [CrossRef]
- Ruiz Carrasco, K.; Maldonado, J.; Biondi, S.; Silva, H. RNA-Seq Analysis of Salt-Stressed Versus Non Salt-Stressed Transcriptomes of Chenopodium quinoa Landrace R49. Genes 2019, 10, 1042. [Google Scholar] [CrossRef] [Green Version]
- Sobota, A.; Swieca, M.; Gesinski, K.; Wirkijowska, A.; Bochnak-Niedźwiecka, J. Yellow-coated Quinoa (Chenopodium quinoa Willd)—Physicochemical, Nutritional, and Antioxidant Properties. J. Sci. Food Agric. 2019, 100, 2035–2042. [Google Scholar] [CrossRef]
- Bose, J.; Rodrigo-Moreno, A.; Lai, D.; Xie, Y.; Shen, W.; Shabala, S. Rapid Regulation of the Plasma Membrane H+-ATPase Activity Is Essential to Salinity Tolerance in Two Halophyte Species, Atriplex lentiformis and Chenopodium quinoa. Ann. Bot. 2015, 115, 481–494. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, A.; Goufo, P.; Barros, A.; Domínguez-Perles, R.; Trindade, H.; Rosa, E.A.S.; Ferreira, L.; Rodrigues, M. Cowpea (Vigna unguiculata L. Walp), a Renewed Multipurpose Crop for a More Sustainable Agri-Food System: Nutritional Advantages and Constraints. J. Sci. Food Agric. 2016; 96. [Google Scholar] [CrossRef]
- Jayathilake, C.; Visvanathan, R.; Deen, A.; Bangamuwage, R.; Jayawardana, B.C.; Nammi, S.; Liyanage, R. Cowpea: An Overview on Its Nutritional Facts and Health Benefits. J. Sci. Food Agric. 2018, 98, 4793–4806. [Google Scholar] [CrossRef]
- Domínguez-Perles, R.; Machado, N.; Abraão, A.S.; Carnide, V.; Ferreira, L.; Rodrigues, M.; Rosa, E.A.D.S.; Barros, A.I.R.N.A. Chemometric Analysis on Free Amino Acids and Proximate Compositional Data for Selecting Cowpea (Vigna unguiculata L.) Diversity. J. Food Compos. Anal. 2016; 53. [Google Scholar] [CrossRef]
- Ehlers, J.D.; Hall, A.E. Cowpea (Vigna unguiculata L. Walp.). Field Crops Res. 1997; 53. [Google Scholar] [CrossRef]
- Amorim, L.L.B.; Ferreira-Neto, J.R.C.; Bezerra-Neto, J.P.; Pandolfi, V.; de Araújo, F.T.; da Silva Matos, M.K.; Santos, M.G.; Kido, E.A.; Benko-Iseppon, A.M. Cowpea and Abiotic Stresses: Identification of Reference Genes for Transcriptional Profiling by QPCR. Plant Methods 2018, 14, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Goufo, P.; Moutinho-Pereira, J.M.; Jorge, T.F.; Correia, C.M.; Oliveira, M.R.; Rosa, E.A.S.; António, C.; Trindade, H. Cowpea (Vigna uunguiculata L. Walp.) Metabolomics: Osmoprotection as a Physiological Strategy for Drought Stress Resistance and Improved Yield. Front. Plant Sci. 2017; 8. [Google Scholar] [CrossRef] [Green Version]
- Hashiguchi, A.; Zhu, W.; Tian, J.; Komatsu, S. Proteomics and Metabolomics-Driven Pathway Reconstruction of Mung Bean for Nutraceutical Evaluation. BBA-Proteins Proteom. 2017. [Google Scholar] [CrossRef]
- Akundabweni, L.S.M.; Maina, D.; Akundabweni, L. Ionomic Variation Characterization in African Leafy Vegetables for Micronutrients Using XRF and HPLC. Afr. J. Food Agric. Nutr. Dev. 2011, 10, 4320–4339. [Google Scholar] [CrossRef]
- Tang, D.; Dong, Y.; Guo, N.; Li, L.; Ren, H. Metabolomic Analysis of the Polyphenols in Germinating Mung Beans (Vigna rradiata) Seeds and Sprouts. J. Sci. Food Agric. 2014, 94, 1639–1647. [Google Scholar] [CrossRef]
- Haider, M.; Hussain, M.; Farooq, M.; Nawaz, A. Zinc Nutrition for Improving the Productivity and Grain Biofortification of Mungbean. J. Soil Sci. Plant Nutr. 2020, 20, 1321–1325. [Google Scholar] [CrossRef]
- Kangama, C.; Xu, R. Introduction of Sorghum (Sorghum bicolor (L.) Moench) into China. Afr. J. Biotechnol. 2005, 4, 575–579.
- Rhodes, D.H.; Hoffmann, L.; Rooney, W.L.; Ramu, P.; Morris, G.P.; Kresovich, S. Genome-Wide Association Study of Grain Polyphenol Concentrations in Global Sorghum (Sorghum bbicolor (L.) Moench) Germplasm. J. Agric. Food Chem. 2014; 62. [Google Scholar] [CrossRef]
- Kulamarva, A.G.; Sosle, V.R.; Raghavan, G.S.V. Nutritional and Rheological Properties of Sorghum. Int. J. Food Prop. 2009, 12, 55–69. [Google Scholar] [CrossRef]
- Kaplan, M.; Kale, H.; Kardes, Y.M.; Karaman, K.; Kahraman, K.; Yılmaz, M.F.; Temizgül, R.; Akar, T. Characterization of Local Sorghum (Sorghum bbicolor L.) Population Grains in Terms of Nutritional Properties and Evaluation by GT Biplot Approach. 2020; 72. [Google Scholar] [CrossRef]
- Johnson, S.M.; Lim, F.-L.; Finkler, A.; Fromm, H.; Slabas, A.R.; Knight, M.R. Transcriptomic Analysis of Sorghum bicolor Responding to Combined Heat and Drought Stress. BMC Genom. 2014, 15, 456–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchanan, C.D.; Lim, S.; Salzman, R.A.; Kagiampakis, I.; Morishige, D.T.; Weers, B.D.; Klein, R.R.; Pratt, L.H.; Cordonnier-Pratt, M.-M.; Klein, P.E.; et al. Sorghum bicolor’s Transcriptome Response to Dehydration, High Salinity and ABA. Plant Mol. Biol. 2005, 58, 699–720. [Google Scholar] [CrossRef] [PubMed]
- Ogbaga, C.; Stepien, P.; Dyson, B.; Rattray, N.; Ellis, D.; Goodacre, R.; Johnson, G. Biochemical Analyses of Sorghum Varieties Reveal Differential Responses to Drought. PLoS ONE 2016, 11, e0154423. [Google Scholar] [CrossRef]
- Swamy, A.K.; Alam, S.; Sengupta, N.; Sarina, R. Differential Proteomic Analysis of Salt Stress Response in Sorghum bbicolor Leaves. Environ. Exp. Bot. 2011, 71, 321–328. [Google Scholar] [CrossRef]
- Ferraro, V.; Piccirillo, C.; Tomlins, K.; Pintado, M.E. Cassava (Manihot esculenta Crantz) and Yam (Dioscorea Spp.) Crops and Their Derived Foodstuffs: Safety, Security and Nutritional Value. Crit. Rev. Food Sci. Nutr. 2016; 56. [Google Scholar] [CrossRef]
- Siriwat, W.; Kalapanulak, S.; Suksangpanomrung, M.; Netrphan, S.; Meechai, A.; Saithong, T. Transcriptomic Data Integration Inferring the Dominance of Starch Biosynthesis in Carbon Utilization of Developing Cassava Roots. Procedia Comput. Sci. 2012, 11, 96–106. [Google Scholar] [CrossRef] [Green Version]
- Salvador, E.; Steenkamp, V.; Mccrindle, C. Production, Consumption and Nutritional Value of Cassava (Manihot esculenta, Crantz) in Mozambique: An Overview. J. Agric. Biotechnol. Sustain. Dev. 2014, 6, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Rabbi, I.; Udoh, L.; Wolfe, M.; Parkes, E.; Gedil, M.; Dixon, A.; Ramu, P.; Jannink, J.-L.; Kulakow, P. Genome-Wide Association Mapping of Correlated Traits in Cassava: Dry Matter and Total Carotenoid Content. Plant Genome 2017, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.J.; Ren, M.Y.; Lu, L.F.; Peng, M.; Guan, X.; Zhou, D.B.; Zhang, M.Y.; Qi, D.F.; Li, K.; Tang, W.; et al. Involvement of Abscisic Acid-Responsive Element-Binding Factors in Cassava (Manihot esculenta) Dehydration Stress Response. Sci. Rep. 2019, 9, 12661. [Google Scholar] [CrossRef]
- Utsumi, Y.; Tanaka, M.; Morosawa, T.; Kurotani, A.; Yoshida, T.; Mochida, K.; Matsui, A.; Umemura, Y.; Ishitani, M.; Shinozaki, K.; et al. Transcriptome Analysis Using a High-Density Oligomicroarray under Drought Stress in Various Genotypes of Cassava: An Important Tropical Crop. DNA Res. 2012, 19, 335–345. [Google Scholar] [CrossRef] [Green Version]
- Lokko, Y.; Anderson, J.V.; Rudd, S.; Raji, A.; Horvath, D.; Mikel, M.A.; Kim, R.; Liu, L.; Hernandez, A.; Dixon, A.G.O.; et al. Characterization of an 18,166 EST Dataset for Cassava (Manihot esculenta Crantz) Enriched for Drought-Responsive Genes. Plant Cell Rep. 2007, 26, 1605–1618. [Google Scholar] [CrossRef]
- Wei, Y.; Shi, H.; Xia, Z.; Tie, W.; Ding, Z.; Yan, Y.; Wang, W.; Hu, W.; Li, K. Genome-Wide Identification and Expression Analysis of the WRKY Gene Family in Cassava. Front. Plant Sci. 2016, 7, 1–18. [Google Scholar] [CrossRef] [Green Version]
- An, F.; Li, G.; Li, Q.; Li, K.; Carvalho, L.; Ou, W.; Chen, S. The Comparatively Proteomic Analysis in Response to Cold Stress in Cassava Plantlets. Plant Mol. Biol. Report. 2016, 34, 1095–1110. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, G.K.; Price, M.L. Amaranth: Grain and Vegetable Types; Echo Technical Note: Bangui, Central Africa, 2008. [Google Scholar]
- Bressani, R. The Proteins of Grain Amaranth. Food Rev. Int. 1989, 5, 13–38. [Google Scholar] [CrossRef]
- Alegbejo, J. Nutritional Value and Utilization of Amaranthus (Amaranthus spp.)—A Review. Bayero J. Pure Appl. Sci. 2014; 6. [Google Scholar] [CrossRef] [Green Version]
- Sunil, M.; Hariharan, A.K.; Nayak, S.; Gupta, S.; Nambisan, S.; Gupta, R.; Panda, B.; Choudhary, B.; Srinivasan, S. The Draft Genome and Transcriptome of Amaranthus hypochondriacus: A C4 Dicot Producing High-Lysine Edible Pseudo-Cereal. DNA Res. 2014, 21, 585–602. [Google Scholar] [CrossRef]
- Huerta-Ocampo, J.Á.; Briones-Cerecero, E.P.; Mendoza-Hernández, G.; De León-Rodríguez, A.; Barba de la Rosa, A.P. Proteomic Analysis of Amaranth (Amaranthus hypochondriacus L.) Leaves under Drought Stress. Int. J. Plant Sci. 2009. [Google Scholar] [CrossRef] [Green Version]
- Lokhande, V.; Nikam, T.; Penna, S. Sesuvium portulacastrum (L.) L. A Promising Halophyte: Cultivation, Utilization and Distribution in India. Genet. Resour. Crop Evol. 2009; 56. [Google Scholar] [CrossRef]
- Zeng, H.-C.; Deng, L.-H.; Zhang, C.-F. Cloning of Salt Tolerance-Related CDNAs from the Mangrove Plant Sesuvium portulacastrum L. J. Integr. Plant Biol. 2006, 48, 952–957. [Google Scholar] [CrossRef]
- Fan, W.; Zhang, Z.; Zhang, Y. Cloning and Molecular Characterization of Fructose-1,6-Bisphosphate Aldolase Gene Regulated by High-Salinity and Drought in Sesuvium portulacastrum. Plant Cell Rep. 2009, 28, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Venkatesalu, V.; Kumar, R.R.; Chellappan, K.P. Growth and Mineral Distribution of Sesuvium portulacastrum l., a Salt Marsh Halophyte, under Sodium Chloride Stress. Commun. Soil Sci. Plant Anal. 1994; 25. [Google Scholar] [CrossRef]
- Mohanraj, R.; Sivasankar, S. Sweet Potato (Ipomoea batatas [L.] Lam)—A Valuable Medicinal Food: A Review. J. Med. 2014; 17. [Google Scholar] [CrossRef]
- Tao, X.; Gu, Y.-H.; Wang, H.-Y.; Zheng, W.; Li, X.; Zhao, C.-W.; Zhang, Y.-Z. Digital Gene Expression Analysis Based on Integrated de Novo Transcriptome Assembly of Sweet Potato (Ipomoea batatas (L.) Lam). 2012; 7. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Li, R.; Ren, L.; Gao, X.; Zhang, Y.; Ma, Z.; Ma, D.; Luo, Y. A Comparative Metabolomics Study of Flavonoids in Sweet Potato with Different Flesh Colors (Ipomoea batatas (L.) Lam). Food Chem. 2018. [Google Scholar] [CrossRef]
- Teow, C.C.; Truong, V.; McFeeters, R.F.; Thompson, R.L.; Pecota, K.V.; Yencho, G.C. Antioxidant Activities, Phenolic and β-Carotene Contents of Sweet Potato Genotypes with Varying Flesh Colours. Food Chem. 2007, 103, 829–838. [Google Scholar] [CrossRef]
- Luo, Y.; Reid, R.; Freese, D.; Li, C.; Watkins, J.; Shi, H.; Zhang, H.; Loraine, A.; Song, B.-H. Salt Tolerance Response Revealed by RNA-Seq in a Diploid Halophytic Wild Relative of Sweet Potato. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Price, E.J.; Bhattacharjee, R.; Lopez-Montes, A.; Fraser, P.D. Metabolite Profiling of Yam (Dioscorea Spp.) Accessions for Use in Crop Improvement Programmes. Metabolomics Off. J. Metab. Soc. 2017; 13. [Google Scholar] [CrossRef] [Green Version]
- Padhan, B.; Panda, D. Potential of Neglected and Underutilized Yams (Dioscorea Spp.) for Improving Nutritional Security and Health Benefits. Front. Pharmacol. 2020; 11. [Google Scholar] [CrossRef] [Green Version]
- Sugihara, Y.; Darkwa, K.; Yaegashi, H.; Natsume, S.; Shimizu, M.; Abe, A.; Hirabuchi, A.; Ito, K.; Oikawa, K.; Oli, M.T.; et al. Genome Analyses Reveal the Hybrid Origin of the Staple Crop White Guinea Yam (Dioscorea rotundata). Proc. Natl. Acad. Sci. USA 2020, 117, 31987–31992. [Google Scholar] [CrossRef]
- Iranshahy, M.; Javadi, B.; Iranshahi, M.; Jahanbakhsh, S.P.; Mahyari, S.; Hassani, F.V.; Karimi, G. A Review of Traditional Uses, Phytochemistry and Pharmacology of Portulaca oleracea L. J. Ethnopharmacol. 2017, 205, 158–172. [Google Scholar] [CrossRef]
- Petropoulos, S.; Karkanis, A.; Martins, N.; Ferreira, I.C.F.R. Phytochemical Composition and Bioactive Compounds of Common Purslane (Portulaca oleracea L.) as Affected by Crop Management Practices. Trends Food Sci. Technol. 2016; 55. [Google Scholar] [CrossRef]
- Adams, S. Purslane Eyed as a Rich Food Source. Agric. Res. 1992, 40, 20–21. [Google Scholar]
- Farag, M.A.; Shakour, Z.T.A. Metabolomics Driven Analysis of 11 Portulaca Leaf Taxa as Analysed via UPLC-ESI-MS/MS and Chemometrics. Phytochemistry 2019, 161, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Howe, P.; Zhou, Y.-F.; Xu, Z.-Q.; Hocart, C.; Zhang, R. Fatty Acids and β-Carotene in Australian Purslane (Portulaca oleracea) Varieties. J. Chromatogr. A 2000, 893, 207–213. [Google Scholar] [CrossRef]
- Shenstone, E.; Lippman, Z.; Van Eck, J. A Review of Nutritional Properties and Health Benefits of Physalis Species. Plant Foods Hum. Nutr. Dordr. Neth. 2020, 75, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Dello-Russo, R. Climatic Stress in the Middle Rio Grande Valley of New Mexico: An Evaluation of Changes in Foraging Behaviors During the Late Archaic / Basketmaker II Period. Ph.D. Thesis, University of New Mexico, Albuquerque, NM, USA, 1999. [Google Scholar]
- Kindscher, K.; Long, Q.; Corbett, S.; Bosnak, K.; Loring, H.; Cohen, M.; Timmermann, B. The Ethnobotany and Ethnopharmacology of Wild Tomatillos, Physalis longifolia Nutt., and Related Physalis Species: A Review. Econ. Bot. 2012; 66. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, X.; Zheng, Q. Metabolomic Profiling of Carotenoid Constituents in Physalis peruviana During Different Growth Stages by LC-MS/MS Technology. J. Food Sci. 2019, 84, 3608–3613. [Google Scholar] [CrossRef]
- Kambhar, S.V. Rumex vesicarius L. (Polygonaceae): An Overview. J. Glob. Ecol. Environ. 2014, 1, 11–14.
- El-Hawary, S.A.; Sokkar, N.M.; Ali, Z.Y.; Yehia, M.M. A Profile of Bioactive Compounds of Rumex vesicarius L. J. Food Sci. 2011, 76, 1195–1202. [Google Scholar] [CrossRef] [PubMed]
- Chippindale, C. Before Scotland: The Story of Scotland before History—Alistair Moffat. J. R. Anthropol. Inst. 2006, 12, 679–680. [Google Scholar] [CrossRef]
- Enescu, C.; Durrant, T.; de Rigo, D.; Caudullo, G. Corylus avellana in Europe: Distribution, Habitat, Usage and Threats. Eur. Atlas For. Tree Species 2016, 54, 86–87. [Google Scholar]
- Alasalvar, C.; Shahidi, F.; Liyanapathirana, C.M.; Ohshima, T. Turkish Tombul Hazelnut (Corylus avellana L.). 1. Compositional Characteristics. J. Agric. Food Chem. 2003; 51. [Google Scholar] [CrossRef]
- Köksal, A.; Artik, N.; Şimşek, A.; Gunes, N. Nutrient Composition of Hazelnut (Corylus avellana L.) Varieties Cultivated in Turkey. Food Chem. 2006; 99. [Google Scholar] [CrossRef]
- Cristofori, V.; Ferramondo, S.; Bertazza, G.; Bignami, C. Nut and Kernel Traits and Chemical Composition of Hazelnut (Corylus avellana L.) Cultivars. J. Sci. Food Agric. 2008; 88. [Google Scholar] [CrossRef]
- Ahmad, M. ; Gul-Zaffar; Dar, Z.; Habib, M. A Review on Oat (Avena sativa L.) as a Dual-Purpose Crop. Sci. Res. 2014; 9. [Google Scholar] [CrossRef] [Green Version]
- Ishida, Y.; Hiei, Y.; Komari, T. High-Efficiency Transformation Techniques. In Applications of Genetic and Genomic Research in Cereals; Miedaner, T., Korzun, V., Eds.; Woodhead Publishing: Sawston, UK, 2019; pp. 97–120. [Google Scholar]
- Ibrahim, M.S.; Ahmad, A.; Sohail, A.; Asad, M.J. Nutritional and Functional Characterization of Different Oat (Avena sativa L.) Cultivars. Int. J. Food Prop. 2020; 23. [Google Scholar] [CrossRef]
- Foresman, B.J.; Oliver, R.E.; Jackson, E.W.; Chao, S.; Arruda, M.P.; Kolb, F.L. Genome-Wide Association Mapping of Barley Yellow Dwarf Virus Tolerance in Spring Oat (Avena sativa L.). 2016; 11. [Google Scholar] [CrossRef] [Green Version]
- Flores, H.E.; Galston, A.W. Osmotic Stress-Induced Polyamine Accumulation in Cereal Leaves: I. Physiological Parameters of the Response. Plant Physiol. 1984, 75, 102–109. [Google Scholar] [CrossRef] [Green Version]
- Pathak, R.; Thakur, V.; Gupta, R.K. Nutritional Analysis of Cereal Bars Formulated Using Morinda citrifolia and Bacopa monnieri. J. Pharmacogn. Phytochem. 2018, 7, 1546–1549. [Google Scholar]
- Prabhudas, S.K.; Natarajan, P. De Novo Assembly of Transcriptome and Draft Chloroplast Genome from RNAseq Data of Bacopa monnieri L. (Bramhi). Can. J. Biotechnol. 2017; 1. [Google Scholar] [CrossRef]
- Debnath, M. Responses of Bacopa monnieri to Salinity and Drought Stress in Vitro. J. Med. Plants Res. 2008, 2, 347–351. [Google Scholar] [CrossRef]
- Fordham, I.; Clevidence, B.; Wiley, E.; Zimmerman, R. Fruit of Autumn Olive: A Rich Source of Lycopene. HortScience 2001, 36, 1136–1137. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Hou, Y.; Hu, H.; Wang, C.; Weilin, Z.; Li, H.; Cheng, Z.; Yang, L. Functional Validation of Phytoene Synthase and Lycopene ε-Cyclase Genes for High Lycopene Content in Autumn Olive Fruit (Elaeagnus umbellata). J. Agric. Food Chem. 2020, 68, 11503–11511. [Google Scholar] [CrossRef]
- Wu, M.-C.; Hu, H.-T.; Yang, L.; Yang, L. Proteomic Analysis of Up-Accumulated Proteins Associated with Fruit Quality during Autumn Olive (Elaeagnus umbellata) Fruit Ripening. J. Agric. Food Chem. 2011, 59, 577–583. [Google Scholar] [CrossRef]
- Sengupta, S.; Majumder, A.L. Insight into the Salt Tolerance Factors of a Wild Halophytic Rice, Porteresia coarctata: A Physiological and Proteomic Approach. Planta 2009, 229, 911–929. [Google Scholar] [CrossRef]
- Ghosh, R.; Mitra, A. Effect of Salinity on Nutritional Value of Saltmarsh Grass (Porteresia coarctata) from Gangetic Delta, Northeast Coast of India. Indian J. Geo-Mar. Sci. 2015, 44, 1043–1052.
- Khalil, J.; Sawaya, W.N.; Hyder, S.Z. Nutrient Composition of Atriplex Leaves Grown in Saudi Arabia. J. Range Manag. USA 1986, 39, 104–107. [Google Scholar] [CrossRef]
- Ohsako, T.; Ohnishi, O. Intra- and Interspecific Phylogeny of Wild Fagopyrum (Polygonaceae) Species Based on Nucleotide Sequences of Noncoding Regions in Chloroplast DNA. Am. J. Bot. 2000, 87, 573–82. [Google Scholar] [CrossRef]
- Pongrac, P.; Vogel-Mikuš, K.; Potisek, M.; Kovačec, E.; Budič, B.; Kump, P.; Regvar, M.; Kreft, I. Mineral and Trace Element Composition and Importance for Nutritional Value of Buckwheat Grain, Groats, and Sprouts. In Molecular Breeding and Nutritional Aspects of Buckwheat, Zhou, M., Kreft, I., Woo, S.-H., Chrungoo, N., Wieslander, G., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 261–271. [Google Scholar]
- Logacheva, M.; Kasianov, A.; Vinogradov, D.; Samigullin, T.; Gelfand, M.; Makeev, V.; Penin, A. De Novo Sequencing and Characterization of Floral Transcriptome in Two Species of Buckwheat (Fagopyrum). BMC Genom. 2011, 12, 1–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Khound, R.; Santra, M.; Santra, D.K. Beyond Bird Feed: Proso Millet for Human Health and Environment. Agriculture 2019, 9, 64. [Google Scholar] [CrossRef] [Green Version]
- Zou, C.; Li, L.; Miki, D.; Li, D.; Tang, Q.; Xiao, L.; Rajput, S.; Deng, P.; Peng, L.; Jia, W.; et al. The Genome of Broomcorn Millet. Nat. Commun. 2019, 10, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Kwon, S.-J.; Yu, J.-H.; Sarker, K.; Cho, K.; Moon, Y.-J.; Jung, T.-W.; Park, C.-H.; Woo, S.-H. Comparison of Protein Profiles of Proso Millet (Panicum miliaceum) Seeds of Various Korean Cultivars. Korean J. Crop Sci. 2017, 62, 40–50. [Google Scholar] [CrossRef]
- Kim, J.K.; Park, S.-Y.; Yeo, Y.; Cho, H.S.; Kim, Y.B.; Bae, H.; Park, C.H.; Lee, J.-H.; Park, S.U. Metabolic Profiling of Millet (Paniciim miliaceum) Using Gas Chromatography—Time-of-Flight Mass Spectrometry (GC-TOFMS) for Quality Assessment. Plant Omics 2013, 6, 73–80. [Google Scholar]
- Gao, Q.-H.; Wu, C.-S.; Wang, M. The Jujube (Ziziphus jujuba Mill.) Fruit: A Review of Current Knowledge of Fruit Composition and Health Benefits. J. Agric. Food Chem. 2013; 61. [Google Scholar] [CrossRef]
- Chang, X; Sun, J. , Liu, L., He, W. Transcriptome Analysis of Differentially Expressed Genes in Wild Jujube Seedlings under Salt Stress. J. Am. Soc. Hortic. Sci. 2020, 1, 1–12.
- Yang, L.; Jin, J.; Fan, D.; Hao, Q.; Niu, J. Transcriptome Analysis of a Jujube (Ziziphus jujuba Mill.) Cultivar Response to Heat Stress. 2021. [Google Scholar] [CrossRef]
- San, B.; Yildirim, A. Phenolic, Alpha-Tocopherol, Beta-Carotene and Fatty Acid Composition of Four Promising Jujube (Ziziphus jujuba Miller) Selections. J. Food Compos. Anal. 2010, 23, 706–710. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, L.; Liu, Z.; Zhao, Z.; Zhao, J.; Wang, Z.; Zhou, G.; Liu, P.; Liu, M. Transcriptome and Metabolome Profiling Unveil the Mechanisms of Ziziphus jujuba Mill. Peel Coloration. Food Chem. 2019, 312, 125903. [Google Scholar] [CrossRef]
- Ramírez, F. Notes about Lulo (Solanum quitoense Lam.): An Important South American Underutilized Plant. Genet. Resour. Crop Evol. 2021; 68. [Google Scholar] [CrossRef]
- Gade, D.W. Ethnobotany of Cañihua (Chenopodium pallidicaule), Rustic Seed Crop of the Altiplano. Econ. Bot. 1970, 24, 55–61. [Google Scholar] [CrossRef]
- Martin, I. Fruits for the Future. 8. Monkey Orange. Strychnos cocculoides. By C. K. Mwamba. Southampton, UK: Southampton Centre for Underutilised Crops (2006), pp. 98, available free on request to national scientists of developing countries. ISBN 0854328416 Exp. Agric. 2007; 43. [Google Scholar] [CrossRef]
- Sebastin, R.; Lee, G.A.; Lee, K.J.; Shin, M.J.; Cho, G.T.; Lee, J.R.; Ma, K.H.; Chung, J.W. The Complete Chloroplast Genome Sequences of Little Millet (Panicum sumatrense Roth ex Roem. and Schult.) (Poaceae). Mitochondrial DNA Part B Resours. 2018. 3, 719-720. [CrossRef] [Green Version]
- Das, R.; Pradhan, S.; Parida, A. De-Novo Transcriptome Analysis Unveils Differentially Expressed Genes Regulating Drought and Salt Stress Response in Panicum sumatrense. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Amaranthus in Flora of North America @ Efloras. Available online: http://www.efloras.org/florataxon.aspx?flora_id=1&taxon_id=101257 (accessed on 7 June 2021).
- Das, S. Amaranthus: A Promising Crop of Future; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Joshi, D.C.; Sood, S.; Hosahatti, R.; Kant, L.; Pattanayak, A.; Kumar, A.; Yadav, D.; Stetter, M.G. From Zero to Hero: The Past, Present and Future of Grain Amaranth Breeding. Theor. Appl. Genet. 2018, 131, 1807–1823. [Google Scholar] [CrossRef]
- Das, S. Domestication, Phylogeny and Taxonomic Delimitation in Underutilized Grain Amaranthus (Amaranthaceae)—A Status Review. Feddes Repert. 2012, 123, 273–282. [Google Scholar] [CrossRef]
- Alemayehu, F.R.; Bendevis, M.A.; Jacobsen, S.-E. The Potential for Utilizing the Seed Crop Amaranth (Amaranthus Spp.) in East Africa as an Alternative Crop to Support Food Security and Climate Change Mitigation. J. Agron. Crop Sci. 2015. [Google Scholar] [CrossRef]
- Venskutonis, R.; Kraujalis, P. Nutritional Components of Amaranth Seeds and Vegetables: A Review on Composition, Properties, and Uses. Compr. Rev. Food Sci. Food Saf. 2013, 12, 381–412. [Google Scholar] [CrossRef] [PubMed]
- Tucker, J.B. Amaranth: The Once and Future Crop. BioScience 1986, 36, 9–13. [Google Scholar] [CrossRef]
- Rastogi, D.A.; Shukla, S. Amaranth: A New Millennium Crop of Nutraceutical Values. Crit. Rev. Food Sci. Nutr. 2013, 53, 109–125. [Google Scholar] [CrossRef]
- Montgomery, J.S.; Giacomini, D.; Waithaka, B.; Lanz, C.; Murphy, B.P.; Campe, R.; Lerchl, J.; Landes, A.; Gatzmann, F.; Janssen, A.; et al. Draft Genomes of Amaranthus tuberculatus, Amaranthus hybridus, and Amaranthus palmeri. Genome Biol. Evol. 2020, 12, 1988–1993. [Google Scholar] [CrossRef]
- Chevarria-Lazo, M.; Bazile, D.; dessauw, D.; Louafi, S.; Trommetter, M.; Hocdé, H. Quinoa and the exchange of genetic resources: Improving the regulation systems. In State of the Art Report on Quinoa around the World in 2013; FAO Regional Office for Latin America and the Caribbean: Rome, Italy, 2015; pp. 83–105. [Google Scholar]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.A.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The Genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golicz, A.A.; Steinfort, U.; Arya, H.; Singh, M.B.; Bhalla, P.L. Analysis of the Quinoa Genome Reveals Conservation and Divergence of the Flowering Pathways. Funct. Integr. Genom. 2020, 20, 245–258. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Duan, X.; Yang, J.; Beeching, J.R.; Zhang, P. Coupled Expression of Cu/Zn-Superoxide Dismutase and Catalase in Cassava Improves Tolerance against Cold and Drought Stresses. Plant Signal. Behav. 2013, 8, e24525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Feng, B.; Xiao, J.; Xia, Z.; Xincheng, Z.; Li, P.; Zhang, W.; Wang, Y.; Møller, B.; Zhang, P.; et al. Cassava Genome from a Wild Ancestor to Cultivated Varieties. Nat. Commun. 2014, 10, 1–9. [Google Scholar] [CrossRef]
- Briones-Labarca, V.; Giovagnoli-Vicuña, C.; Figueroa-Alvarez, P.; Quispe, I.; Pérez-Won, M. Extraction of β -Carotene, Vitamin C and Antioxidant Compounds from Physalis peruviana (Cape Gooseberry) Assisted by High Hydrostatic Pressure. Food Nutr. Sci. 2013, 4, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.-G.; Jiang, W.; Mantri, N.; Bao, X.-Q.; Chen, S.-L.; Tao, Z.-M. Transciptome Analysis Reveals Flavonoid Biosynthesis Regulation and Simple Sequence Repeats in Yam (Dioscorea alata L.) Tubers. BMC Genom. 2015; 16. [Google Scholar] [CrossRef] [Green Version]
- Garzón-Martínez, G.A.; Zhu, Z.I.; Landsman, D.; Barrero, L.S.; Mariño-Ramírez, L. The Physalis peruviana Leaf Transcriptome: Assembly, Annotation and Gene Model Prediction. BMC Genom. 2012, 13, 151. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Li, J.; Wang, L.; Zhang, J.; He, C. Transcriptomic Variation of the Flower-Fruit Transition in Physalis and Solanum. Planta 2020, 252, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Maathuis, F.; Diatloff, E. Roles and Functions of Plant Mineral Nutrients. Methods Mol. Biol. 2013, 953, 1–21. [Google Scholar] [CrossRef]
- Singh, B.; Schulze, D.G.; Soil Minerals and Plant Nutrition. Nature Education Knowledge. Available online: https://www.nature.com/scitable/knowledge/library/soil-minerals-and-plant-nutrition-127881474/ (accessed on 5 July 2021).
- Chrispeels, M.J.; Crawford, N.M.; Schroeder, J.I. Proteins for Transport of Water and Mineral Nutrients across the Membranes of Plant Cells. Plant Cell 1999, 11, 661–676. [Google Scholar] [CrossRef] [Green Version]
- Foy, C.D.; Chaney, R.L.; White, M.C. The Physiology of Metal Toxicity in Plants. Annu. Rev. Plant Physiol. 1978, 29, 511–566. [Google Scholar] [CrossRef]
- Gill, M. Heavy Metal Stress in Plants: A Review. Int. J. Adv. Res. 2014, 2, 1043–1055. [Google Scholar]
- Schilter, B.; Andersson, C.; Anton, R.; Constable, A.; Kleiner, J.; O’Brien, J.; Renwick, A.G.; Korver, O.; Smit, F.; Walker, R.; et al. Guidance for the Safety Assessment of Botanicals and Botanical Preparations for Use in Food and Food Supplements. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2003, 41, 1625–1649. [Google Scholar] [CrossRef]
- Salt, D.; Baxter, I.; Lahner, B. Ionomics and the Study of the Plant Ionome. Annu. Rev. Plant Biol. 2008, 59, 709–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.-Y.; Salt, D.E. Plant Ionomics: From Elemental Profiling to Environmental Adaptation. Mol. Plant 2016, 9, 787–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salt, D.E. Update on Plant Ionomics. Plant Physiol. 2004, 136, 2451–2456. [Google Scholar] [CrossRef] [Green Version]
- Migeon, A.; Blaudez, D.; Wilkins, O.; Montanini, B.; Campbell, M.M.; Richaud, P.; Thomine, S.; Chalot, M. Genome-Wide Analysis of Plant Metal Transporters, with an Emphasis on Poplar. Cell. Mol. Life Sci. 2010, 67, 3763–3784. [Google Scholar] [CrossRef] [PubMed]
- Nandal, U.; Bhardwaj, R.L. The Role of Underutilized Fruits in Nutritional and Economic Security of Tribals: A Review. Crit. Rev. Food Sci. Nutr. 2014, 54, 880–890. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, S.; Datta, B.K.; Saha, A. Determination of Mineral Content and Heavy Metal Content of Some Traditionally Important Aquatic Plants of Tripura, India Using Atomic Absorption Spectroscopy. J. Agric. Technol. 2012, 8, 1467–1476. [Google Scholar]
- Chacha, J.; Laswai, H. Micronutrients Potential of Underutilized Vegetables and Their Role in Fighting Hidden Hunger. Int. J. Food Sci. 2020, 2020, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satismruti, K.; Natesan, S.; Sampathrajan, V.; Raja, R.; Muthurajan, R. Plant Ionomics: A Platform for Identifying Novel Gene Regulating Plant Mineral Nutrition. Am. J. Plant Sci. 2013, 447162, 1309–1315. [Google Scholar] [CrossRef] [Green Version]
- Elshamy, M.M.; Heikal, Y.M.; Bonanomi, G. Phytoremediation Efficiency of Portulaca oleracea L. Naturally Growing in Some Industrial Sites, Dakahlia District, Egypt. 2019. [Google Scholar] [CrossRef]
- Amirul Alam, M.; Juraimi, A.S.; Rafii, M.Y.; Hamid, A.A.; Kamal Uddin, M.; Alam, M.Z.; Latif, M.A. Genetic Improvement of Purslane (Portulaca oleracea L.) and Its Future Prospects. Mol. Biol. Rep. 2014; 41. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Sreedharan, S.; Achigan-Dako, E.G.; Singh, P.; Ramchiary, N. Improvement of a Traditional Orphan Food Crop, Portulaca oleracea L. (Purslane) Using Genomics for Sustainable Food Security and Climate Resilient Agriculture. Front. Sustain. Food Syst. 2021. [Google Scholar] [CrossRef]
- Xing, J.; Zhao, B.; Dong, J.; Liu, C.; Wen, Z.; Zhu, X.; Ding, H.; He, T.; Yang, H.; Wang, M.; et al. Transcriptome and Metabolome Profiles Revealed Molecular Mechanisms Underlying Tolerance of Portulaca oleracea to Saline Stress. Russ. J. Plant Physiol. 2020, 67, 146–152. [Google Scholar] [CrossRef]
- Patel, S. Plant Genus Elaeagnus: Underutilized Lycopene and Linoleic Acid Reserve with Permaculture Potential. Fruits 2015, 70, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Mustafa, A.; Ahmed, A.; Tantray, A.; Parry, P. Ethnopharmacological Potential and Medicinal Uses of Miracle Herb Dioscorea Spp. J. Ayurvedic Herb. Med. 2018, 4, 79–85. [Google Scholar]
- Akoroda, M.O. Yams: Dioscorea spp. In Genetic Improvement of Vegetable Crops; Kalloo, G., Bergh, B.O., Eds.; Pergamon: Amsterdam, The Netherlands, 1993; pp. 717–733. [Google Scholar]
- Sharma, S.; Deswal, R. Genomic and Proteomic Tools for Understanding Mysterious Protein Dioscorin from Dioscorea Tuber. In; Plant Omics and Crop Breeding, Zargar, S.M., Rai, V., Eds.; Apple Academic Academic Press: Boca Raton, USA, 2016; pp. 97–114. [Google Scholar]
- Nakayasu, M.; Kawasaki, T.; Lee, H.; Sugimoto, Y.; Onjo, M.; Muranaka, T.; Mizutani, M. Identification of Furostanol Glycoside 26-O-β-Glucosidase Involved in Steroidal Saponin Biosynthesis from Dioscorea esculenta. Plant Biotechnol. 2015, 32, 1015–1023. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Li, B.; Li, L.; Ma, W.; Liu, Y.; Feng, S.; Wang, Z. Genome Survey Sequencing of Dioscorea zingiberensis. Genome 2018, 61, 567–574. [Google Scholar] [CrossRef]
- Valdivia, M.; Tecante, A. Chia (Salvia hispanica): A Review of Native Mexican Seed and Its Nutritional and Functional Properties. Adv. Food Nutr. Res. 2015, 75, 53–75. [Google Scholar] [CrossRef]
- Tacer-Caba, Z. The concept of superfoods in diet. In The Role of Alternative and Innovative Food Ingredients and Products in Consumer Wellness; Galanakis, C.M., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 73–101. [Google Scholar]
- Orona-Tamayo, D.; Valverde, M.; Paredes-Lopez, O. Chia-The New Golden Seed for the 21st Century: Nutraceutical Properties and Technological Uses. In Sustainable Protein Sources, 1st ed.; Academic Press: London, UK, 2016; pp. 265–281. [Google Scholar]
- Melo, D.; Machado, T.; Oliveira, M. Chia Seeds: An Ancient Grain Trending in Modern Human Diets. Food Funct. 2019, 10, 3068–3089. [Google Scholar] [CrossRef]
- Hao, D.-C.; Ge, G.-B.; Xiao, P.-G. Anticancer Drug Targets of Salvia Phyto metabolites: Chemistry, Biology and Omics. Curr. Drug Targets 2018, 19, 1–20. [Google Scholar] [CrossRef]
- Ullah, R.; Nadeem, M.; Khalique, A.; Imran, M.; Mehmood, S.; Javid, A.; Hussain, J. Nutritional and Therapeutic Perspectives of Chia (Salvia hispanica L.): A Review. J. Food Sci. Technol. 2015; 53. [Google Scholar] [CrossRef] [Green Version]
- Parker, J.; Schellenberger, A.N.; Roe, A.L.; Oketch-Rabah, H.; Calderón, A.I. Therapeutic Perspectives on Chia Seed and Its Oil: A Review. Planta Med. 2018, 84, 606–612. [Google Scholar] [CrossRef] [Green Version]
- Peláez, P.; Orona-Tamayo, D.; Montes-Hernández, S.; Valverde, M.; Paredes-Lopez, O.; Cibrian, A. Comparative Transcriptome Analysis of Cultivated and Wild Seeds of Salvia hispanica (Chia). Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Sreedhar, R.V.; Kumari, P.; Rupwate, S.; Rajasekharan, R.; Srinivasan, M. Exploring Triacylglycerol Biosynthetic Pathway in Developing Seeds of Chia (Salvia hispanica L.): A Transcriptomic Approach. 2015; 10. [Google Scholar] [CrossRef]
- Wiehle, M.; Prinz, K.; Kehlenbeck, K.; Goenster, S.; Mohamed, S.A.; Finkeldey, R.; Buerkert, A.; Gebauer, J. The African Baobab (Adansonia digitata, Malvaceae): Genetic Resources in Neglected Populations of the Nuba Mountains, Sudan. Am. J. Bot. 2014, 101, 1498–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahul, J.; Jain, M.K.; Singh, S.P.; Kamal, R.K. ; Anuradha; Naz, A.; Gupta, A.K.; Mrityunjay, S.K. Adansonia digitata L. (Baobab): A Review of Traditional Information and Taxonomic Description. Asian Pac. J. Trop. Biomed. 2015; 5. [Google Scholar] [CrossRef] [Green Version]
- Chládová, A.; Kalousová, M.; Mandák, B.; Kehlenbeck, K.; Prinz, K.; Šmíd, J.; Van Damme, P.; Lojka, B. Genetic Diversity and Structure of Baobab (Adansonia digitata L.) in Southeastern Kenya. R. Soc. Open Sci. 2019; 6. [Google Scholar] [CrossRef] [Green Version]
- Dillon, S.L.; Shapter, F.M.; Henry, R.J.; Cordeiro, G.; Izquierdo, L.; Lee, L.S. Domestication to Crop Improvement: Genetic Resources for Sorghum and Saccharum (Andropogoneae). Ann. Bot. 2007, 100, 975–989. [Google Scholar] [CrossRef] [Green Version]
- Meyer, R.; Purugganan, M. Evolution of Crop Species: Genetics of Domestication and Diversification. Nat. Rev. Genet. 2013, 14, 840–52. [Google Scholar] [CrossRef] [PubMed]
- Smykal, P.; Nelson, M.; Berger, J.; Wettberg, E. The Impact of Genetic Changes during Crop Domestication. Agronomy 2018, 8, 119. [Google Scholar] [CrossRef] [Green Version]
- Poncet, V.; Robert, T.; Sarr, A.; Gepts, P. Quantitative Trait Locus Analyses of the Domestication Syndrome and Domestication Process. Encycl. Plant Crop Sci. 2004, 1069, 1069–1073. [Google Scholar] [CrossRef]
- Simons, K.; Fellers, J.; Trick, H.; Zhang, Z.; Tai, Y.-S.; Gill, B.; Faris, J. Molecular Characterization of the Major Wheat Domestication Gene Q. Genetics 2006, 172, 547–555. [Google Scholar] [CrossRef] [Green Version]
- Salentijn, E.M.J.; Pereira, A.B.; Angenent, G.C.; Linden, C.G. van der; Krens, F.A.; Smulders, M.J.M.; Vosman, B. Plant Translational Genomics: From Model Species to Crops. Mol. Breed. 2007, 20, 1–13. [Google Scholar] [CrossRef]
- Fraser, P.D.; Aharoni, A.; Hall, R.D.; Huang, S.; Giovannoni, J.J.; Sonnewald, U.; Fernie, A.R. Metabolomics Should Be Deployed in the Identification and Characterization of Gene-Edited Crops. Plant J. Cell Mol. Biol. 2020, 102, 897–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Fernie, A.R.; Yan, J. De Novo Domestication: An Alternative Route toward New Crops for the Future. Mol. Plant 2019, 12, 615–631. [Google Scholar] [CrossRef] [Green Version]
- Zsögön, A.; Cermak, T.; Naves, E.; Notini, M.; Edel, K.; Weinl, S.; Freschi, L.; Voytas, D.; Kudla, J.; Peres, L. De Novo Domestication of Wild Tomato Using Genome Editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [Green Version]
- Lemmon, Z.; Reem, N.; Dalrymple, J.; Soyk, S.; Swartwood, K.; Rodriguez-Leal, D.; Eck, J.; Lippman, Z. Rapid Improvement of Domestication Traits in an Orphan Crop by Genome Editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef]
- Ahmar, S.; Saeed, S.; Khan, M.H.; Ullah Khan, S.; Mora-Poblete, F.; Kamran, M.; Faheem, A.; Maqsood, A.; Rauf, M.; Saleem, S.; et al. A Revolution toward Gene-Editing Technology and Its Application to Crop Improvement. Int. J. Mol. Sci. 2020, 21, 5665. [Google Scholar] [CrossRef] [PubMed]
- Maher, M.; Nasti, R.; Vollbrecht, M.; Starker, C.; Clark, M.; Voytas, D. Plant Gene Editing through de Novo Induction of Meristems. Nat. Biotechnol. 2020, 38, 1–6. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Zhu, H. CRISPR Technology Is Revolutionizing the Improvement of Tomato and Other Fruit Crops. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Van Eck, J. Applying Gene Editing to Tailor Precise Genetic Modifications in Plants. J. Biol. Chem. 2020, 295, 13267–13276. [Google Scholar] [CrossRef]
- Hille, F.; Charpentier, E. CRISPR-Cas: Biology, Mechanisms and Relevance. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 1–16. [Google Scholar] [CrossRef]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 System for Plant Genome Editing and Beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Zhang, J.-P.; Li, X.-L.; Li, G.-H.; Chen, W.; Arakaki, C.; Botimer, G.; Baylink, D.; Zhang, L.; Wen, W.; Fu, Y.-W.; et al. Efficient Precise Knockin with a Double Cut HDR Donor after CRISPR/Cas9-Mediated Double-Stranded DNA Cleavage. Genome Biol. 2017, 18, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, D.; Bhattacharjee, O.; Mandal, D.; Sen, M.K.; Dey, D.; Dasgupta, A.; Kazi, T.A.; Gupta, R.; Sinharoy, S.; Acharya, K.; et al. CRISPR-Cas9 System: A New-Fangled Dawn in Gene Editing. Life Sci. 2019, 232, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Syombua, E.D.; Zhang, Z.; Tripathi, J.N.; Ntui, V.O.; Kang, M.; George, O.O.; Edward, N.K.; Wang, K.; Yang, B.; Tripathi, L. A CRISPR/Cas9-Based Genome-Editing System for Yam (Dioscorea Spp.). Plant Biotechnol. J. 2021; 19. [Google Scholar] [CrossRef]
- Park, J.; Bae, S.; Kim, J.-S. Cas-Designer: A Web-Based Tool for Choice of CRISPR-Cas9 Target Sites. Bioinformatics 2015, 31, 4014–4016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkins, P.A.; Voytas, D.F. Overcoming Bottlenecks in Plant Gene Editing. Genome Stud. Mol. Genet. 2020, 54, 79–84. [Google Scholar] [CrossRef]
- Mushtaq, M.; Sakina, A.; Wani, S.H.; Shikari, A.B.; Tripathi, P.; Zaid, A.; Galla, A.; Abdelrahman, M.; Sharma, M.; Singh, A.K.; et al. Harnessing Genome Editing Techniques to Engineer Disease Resistance in Plants. Front. Plant Sci. 2019, 10, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soundararajan, P.; Won, S.; Kim, J. Insight on Rosaceae Family with Genome Sequencing and Functional Genomics Perspective. BioMed Res. Int. 2019, 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Iwata, H.; Gaston, A.; Remay, A.; Thouroude, T.; Jeauffre, J.; Kawamura, K.; Oyant, L.H.-S.; Araki, T.; Denoyes, B.; Foucher, F. The TFL1 Homologue KSN Is a Regulator of Continuous Flowering in Rose and Strawberry. Plant J. Cell Mol. Biol. 2012, 69, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Koskela, E.A.; Mouhu, K.; Albani, M.C.; Kurokura, T.; Rantanen, M.; Sargent, D.J.; Battey, N.H.; Coupland, G.; Elomaa, P.; Hytönen, T. Mutation in TERMINAL FLOWER1 Reverses the Photoperiodic Requirement for Flowering in the Wild Strawberry Fragaria vesca. Plant Physiol. 2012, 159, 1043–1054. [Google Scholar] [CrossRef] [Green Version]
- Ferchichi, S.; Hessini, K.; Dell Aversana, E.; D Amelia, L.; Woodrow, P.; Ciarmiello, L.F.; Fuggi, A.; Carillo, P. Hordeum vulgare and Hordeum maritimum Respond to Extended Salinity Stress Displaying Different Temporal Accumulation Pattern of Metabolites. Funct. Plant Biol. FPB 2018, 45, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- von Bothmer, R.; Jacobsen, N.; Baden, C.; Jørgensen, R.B.; Linde-Laursen, I. An ecogeographical study of the genus Hordeum. 2nd ed. Systematic and Ecogeographic Studies on Crop Genepools 7. International Plant Genetic Resources Institute: Rome, 1995; pp. 129 ISBN 92-9043-229-2.
- Yu, S.; Long, H.; Deng, G.; Pan, Z.; Liang, J.; Zeng, X.; Tang, Y.; Tashi, N.; Yu, M. A Single Nucleotide Polymorphism of Nud Converts the Caryopsis Type of Barley (Hordeum vulgare L.). Plant Mol. Biol. Report. 2015; 34. [Google Scholar] [CrossRef] [Green Version]
- Komatsuda, T.; Pourkheirandish, M.; He, C.; Azhaguvel, P.; Kanamori, H.; Perovic, D.; Stein, N.; Graner, A.; Wicker, T.; Tagiri, A.; et al. Six-Rowed Barley Originated from a Mutation in a Homeodomain-Leucine Zipper I-Class Homeobox Gene. Proc. Natl. Acad. Sci. 2007, 104, 1424–1429. [Google Scholar] [CrossRef] [Green Version]
- Digel, B.; Tavakol, E.; Verderio, G.; Tondelli, A.; Xu, X.; Cattivelli, L.; Rossini, L.; von Korff, M. Photoperiod-H1 (Ppd-H1) Controls Leaf Size. Plant Physiol. 2016, 172, 405–415. [Google Scholar] [CrossRef] [Green Version]
- Genger, R.; Williams, K.; Raman, H.; Read, B.; Wallwork, H.; Burdon, J.; Brown, A. Leaf Scald Resistance Genes in Hordeum vulgare and Hordeum vulgare Ssp. Spontaneum: Parallels between Cultivated and Wild Barley. Aust. J. Agric. Res. 2003; 54. [Google Scholar] [CrossRef]
- Herrero, E.; Kolmos, E.; Bujdoso, N.; Yuan, Y.; Wang, M.; Berns, M.C.; Uhlworm, H.; Coupland, G.; Saini, R.; Jaskolski, M.; et al. EARLY FLOWERING4 Recruitment of EARLY FLOWERING3 in the Nucleus Sustains the Arabidopsis Circadian Clock. Plant Cell 2012, 24, 428–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramsay, L.; Comadran, J.; Druka, A.; Marshall, D.F.; Thomas, W.T.B.; Macaulay, M.; MacKenzie, K.; Simpson, C.; Fuller, J.; Bonar, N.; et al. INTERMEDIUM-C, a Modifier of Lateral Spikelet Fertility in Barley, Is an Ortholog of the Maize Domestication Gene TEOSINTE BRANCHED 1. Nat. Genet. 2011, 43, 169–172. [Google Scholar] [CrossRef]
- Bortolotto, I.M.; Amorozo, M.C.d.M.; Neto, G.G.; Oldeland, J.; Damasceno-Junior, G.A. Knowledge and Use of Wild Edible Plants in Rural Communities along Paraguay River, Pantanal, Brazil. J. Ethnobiol. Ethnomedicine 2015, 11, 46–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.; Li, X.; Liu, F.; Sun, X.; Li, C.; Zhu, Z.; Fu, Y.; Cai, H.; Wang, X.; Xie, D.; et al. Control of a Key Transition from Prostrate to Erect Growth in Rice Domestication. Nat. Genet. 2008, 40, 1360–1364. [Google Scholar] [CrossRef]
- Li, C.; Zhou, A.; Sang, T. Rice Domestication by Reducing Shattering. Science 2006, 311, 1936–1939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Griffith, M.E.; Li, X.; Zhu, Z.; Tan, L.; Fu, Y.; Zhang, W.; Wang, X.; Xie, D.; Sun, C. Origin of Seed Shattering in Rice (Oryza sativa L.). 2007; 20. [Google Scholar] [CrossRef]
- Zhu, B.-F.; Si, L.; Wang, Z.; Zhou, Y.; Zhu, J.; Shangguan, Y.; Lu, D.; Fan, D.; Li, C.; Lin, H.; et al. Genetic Control of a Transition from Black to Straw-White Seed Hull in Rice Domestication. Plant Physiol. 2011, 155, 1301–1311. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, M.T.; Thomson, M.J.; Pfeil, B.E.; McCouch, S. Caught Red-Handed: Rc Encodes a Basic Helix-Loop-Helix Protein Conditioning Red Pericarp in Rice. Plant Cell 2006, 18, 283–294. [Google Scholar] [CrossRef] [Green Version]
- Filiz, E.; Akbudak, M.A. Ammonium Transporter 1 (AMT1) Gene Family in Tomato (Solanum lycopersicum L.): Bioinformatics, Physiological and Expression Analyses under Drought and Salt Stresses. 2020. [Google Scholar] [CrossRef]
- Lee, J.; Park, J.J.; Kim, S.; Yim, J.; An, G. Mutations in the Rice Liguleless Gene Result in a Complete Loss of the Auricle, Ligule, and Laminar Joint. Plant Mol. Biol. 2007, 65, 487–499. [Google Scholar] [CrossRef]
- Baicharoen, A.; Vijayan, R.; Pongprayoon, P. Structural Insights into Betaine Aldehyde Dehydrogenase (BADH2) from Oryza sativa Explored by Modeling and Simulations. Sci. Rep. 2017, 8, 12892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and Initial Characterization of GW5, a Major QTL Associated with Rice Grain Width and Weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef]
- Merida, A.; Rodriguez-Galan, J.; Vincent, C.; Romero, J. Expression of the Granule-Bound Starch Synthase I (Waxy) Gene from Snapdragon Is Developmentally and Circadian Clock Regulated. Plant Physiol. 1999, 120, 401–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, H.; Yang, C.; Zhu, J.; Zhang, L.; Bai, Y.; Li, E.; Gilbert, R.G. Competition between Granule Bound Starch Synthase and Starch Branching Enzyme in Starch Biosynthesis. Rice 2019, 12, 96–96. [Google Scholar] [CrossRef] [Green Version]
- Takano-Kai, N.; Jiang, H.; Kubo, T.; Sweeney, M.; Matsumoto, T.; Kanamori, H.; Padhukasahasram, B.; Bustamante, C.; Yoshimura, A.; Doi, K.; et al. Evolutionary History of a Gene Conferring Grain Length in Rice. Genetics 2009, 182, 1323–1334. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Shimamoto, K. Heading Date 1 (Hd1), an Ortholog of Arabidopsis CONSTANS, Is a Possible Target of Human Selection during Domestication to Diversify Flowering Times of Cultivated Rice. Genes Genet. Syst. 2011, 86, 175–182. [Google Scholar] [CrossRef] [Green Version]
- Konishi, S.; Izawa, T.; Lin, S.Y.; Ebana, K.; Fukuta, Y.; Sasaki, T.; Yano, M. An SNP Caused Loss of Seed Shattering during Rice Domestication. Science 2006, 312, 1392–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulbert, S.H.; Richter, T.E.; Axtell, J.D.; Bennetzen, J.L. Genetic Mapping and Characterization of Sorghum and Related Crops by Means of Maize DNA Probes. Proc. Natl. Acad. Sci. USA 1990, 87, 4251–4255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorweiler, J.E.; Doebley, J. Developmental Analysis of Teosinte Glume Architecture1: A Key Locus in the Evolution of Maize (Poaceae). Am. J. Bot. 1997, 84, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.; Stec, A.; Gustus, C. Teosinte Branched1 and the Origin of Maize: Evidence for Epistasis and the Evolution of Dominance. Genetics 1995, 141, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Nussbaum-Wagler, T.; Li, B.; Zhao, Q.; Vigouroux, Y.; Faller, M.; Bomblies, K.; Lukens, L.; Doebley, J.F. The Origin of the Naked Grains of Maize. Nature 2005, 436, 714–719. [Google Scholar] [CrossRef]
- Wang, H.; Studer, A.J.; Zhao, Q.; Meeley, R.; Doebley, J.F. Evidence That the Origin of Naked Kernels During Maize Domestication Was Caused by a Single Amino Acid Substitution in Tga1. Genetics 2015, 200, 965–974. [Google Scholar] [CrossRef] [Green Version]
- Yang, X. Study of RAMOSA1 Function during Maize Inflorescence Development. Ph.D. Thesis, Iowa State University, Ames, IA, USA, 2011. [CrossRef] [Green Version]
- Hubbard, L.; McSteen, P.; Doebley, J.; Hake, S. Expression Patterns and Mutant Phenotype of Teosinte Branched1 Correlate with Growth Suppression in Maize and Teosinte. Genetics 2002, 162, 1927–1935. [Google Scholar] [CrossRef] [PubMed]
- Gallavotti, A.; Zhao, Q.; Kyozuka, J.; Meeley, R.B.; Ritter, M.K.; Doebley, J.F.; Pè, M.E.; Schmidt, R.J. The Role of Barren Stalk1 in the Architecture of Maize. Nature 2004, 432, 630–635. [Google Scholar] [CrossRef]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS Gene of Arabidopsis Promotes Flowering and Encodes a Protein Showing Similarities to Zinc Finger Transcription Factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xu, M. CCT Family Genes in Cereal Crops: A Current Overview. Crop J. 2017, 5, 449–458. [Google Scholar] [CrossRef]
- Han, J.-J.; Jackson, D.; Martienssen, R. Pod Corn Is Caused by Rearrangement at the Tunicate1 Locus. Plant Cell 2012, 24, 2733–2744. [Google Scholar] [CrossRef] [Green Version]
- James, M.G.; Robertson, D.S.; Myers, A.M. Characterization of the Maize Gene Sugary1, a Determinant of Starch Composition in Kernels. Plant Cell 1995, 7, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Dinges, J.R.; Colleoni, C.; Myers, A.M.; James, M.G. Molecular Structure of Three Mutations at the Maize Sugary1 Locus and Their Allele-Specific Phenotypic Effects. Plant Physiol. 2001, 125, 1406–1418. [Google Scholar] [CrossRef] [Green Version]
- Smartt, J. Evolution of genetic resources. In Grain legumes; Smartt, J., Ed.; Cambridge University Press: Cambridge, 1990; pp. 140–175. [Google Scholar]
- Weeden, N.F. Genetic Changes Accompanying the Domestication of Pisum Sativum: Is There a Common Genetic Basis to the “domestication Syndrome” for Legumes? Ann. Bot. 2007, 100, 1017–1025. [Google Scholar] [CrossRef] [Green Version]
- de Wet, J.M.J.; Oestry-Stidd, L.L.; Cubero, J.I. Origins and Evolution of Foxtail Millets (Setaria Italica). J. Tradit. Agric. Appl. Bot. 1979, 26, 53–64. [Google Scholar] [CrossRef]
- Fukunaga, K.; Kawase, M.; Kato, K. Structural Variation in the Waxy Gene and Differentiation in Foxtail Millet [Setaria italica (L.) P. Beauv.]: Implications for Multiple Origins of the Waxy Phenotype. Mol. Genet. Genom. 2002. [Google Scholar] [CrossRef]
- Seung, D. Amylose in Starch: Towards an Understanding of Biosynthesis, Structure and Function. New Phytol. 2020, 228, 1490–1504. [Google Scholar] [CrossRef]
- Mueller, L.A.; Solow, T.H.; Taylor, N.; Skwarecki, B.; Buels, R.; Binns, J.; Lin, C.; Wright, M.H.; Ahrens, R.; Wang, Y.; et al. The SOL Genomics Network. A Comparative Resource for Solanaceae Biology and Beyond. Plant Physiol. 2005, 138, 1310–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, G.R.; Muños, S.; Anderson, C.; Sim, S.-C.; Michel, A.; Causse, M.; Gardener, B.B.M.; Francis, D.; van der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the Tomato Germplasm and the Relationship to Fruit Shape Diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paterson, A.H.; Lander, E.S.; Hewitt, J.D.; Peterson, S.; Lincoln, S.E.; Tanksley, S.D. Resolution of Quantitative Traits into Mendelian Factors by Using a Complete Linkage Map of Restriction Fragment Length Polymorphisms. Nature 1988, 335, 721–726. [Google Scholar] [CrossRef]
- Li, B.; Sun, S.; Gao, X.; Wu, M.; Deng, Y.; Zheng, Q.; Li, X.; Xiao, J.; Ke, Y.; Wang, S. Overexpression a “Fruit-Weight 2.2-like” Gene OsFWL5 Improves Rice Resistance. 2019; 12. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Ellis, B. Overview of OVATE FAMILY PROTEINS, A Novel Class of Plant-Specific Growth Regulators. Front. Plant Sci. 2016, 7, 417. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Xiao, H.; Cabrera, A.; Meulia, T.; van der Knaap, E. SUN Regulates Vegetative and Reproductive Organ Shape by Changing Cell Division Patterns. Plant Physiol. 2011, 157, 1175–1186. [Google Scholar] [CrossRef] [Green Version]
- Rossetto, M.; Jackes, B.; Scott, K.; Henry, R. Intergeneric Relationships in the Australian Vitaceae: New Evidence from CpDNA Analysis. Genet. Resour. Crop Evol. 2001, 48, 307–314. [Google Scholar] [CrossRef]
- Péros, J.-P.; Launay, A.; Berger, G.; Lacombe, T.; This, P. MybA1 Gene Diversity across the Vitis Genus. Genetica 2015, 143, 373–384. [Google Scholar] [CrossRef] [PubMed]
- USDA, GRIN and NRCS DATABASE. Plants. Available online: https://plants.sc.egov.usda.gov/home (accessed on 7 June 2021).
- Dawson, I.; Powell, W.; Hendre, P.; Bančič, J.; Hickey, J.; Kindt, R.; Hoad, S.; Hale, I.; Jamnadass, R. The Role of Genetics in Mainstreaming the Production of New and Orphan Crops to Diversify Food Systems and Support Human Nutrition. New Phytol. 2019, 224, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted Mutagenesis, Precise Gene Editing, and Site-Specific Gene Insertion in Maize Using Cas9 and Guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alagoz, Y.; Gurkok, T.; Zhang, B.; Unver, T. Manipulating the Biosynthesis of Bioactive Compound Alkaloids for Next-Generation Metabolic Engineering in Opium Poppy Using CRISPR-Cas 9 Genome Editing Technology. Sci. Rep. 2016, 6, 30910. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of Broad Virus Resistance in Non-Transgenic Cucumber Using CRISPR/Cas9 Technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef] [Green Version]
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, S.; Kim, J.-S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef] [PubMed]
- Jyoti, A.; Kaushik, S.; Srivastava, V.K.; Datta, M.; Kumar, S.; Yugandhar, P.; Kothari, S.L.; Rai, V.; Jain, A. The Potential Application of Genome Editing by Using CRISPR/Cas9, and Its Engineered and Ortholog Variants for Studying the Transcription Factors Involved in the Maintenance of Phosphate Homeostasis in Model Plants. Semin. Cell Dev. Biol. 2019; s9. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/SgRNA-Mediated Targeted Gene Modification in Arabidopsis, Tobacco, Sorghum and Rice. Nucleic Acids Res. 2013, 41, 648–657. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the Four Yield-Related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Front. Plant Sci. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Simmonds, J.; Pan, Q.; Davidson, D.; He, F.; Battal, A.; Akhunova, A.; Trick, H.; Uauy, C.; Akhunov, E. Gene Editing and Mutagenesis Reveal Inter-Cultivar Differences and Additivity in the Contribution of TaGW2 Homoeologues to Grain Size and Weight in Wheat. Theor. Appl. Genet. 2018, 131, 2463–2475. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.Z.; Henry, I.M.; Lynagh, P.G.; Comai, L.; Cahoon, E.B.; Weeks, D.P. Significant Enhancement of Fatty Acid Composition in Seeds of the Allohexaploid, Camelina sativa, Using CRISPR/Cas9 Gene Editing. Plant Biotechnol. J. 2017, 15, 648–657. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Chen, L.; Liu, X.; Guo, C.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-Mediated Targeted Mutagenesis of GmFT2a Delays Flowering Time in Soya Bean. Plant Biotechnol. J. 2018, 16, 176–185. [Google Scholar] [CrossRef] [Green Version]
- Qi, W.; Zhu, T.; Tian, Z.; Li, C.; Zhang, W.; Song, R. High-Efficiency CRISPR/Cas9 Multiplex Gene Editing Using the Glycine tRNA-Processing System-Based Strategy in Maize. BMC Biotechnol. 2016, 16, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.; Kudapa, H.; Pazhamala, L.T.; Chitikineni, A.; Thudi, M.; Gaur, P.; Pasupuleti, J.; Fikre, A.; Kimurto, P.; Ellis, N. Translational Genomics in Agriculture: Some Examples in Grain Legumes. Crit. Rev. Plant Sci. 2015, 34, 169–194. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Zhang, C.; Sun, Z.; Wang, L.; Duanmu, D.; Fan, Q. Genome Editing in Cowpea Vigna unguiculata Using CRISPR-Cas9. Int. J. Mol. Sci. 2019, 20, 2471. [Google Scholar] [CrossRef] [Green Version]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-Frequency, Precise Modification of the Tomato Genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid Generation of a Transgene-Free Powdery Mildew Resistant Tomato by Genome Deletion. Sci. Rep. 2017, 7, 482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, N.M.; Baltes, N.J.; Voytas, D.F.; Douches, D.S. Geminivirus-Mediated Genome Editing in Potato (Solanum tuberosum L.) Using Sequence-Specific Nucleases. Front. Plant Sci. 2016; 7. [Google Scholar] [CrossRef] [Green Version]
- Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.-S.; Ohlsson, P.; Gonzalez, M.N.; Samuelsson, M.; Hofvander, P. Genome Editing in Potato via CRISPR-Cas9 Ribonucleoprotein Delivery. Physiol. Plant. 2018, 164, 378–384. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 Variants Generated by CRISPR-Cas9 Improve Maize Grain Yield under Field Drought Stress Conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Li, S.; Li, Z.; Li, H.; Song, W.; Zhao, H.; Lai, J.; Xia, L.; Li, D.; Zhang, Y. A Barley Stripe Mosaic Virus-Based Guide RNA Delivery System for Targeted Mutagenesis in Wheat and Maize. Mol. Plant Pathol. 2019, 20, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted Mutagenesis in Zea Mays Using TALENs and the CRISPR/Cas System. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Xu, F.; Liu, L.; Char, S.N.; Ding, Y.; Je, B.I.; Schmelz, E.; Yang, B.; Jackson, D. The Maize Heterotrimeric G Protein β Subunit Controls Shoot Meristem Development and Immune Responses. Proc. Natl. Acad. Sci. USA 2020, 117, 1799–1805. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Gadlage, M.J.; Lafitte, H.R. Superior Field Performance of Waxy Corn Engineered Using CRISPR–Cas9. Nat. Biotechnol. 2020, 38, 579–581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Chen, R.; Yang, L.; Fan, K.; Liu, Y.; Wang, G.; Ren, Z.; Liu, Y. Generation of Transgene-Free Semidwarf Maize Plants by Gene Editing of Gibberellin-Oxidase 20-3 Using CRISPR/Cas9. Front. Plant Sci. 2020, 11, 1048. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, X.; Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L. Engineering Herbicide-Resistant Rice Plants through CRISPR/Cas9-Mediated Homologous Recombination of Acetolactate Synthase. Mol. Plant 2016, 9, 628–631. [Google Scholar] [CrossRef] [Green Version]
- Begemann, M.B.; Gray, B.N.; January, E.; Gordon, G.C.; He, Y.; Liu, H.; Wu, X.; Brutnell, T.P.; Mockler, T.C.; Oufattole, M. Precise Insertion and Guided Editing of Higher Plant Genomes Using Cpf1 CRISPR Nucleases. Sci. Rep. 2017, 7, 11606. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhang, X.; Sun, Y.; Zhang, J.; Du, W.; Guo, X.; Li, S.; Zhao, Y.; Xia, L. Efficient Allelic Replacement in Rice by Gene Editing: A Case Study of the NRT1. 1B Gene. J. Integr. Plant Biol. 2018, 60, 536–540. [Google Scholar] [CrossRef] [Green Version]
- Toda, E.; Koiso, N.; Takebayashi, A.; Ichikawa, M.; Kiba, T.; Osakabe, K.; Osakabe, Y.; Sakakibara, H.; Kato, N.; Okamoto, T. An Efficient DNA- and Selectable-Marker-Free Genome-Editing System Using Zygotes in Rice. Nat. Plants 2019, 5, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Chen, K.; Li, T.; Zhang, Y.; Wang, Y.; Zhao, Q.; Liu, J.; Zhang, H.; Liu, C.; Ran, Y.; et al. Efficient DNA-Free Genome Editing of Bread Wheat Using CRISPR/Cas9 Ribonucleoprotein Complexes. Nat. Commun. 2017, 8, 14261. [Google Scholar] [CrossRef]
- Murovec, J.; Guček, K.; Bohanec, B.; Avbelj, M.; Jerala, R. DNA-Free Genome Editing of Brassica oleracea and B. rapa Protoplasts Using CRISPR-Cas9 Ribonucleoprotein Complexes. Front. Plant Sci. 2018; 9. [Google Scholar] [CrossRef]
- Hu, B.; Li, D.; Liu, X.; Qi, J.; Gao, D.; Zhao, S.; Huang, S.; Sun, J.; Yang, L. Engineering Non-Transgenic Gynoecious Cucumber Using an Improved Transformation Protocol and Optimized CRISPR/Cas9 System. Mol. Plant 2017, 10, 1575–1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Lee, E.; Heo, J.; Kim, D.H.; Chun, H.J.; Kim, M.C.; Bang, W.Y.; Lee, Y.K.; Park, S.J. Rapid Generation of Transgenic and Gene-Edited Solanum nigrum Plants Using Agrobacterium-Mediated Transformation. Plant Biotechnol. Rep. 2020, 14, 497–504. [Google Scholar] [CrossRef]
- Wu, S.; Zhu, H.; Liu, J.; Yang, Q.; Shao, X.; Bi, F.; Hu, C.; Huo, H.; Chen, K.; Yi, G. Establishment of a PEG-Mediated Protoplast Transformation System Based on DNA and CRISPR/Cas9 Ribonucleoprotein Complexes for Banana. BMC Plant Biol. 2020, 20, 425. [Google Scholar] [CrossRef]
- Lin, C.-S.; Hsu, C.-T.; Yang, L.-H.; Lee, L.-Y.; Fu, J.-Y.; Cheng, Q.-W.; Wu, F.-H.; Hsiao, H.C.-W.; Zhang, Y.; Zhang, R.; et al. Application of Protoplast Technology to CRISPR/Cas9 Mutagenesis: From Single-Cell Mutation Detection to Mutant Plant Regeneration. Plant Biotechnol. J. 2018, 16, 1295–1310. [Google Scholar] [CrossRef] [Green Version]
- Weiss, T.; Wang, C.; Kang, X.; Zhao, H.; Elena Gamo, M.; Starker, C.G.; Crisp, P.A.; Zhou, P.; Springer, N.M.; Voytas, D.F.; et al. Optimization of Multiplexed CRISPR/Cas9 System for Highly Efficient Genome Editing in Setaria viridis. Plant J. 2020, 104, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Mamidi, S.; Healey, A.; Huang, P.; Grimwood, J.; Jenkins, J.; Barry, K.; Sreedasyam, A.; Shu, S.; Lovell, J.T.; Feldman, M.; et al. A Genome Resource for Green Millet Setaria viridis Enables Discovery of Agronomically Valuable Loci. Nat. Biotechnol. 2020, 38, 1203–1210. [Google Scholar] [CrossRef]
- Curtin, S.J.; Xiong, Y.; Michno, J.-M.; Campbell, B.W.; Stec, A.O.; Čermák, T.; Starker, C.; Voytas, D.F.; Eamens, A.L.; Stupar, R.M. CRISPR/Cas9 and TALENs Generate Heritable Mutations for Genes Involved in Small RNA Processing of Glycine max and Medicago truncatula. Plant Biotechnol. J. 2018, 16, 1125–1137. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Hou, Y.; Wang, H.; Ji, R.; Liu, B.; Wen, J.; Niu, L.; Lin, H. Targeted Mutagenesis by CRISPR/Cas9 System in the Model Legume Medicago truncatula. Plant Cell Rep. 2017, 36, 371–374. [Google Scholar] [CrossRef]
- Che, P.; Chang, S.; Simon, M.K.; Zhang, Z.; Shaharyar, A.; Ourada, J.; O’Neill, D.; Torres-Mendoza, M.; Guo, Y.; Marasigan, K.M.; et al. Developing a Rapid and Highly Efficient Cowpea Regeneration, Transformation and Genome Editing System Using Embryonic Axis Explants. Plant J. 2021, 106, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Badhan, S.; Ball, A.S.; Mantri, N. First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts. Int. J. Mol. Sci. 2021, 22, 396. [Google Scholar] [CrossRef]
- Mao, Y.; Botella, J.; Liu, Y.; Zhu, J.-K. Gene Editing in Plants: Progress and Challenges. Natl. Sci. Rev. 2019, 6, 421–437. [Google Scholar] [CrossRef] [Green Version]
- Jansing, J.; Schiermeyer, A.; Schillberg, S.; Fischer, R.; Bortesi, L. Genome Editing in Agriculture: Technical and Practical Considerations. Int. J. Mol. Sci. 2019, 20, 2888. [Google Scholar] [CrossRef] [Green Version]
- Yang, B. Grand Challenges in Genome Editing in Plants. Front. Genome Ed. 2020, 2, 2. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, S. ; Komal; Ramchiary, N.; Singh, P. Role of Traditional Ethnobotanical Knowledge and Indigenous Communities in Achieving Sustainable Development Goals. 2021; 13. [Google Scholar] [CrossRef]
- Evans, P.; Quinoa Boom Offers Hard Lesson in Food Economics. CBC News, 13 January 2013. Available online: https://www.cbc.ca/news/business/quinoa-boom-offers-hard-lesson-in-food-economics-1.1358699 (accessed on 7 June 2021).
- FAO. The Impact of the Quinoa Boom on Bolivian Family Farmers. Available online: http://www.fao.org/resources/infographics/infographics-details/en/c/225070/ (accessed on 7 July 2021).




| Sl. No. | Traditional Food Plant | Occurrence and Traditional Use | Important Nutritional and Stress Resilient Traits |
|---|---|---|---|
| 1 | Lolium perenne (Perennial ryegrass, Poaceae) | Used as a cereal in North America, Southern countries of Europe, North Africa, Middle East and towards the eastern sides of Central Asia [84]. | The seed has a nutritional value similar to oats (Avena sativa) and contains gluten which is an important trait of baked food [84]. |
| 2 | Cleome gynandra (Stinkweed, Capparaceae) | It is an important vegetable in rural areas of several African countries [85]. | Rich in linoleic acid and amino acids content such as glutamic acid, aspartic acid, arginine, tyrosine, histidine and lysine [85]. The C4 photosynthetic pathway helps them to survive in dry and hot conditions [86]. Adapted to several types of soils and can grow in humid, semiarid and arid climates [87]. |
| 3 | Basella alba (Vine spinach, Basellaceae) | Used throughout temperate regions and the tropics [88]. | Leaves are rich in calcium, fiber, fat, protein and carbohydrates [89]. They are extremely heat tolerant and are also adapted to a variety of soils and climates [90]. |
| 4 | Vigna subterranea (Bambara groundnut, Fabaceae) | An important indigenous crop in sub-Saharan African countries such as South Africa, Senegal and Kenya, and Madagascar [91]. | Drought and pest resistant, able to survive in poor soils. Rich in protein whereas fat content is low [92]. Rich in essential sulfur-containing amino acids such as Methionine and provides a good amount of fiber, iron, potassium and calcium [93]. |
| 5 | Chlorophytum comosum (Spider plant, Asparagaceae) | Iran [94]. | Tubers are rich in carbohydrates, fiber and calcium [94]. |
| 6 | Corchorus spp. (Mallow, Malvaceae) | In India, Africa and the Middle East, it has been a popular vegetable since ancient times [95]. | The leaves are a good source of calcium, iron, beta carotene, vitamin C and α-tocopherol. Plants also show antioxidant activity [96]. |
| 7 | Macrotyloma uniflorum (Horse gram, Fabaceae) | Cultivated in Asian countries, especially India and Myanmar, and African countries [97]. | Adapted to drought and poor fertile solid conditions. A potential source of nutrients such as protein, iron and calcium [97]. |
| 8 | Fagopyrum tataricum, F. esculentum (Buckwheat, Polygonaceae) | Found on a large scale in Asian and Southeast Asian countries. It was spread from China to Japan and Korea. It is also consumed in Russia, Sweden, Europe and North America [98]. | Proteins are rich in essential amino acid lysine [98]. |
| 9 | Brassica carinata (Ethiopian mustard, Brassicaceae) | Consumed all over the world and considered important food crops in European countries, India, Japan and China [99]. It is an important green leafy vegetable in Zambia and in most parts of tropical Africa [100]. | High levels of glutamic acid, arginine and proline [99]. |
| 10 | Colocasia esculenta (Taro, Araceae) | It is found all over the Pacific islands and other parts of the world. Africa is the bulk producer of taro, followed by Asia and Oceania [101]. | Rich in small starch grains and proteins. Nutritive than other tubers and rich in vitamins (thiamine, vitamin C, niacin and riboflavin) and minerals (iron, phosphorus and calcium). Taro corms have a high quantity of magnesium and potassium; also a good source of carotene [102]. |
| 11 | Boscia senegalensis (Aizen plant, Capparaceae) | Native to the Sahel region of Africa [103]. | Protein contains a considerable quantity of tryptophan and arginine. Zinc and iron are present at a relatively high level [104]. High degree of drought resistance [105]. It is highly drought tolerant and can perform very well poor soil conditions [103]. |
| 12 | Sphenostylis stenocarpa (African yam bean, Fabaceae) | Cultivated in different regions of African countries [106]. | The legume and tuber of the plant is edible. Adapted to wide range of climatic, geographical and edaphic conditions [106]. They have a short growing period [107]. |
| 13 | Telfairia occidentalis (Fluted guard, Cucurbitaceae) | The crop is extensively cultivated in southern Nigeria [108]. | Leafy vegetable with oil-rich leaves. Its nutritious seeds are also consumed as they are a good source of minerals and proteins [108]. |
| 14 | Digitaria exilis (Fonio millet, Poaceae) | Cultivated throughout West Africa [109]. | Rich in minerals, vitamins, carbohydrates, protein, fiber and iron. Another advantage is that it is gluten free [110]. Grows in poor-fertile soil and rain-deficient areas [111]. Long storage life without preservatives [109]. |
| 15 | Crotalaria brevidens (Rattle pod, Fabaceae) | Widely consumed and cultivated in East Africa and West Africa [112]. | Good source of β-carotene, ascorbate, folic acid, riboflavin, iron, calcium and magnesium [59]. They have nitrogen fixing capacity, drought tolerance, produce seeds under tropical conditions and are suitable for intercropping [112]. |
| 16 | Dacryodes edulis (African pear, Burseraceae) | Cultivated in Guinea and widely in other tropical parts of Africa [113]. | Edible fruits contain lipid, protein, vitamins and minerals such as potassium, calcium, magnesium, iron, zinc, copper and selenium [113,114,115]. |
| 17 | Treculia africana (African breadfruit, Moraceae) | Cultivated in Nigeria and Africa as a whole [116]. | Seeds are highly nutritious because of the presence of minerals such as potassium, magnesium and calcium, vitamins, fats, proteins and carbohydrates [117]. Grows in marginal areas where other species may not be able to grow [116]. |
| 18 | Momordica balsamina (Balsam apple, Cucurbitaceae) | Indegenous to the countries of tropical Africa, Arabia, Asia and Australia. Widely distributed in Swaziland, Namibia, Botswana and the provinces of South Africa [118]. | Leaves are rich in protein and fat. They have higher values of minerals such as calcium, magnesium and iron [119]. Leaves also contain 17 amino acids [118]. |
| 19 | Adansonia digitata (Baobab, Malvaceae) | Distributed throughout the drier parts of Africa, Namibia, Ethiopia, Sudan and Sahara [120]. | Contains vitamin B2/Riboflavin, calcium, phosphorus, iron, vitamin A and vitamin C. It contains almost 10 times more vitamin C than oranges [121]. It is drought tolerant and can tolerate various ranges of pH. It can also grow in calcareous soils and rocky hillsides [120]. |
| 20 | Berchemia discolour (Bird plum, Rhamnaceae) | Indigenous Southern African fruit tree species. Widely distributed in the regions of northern, eastern, central and southern Africa [122]. | The dry pulp is a rich source of calcium, carbohydrates, iron, sodium, potassium and magnesium [122]. |
| 21 | Heinsia crinita (Bush apple, Rubiaceae) | Indigenous to West Africa, especially the southern part of Nigeria [123]. | Rich in calcium, magnesium, potassium, iron and zinc [123]. |
| 22 | Psophocarpus tetragonolobus (Winged beans, Fabaceae) | It grows widely in Malaysia, Indonesia, the Philippines, Bangladesh, Thailand, Sri Lanka, India, Myanmar and African countries [124]. | Seeds, pods, tubers, foliage and flowers are nutritious [124] and contain higher crude protein [125]. It has adequate quantity of minerals such as P, K, Ca, S, Na, Mg, Mn, Fe, B, Sr, Zn, Ba, Cu and Cr, and vitamins such as vitamin A, vitamin B1, vitamin B2, vitamin B3, vitamin B6, vitamin B9, vitamin C and vitamin E [126]. It is suitable to be grown in hot, humid conditions and possess nitrogen fixation capacity [127]. |
| 23 | Tropaeolum tuberosum (Mashua, Tropaeolaceae) | Traditional subsistence tuber crops indigenous to the Andean highlands [128]. | It can be grown in poor soils without pesticides and fertilizers [128]. They have a high level of protein with an ideal balance of essential amino acids. More content of vitamin C and provitamin A (equivalents of Retinol) than other Andean tubers. Rich in magnesium, phosphorus, iron and zinc [129]. |
| 24 | Oxalis tuberosa (Oca, Oxalidaceae) | Second important tuber crop in Bolivia and Peru. Cultivated as an important crop in Central Andes, Chile, Argentina, Ecuador, Bolivia and Peru [130]. | Iron- and calcium-rich tubers [131]. Notable quantities of fructo-oligosaccharides reported [130]. |
| 25 | Smallanthus sonchifolius (Yacon, Asteraceae) | Cultivated in Bolívia, Peru, Czech Republic, Argentina, Italy, Brazil, Ecuador, Korea, Japan, New Zealand and the United States [132]. | Rich in fructooligosaccharides that are good for colon health. They are extremely hardy plants and adapted to cold and hot conditions [133]. |
| 26 | Chenopodium pallidicaule (Cañiwa, Amaranthaceae) | Majorly grown in Bolivian and Peruvian Altiplano [134]. | Exceptional protein quantity and quality and grains are enriched with micronutrients such as calcium and iron [134]. The nutritional value is equivalent to milk proteins [135]. Gross et al. [136] recognized that it has a balanced amino acid composition and 15.3% protein content. It does not have saponins, which gives a bitter taste and hence it is possible to consume directly without washing. Drought- and frost-resistant plants, well adapted to rocky and poor nutrient soil [134]. |
| 27 | Lablab purpureus (Hyacinth bean, Fabaceae) | Third high priority vegetable in the south-western and central regions of Bangladesh [137]. Cultivated as a minor crop in tropical regions of Asia and Africa [138] | Extremely resilient to drought-prone areas. A good source of vegetable protein and also a potent source of fats, carbohydrates, fibers and minerals such as phosphorus, calcium and iron [139]. |
| 28 | Sclerocarya birrea (Marula, Anacardiaceae) | African fruit tree [140]. | Seeds contain sufficient amounts of calcium, phosphorus, magnesium, iron, potassium and copper. Seed edible part has 36.4% of protein, with high levels of cysteine and methionine. Fruits are rich in ascorbic acid and juice extracts contain 33 types of sesquiterpene hydrocarbons [140]. |
| 29 | Amorphophallus paeoniifolius (Elephant foot yam, Araceae) | Cultivated in Southeast Asian countries such as Malaysia, the Philippines and Indonesia [141]. | Multiple edible parts such as leaves, rhizomes and petioles. Immunity booster and rich in carbohydrates, phenols, alkaloids, tannins, flavones, steroids, coumarins, vitamins, minerals and antioxidants [142]. |
| 30 | Solanum quitoense (Lulo, Solanaceae) | Majorly cultivated and consumed in Columbia, Ecuador and Central America [143]. | Carotenoid content of fruit is high. Very low fat content but rich in proteins [143]. |
| 31 | Senna tora (Sickle pod, Caesalpiniaceae) | India [144]. | Its leaves consist of lipids, crude fiber, crude protein and minerals (iron, calcium, cobalt sodium, zinc, magnesium, manganese and potassium) [144]. Sickle pods hold great potential as a source of medicine, minerals. They exhibit drought tolerance [145]. |
| 32 | Ziziphus jujuba (Buckthorns, Rhamnaceae) | Widely distributed in Europe, Southern and Eastern Asia and Australia [146]. | They grow in different soils and are resistant to alkalinity and salinity, and better adapted to arid regions. They contain high amounts of fructose and fiber. Jujube fruit is rich in unsaturated fatty acids especially linoleic acid (omega-6). They are rich in vitamin C also. Excellent source of magnesium, phosphorus, potassium, sodium and zinc [146,147]. |
| 33 | Pyrus pyrifolia (Asian pear, Rosaceae) | It is cultivated throughout Central and South China, Russia, Korea, Japan, Vietnam, Thailand, India, Indonesia and the Philippines. As of recently, it is also cultivated in Australia, New Zealand, the USA and Europe (Italy, France) [148]. | Abundant in vitamin B and minerals [148]. |
| 34 | Achyranthes bidentata (Ox knee, Amaranthaceae) | Grown as cereal in Korea, Vietnam and China. In India and China, leaves and seeds are consumed [149]. | Seeds are rich in proteins and minerals such as iron, calcium, phosphorus, potassium and magnesium. It contains 1.6 times higher quantity of vitamin E than Amaranthus seeds [149]. |
| 35 | Setaria italica (Foxtail millet, Poaceae) | China, India and other Asian countries [150]. | Great tolerance to drought and can grow in arid and barren lands [150]. |
| 36 | Grewia asiatica (Phalsa, Malvaceae) | Various parts of South Asia including Cambodia, Philippines and Laos [151]. | Rich in vitamin A, vitamin C, minerals and fiber. Can grow nicely under water-deficient conditions [152]. |
| 37 | Aegle marmelos (Bael, Rutaceae) | Cultivated throughout India, Nepal, Tibet, Sri Lanka, Laos, Thailand, Malaysia, Phillipines, Vietnam and Myanmar [153]. | Potent source of vitamins (A, B, C, folate) and minerals, antioxidants, dietary fiber, amino acids and bioactive compounds [153]. They are adapted to high salinity conditions [154]. |
| 38 | Carissa carandas (Koranda, Apocynaceae) | India [155]. | Rich source of vitamin C, iron, calcium and phosphorus [155]. They are xerophytic and suitable for growing in dry land [156]. |
| 39 | Artocarpus heterophyllus (Jackfruit, Moraceae) | Majorly cultivated in tropical regions of Burma, Sri Lanka, Indonesia, Malaysia, Jamaica, India, Mauritius, Brazil, East Africa, Seychelles and Rodrigues Island [157]. | Fruits are rich in carbohydrates and vitamins such as A, C and folic acid. Rich in calcium and magnesium [158]. Tolerant to water deficit conditions [157]. |
| 40 | Ullucus tuberosus (Olluco, Basellaceae) | Peru, Ecuador, Colombia, Venezuela and northwestern Argentina [159]. | Resistant against frost and drought and can perform in poor soils. Lower in fat than corn [159]. |
| 41 | Arracacia xanthorrhiza (Arracacha, Apiaceae) | It is found in South American Countries such as Ecuador, Colombia, Brazil and Venezuela [160]. | Adapted to mesothermic, montane, day length regimes and tropical frost-free conditions [160]. |
| 42 | Morinda citrifolia (Indian mulberry, Rubiaceae) | Native to Southeast Asia and Australia and widely distributed globally [161]. | Vitamins such as ascorbic acid and provitamin A, amino acids such as aspartic acid, mineral and an alkaloid, xeronine, are detected in its fruits [162]. The plant shows tolerance to a number of stresses such as drought, water logging and salinity [161]. |
| 43 | Canavalia gladiata (Sword bean, Leguminosae) | They are cultivated on a limited scale in Asia, West Indies, Africa and South America [163]. | Seed coat of the sword bean is rich in gallic acid and other derivatives [164]. Seeds are a rich source of sodium, potassium and calcium [165]. The crude protein content of sword beans is high. Some cultivars are fairly resistant to drought [163]. |
| 44 | Lupinus mutabilis (Tarwi, Leguminosae) | Distributed widely in the Andes, Venezuela, Colombia, Ecuador, Peru and Bolivia, Australia, Germany, New Zealand, Poland and the United Kingdom [166]. | Seeds have high protein and lipid content whereas fiber and carbohydrate content are lower compared to other lupin species [167]. It has adaptability to temperate and cold climates. It can grow on marginal land and low fertility soils [168]. |
| 45 | Limonia acidissima (Wood Apple, Rutaceae) | Native to India but also cultivated in Bangladesh, Pakistan and Sri Lanka [169]. | The fruits are rich in β-carotene, vitamin B, vitamin C, thiamine and riboflavin. Fruit pulp is enriched with citric acid, other fruit acids, mucilage and minerals. Other compounds such as alkaloids, coumarins, fatty acids and sterols are also detected in its fruits [169]. It is well adapted to drier conditions and thus shows a greater stress tolerance [170]. |
| 46 | Cordia myxa (Indian Cherry, Boraginaceae) | It is found globally especially in the tropics. It grows naturally in India, Myanmar and Afghanistan [171]. | It displays drought tolerance and because of that it can easily grow in arid and semi-arid regions [171]. |
| 47 | Carissa carandas (Karonda, Apocynaceae) | The plant is distributed in various parts of the world such as Nepal, Afghanistan, India, Sri Lanka, Java, Malaysia, Myanmar, Pakistan, Australia and South Africa [172]. | Fruits are rich in calcium, iron, vitamin C, vitamin A [173]. The plant shows drought tolerance [172]. |
| 48 | Lepidium meyenii (Maca, Brassicaceae) | Nutritionally highly valuable and is native to Peru [174]. | It contains good quantities of fiber, essential amino acids, fatty acids, vitamin C and minerals such as copper, iron and calcium [175]. |
| 49 | Pastinaca sativa (Parsnips, Apiaceae) | It is commonly found in old fields, roadsides and woodland edges in North America [176]. | Rich in vitamins and minerals; particularly rich in potassium [176]. It shows drought tolerance [177]. |
| 50 | Xanthosoma sagittifolium (American taro, Araceae) | Traditionally used as a tuber crop, native to Nigeria and tropical Africa [178]. | Good source of carbohydrates and starch. Superior in terms of their protein digestibility and mineral composition such as calcium, phosphorus and magnesium [178]. |
| 51 | Colocasia antiquorum (Taro, Araceae) | Widely consumed throughout the world especially Africa, Asia, the West Indies and South America [179]. | The corms are full of anthocyanins [179]. They are salt tolerant [180]. |
| 52 | Nelumbo nucifera (Lotus, Nymphaeaceae) | Creeping rhizomes are found throughout India; also found in China and Japan [181]. | It is a good source of protein and total carbohydrates and possesses high calorific value. It also contains higher quantities of essential minerals such as Na, K, Mg, Fe, Co, Zn and P [182]. Exhibits flooding tolerance [183]. |
| 53 | Plectranthus rotundifolius (Spreng, Lamiaceae) | Eaten for its edible tubers, native to tropical Africa. Grown in Africa and South East Asia [184]. | It contains higher mineral content than potato, sweet potato and cassava [185]. Highly tolerant to drought [186]. |
| 54 | Triticum monococcum (Einkorn wheat, Poaceae) | It has been an ancient staple food crop for many years. However, it is presently cultivated only in the Mediterranean region and continental Europe [187]. | Not very good in dietary fiber but it contains good amounts of proteins, unsaturated fatty acids, zinc and iron. It contains antioxidant compounds such as carotenoids, tocols and conjugated polyphenols [187]. They exhibit tolerance to salinity and frost [188]. |
| 55 | Triticum dicoccon (Emmer wheat, Poaceae) | Used as a cereal crop in the Middle- East, Central and West Asia and Europe [189]. | Rich in proteins, carbohydrates and minerals, poor in fats [189]. Shows drought tolerance [190]. |
| 56 | Triticum spelta (Dinkel wheat, Poaceae) | It has been an important staple food in parts of Europe in the ancient past [191]. | High vitamin content [191] and rich source of iron, zinc, copper, magnesium, potassium, sodium and selenium [192]. They have high flooding tolerance [193]. |
| 57 | Eleusine coracana (Finger millet, Poaceae) | It is produced in India, Niger, Mali, Burkina Faso, Chad and China [194]. | It is rich in calcium, dietary fiber, protein, minerals, phenolics and vitamins such as thiamine and riboflavin. It contains a good quantity of iron and amino acids such as methionine, isoleucine, leucine and phenylalanine [194]. They are tolerant to drought, pests and pathogens [195]. |
| 58 | Panicum sumatrense (Little millet, Poaceae) | Found in the Caucasus, China, India and Malaysia [196]. | Rich in micronutrients such as calcium and iron. They also contain high dietary fiber content and essential amino acids and have low glycemic index [196]. It also shows considerable tolerance against drought, salinity stresses and diseases. |
| 59 | Panicum miliaceum (Proso millet, Poaceae) | Produced in China, Russia, India and some countries of Eastern Europe and North America [197]. | The protein contains essential amino acids such as leucine, isoleucine and methionine than wheat [197]. They are drought tolerant [198]. |
| 60 | Pennisetum glaucum (Pearl millet, Poaceae) | An important cereal in arid and semiarid regions of Asia and Africa [199]. | It has high levels of calcium, iron, zinc, lipids and amino acids. Contains omega-9, omega-6 and omega-3 fatty acids. The tannins and phytates act as strong antioxidants [200,201]. It has a low glycemic index and it is a gluten-free crop. They are extremely drought-tolerant [202]. |
| 61 | Brosimum alicastrum (Breadnut, Moraceae) | Grown in southern Mexico [203]. | The flour obtained from the seeds is characterized by high protein, dietary fiber and micronutrient content. They are drought tolerant [204]. |
| 62 | Artocarpus altilis (Breadfruit, Moraceae) | It is an important food in the Pacific [205]. | Rich in fiber, protein, magnesium, potassium, phosphorus, thiamine (B1) and niacin (B3). They have tolerance to salinity and can grow on coralline soils and atolls [206]. |
| 63 | Mucuna pruriens (Velvet bean, Fabaceae) | Cultivated in Southeast Asian countries, including India and Sri Lanka, and Central South American countries as a legume for its seeds [207]. | The seeds are rich in dietary fiber and proteins [207]. They grow well in less fertile soil and show adaptation to drought conditions and acidified soils [208]. |
| 64 | Pachira aquatica (Malabar Chestnut, Bombacaceae) | Native to Southern Mexico, Guyana and Northeastern Brazil and introduced in other areas such as Guangdong, Southern Yunnan and Taiwan as a cultivated plant [209]. | Seeds contain a high amount of lipids, proteins with high amounts of essential amino acids such as tryptophan, threonine and phenylalanine/tyrosine [210]. Seeds contain more phosphate, magnesium, zinc, iron and copper than some fruits and other starchy foods [209]. |
| 65 | Strychnos cocculoides (Monkey orange, Loganiaceae) | The species is native to Botswana, Kenya, Namibia, South Africa, Tanzania, Uganda, Zambia and Zimbabwe [211]. | Adapted to drought prone and semi-arid areas. The vitamin C content of the fruits varies from 34.2 mg/100 g to 88 mg/100 g. Considered an essential source of iron [212]. |
| Sl. No. | Traditional Food Plant | Distribution | Important Nutritional and Stress Resilient Traits | Exceptionally Notable Character | Applications of Different Omics Technologies |
|---|---|---|---|---|---|
| 1. | Eleusine coracana (L.) Gaertn. (Finger millets, Poaceae) | Majorly produced in Mali, Niger, India, Burkina Faso and China [194]. | Tolerant to pathogens and pests. Drought resistant. Rich in minerals such as calcium and iron, vitamins, protein, dietary fiber and phenolics [194,195]. | Minerals and micronutrients are superior to rice and wheat [268]. | 1. Using genomics tools, Nirgude et al. [269] reported higher expression of opaque2 (regulate seed storage proteins), calcium transporters and calmodulin gene (calcium storage) and Kumar et al. [270] discussed allele mining strategies for PiKh and Pi21 genes that show resistance against Pyricularia oryzae blast disease. 2. Using transcriptomics, expression of several genes such as calcium transporters (CaX, CDPKs, CBPs) are reported [271]. Several transcription factors such as MYB, MYC, WRKY and ZFD were detected during drought stress [195]. 3. Proteomics study led to the identification of a calcium-binding protein, calreticulin [272]. Anatala et al. [273] reported heat shock proteins (HSPs), storage proteins and late embryogenesis abundant (LEA) during drought stress. |
| 2 | Setaria italica (L.) P. Beauv. (Foxtail millet, Poaceae) | Majorly cultivated in Asian countries such as India and China [150]. | Great drought tolerant potential and grows well in barren and arid land [150]. | Rich in essential amino acids, vitamin B, protein and micro elements [274]. | 1. Lata et al. [275] and Shi et al. [276] reported POD precursors, late embryogenesis abundant (LEAs) and aquaporins for drought tolerance by using transcriptomics. Phospholipid hydroperoxide glutathione peroxidase (PHGPX), ascorbate peroxidase (APX) and catalase 1 (CAT1) during salinity tolerance were reported using transcriptomics by Sreenivasulu et al. [277]. |
| 3. | Moringa oleifera Lam. (Drumstick, Moringaceae) | Distributed mainly in Middle Eastern, African and Asian countries [278]. | It has high micronutrient and vitamin content. It also shows antioxidant and medicinal activities. They can withstand occasional waterlogged conditions and adapt to hot and semi-arid conditions [279]. They are tolerant to heat, cold, salinity, nutrient starvation, variable light conditions and water deficiency [280]. | Rich in micronutrients and vitamin A [279]. | 1. WRKY transcription factors for various abiotic stress tolerance and copies of Cys2His2 zinc finger motifs (C2H2), APETALA2/ethylene-responsive element-binding protein (AP2-EREBP), C3H transcription factors for drought and cold resistance were reported [280]. High-throughput sequencing technology reported microRNAs related to biotic and abiot stress tolerance [281]. Nutritional component-related genes such as Vacuolar iron transporters (VIT), calreticulin for calcium storage, Zinc transporters, magnesium transporter and genes for vitamin C biosynthesis recognised [282]. 2. Flavonoid compounds and rutinoside sugar compounds were detected using metabolomics by Makita [283]. |
| 4. | Chenopodium quinoa Willd. (Quinoa, Amaranthaceae) | Cultivated as an important crop since ancient times in various parts of North-Altiplano, South and Central Chile [284]. | Rich source of minerals such as magnesium, iron, calcium, copper, potassium, zinc and phosphorus [66]. They have antioxidant activity (e.g., polyphenols) and rich in vitamins such as Vit. A, B1, B2, B9, C and E, lipids, proteins rich in essential amino acids particularly methionine and lysine, dietary fiber and carbohydrates [285]. They have extreme agro-ecological adaptability [286]. | Higher mineral content than maize and barley including calcium, magnesium, iron, copper, potassium, phosphorus and zinc [66]. | 1. Draft gene sequence and genes related to abiotic stress and nutrients were identified [287]. 2. Xyloglucan endotransglucosylase genes, an expansion A7-like gene and Ethylene Responsive Factor (ERF) genes were found to be downregulated in salt-tolerant plants [288]. 3. Sobota et al. [289] reported albumin and globulins through proteomics. 4. Root cell membrane’s potential, net H+, Na+ and K+ fluxes during salinity adaptation through ionomics study [290]. |
| 5. | Vigna unguiculata (L.) Walp. (Cow pea, Fabaceae) | Cultivated across Africa, Southeast Asia, Latin Southern and the United States of America. It is not widely cultivated in Europe but used in some Mediterranean countries [291]. | Rich in proteins and carbohydrates [292]. Proteins are rich in lysine and tryptophan amino acids [293]. Shows considerable adaptation to the warm climate with adequate rainfall [292]. | High quantity of folic acid and low quantity of antinutrients [294]. | 1. Up-regulated expression of chalcone isomerase and chalcone synthase in the salt-tolerant plants were reported [295]. 2. Sugars, proline, galactinol and quercetin were identified as osmolytes during osmotic stress using metabolomics [296]. 3. Identified amino acids which are related to glycolysis and tricarboxylic acid cycle [297]. 4. Lutein and beta carotene were reported using metabolomics [298]. |
| 6. | Vigna radiata (L.) R. Wilczek (Mungbean, Leguminosae) | African regions, South and Southeast Asia [299]. | Drought resistant. Higher iron and folate content [299]. | Rich in digestible protein quantity than other pulses [300]. | 1. Eight flavonoids (vitexin, isovitexin, rutin, kaempferol 3-O-rutinoside, isoquercitrin, genistein, daidzein and isorhamnetin) and two phenolics were reported using metabolomics [299]. |
| 7. | Sorghum bicolor (L.) Moench(Sorghum, Poaceae) | Major food in semi-arid tropical temperatures of African and Asian regions [301]. | Suitable for cultivation in dry areas and poor soil conditions [302]. Gluten-free cereal that is rich in antioxidants and phenolic compounds [303]. | Gluten-free grains [303,304]. | 1. Quantitative trait loci for sorghum polyphenols were recognized [302]. 2. Increased expression of Late Embryogenesis Abundant (LEA), delta 1-pyrroline-5-carboxylate synthase 2 (P5CS2) and high-affinity K+ transporter 1 (HKT1) for drought tolerance [305]. Salinity and osmotic stress tolerance genes reported [306]. 3. Presence of fructose, galactose, lactose, cellobiose and sedoheptulose as an osmotic protectant were detected using metabolomics [307]. 4. Glutathione-S transferases and l-ascorbate peroxidase during salinity stress identified [308]. |
| 8. | Manihot esculenta Crantz. (Cassava, Euphorbiaceae) | Used by different communities all over the world, mainly tropical and subtropical areas [309]. | Adapted to marginal soil conditions and erratic rain. Carbohydrate and protein rich [310]. | Rich source of energy [311]. | 1. Using genomics, carotenoid markers on chromosome 1 and candidate genes for carotenoid (phytoene synthase) and starch biosynthesis were reported [312]. 2. Identification of starch biosynthesis genes [310]. Expression profiling and characterization of drought responsive Abscisic acid (ABA)-responsive element (ABRE)-binding factors (ABFs) [313]. Upregulation of 1300 genes during drought stress [314]. Transcription factors related to heat stress (A3, heat-shock transcription factor 21 and a homeobox-leucine zipper protein ATHB12) and dehydration tolerance (ERD1, RD19, RD22 precursor, drought-induced protein Di19-like) were reported [315]. WRKY genes related to abiotic stress tolerance [316]. 3. Proteomics—ATP synthase subunit beta, Rubisco activase (RCA), Rubisco, phosphoglycerate, chaperone peroxiredoxin, heat shock protein, glutathione transferase profiling during cold stress [317]. |
| 9. | Amaranthus hypochondriacus L., Amaranthus viridis L. (Amaranth, Amaranthaceae) | Consumed in China since ancient times. Central America, South America. It is also used in Africa and Caribbean [318]. | Leaves and seeds are rich in quality proteins and its quantity is higher than maize. Proteins contain higher amounts of amino acid lysine and sulfur containing amino acids [319]. Amaranth oil contains unsaturated linolenic fatty acid which is good for human health [320]. | High quality protein with rich lysine content in leaves and seed [319]. | 1. Gene annotation of lysine biosynthetic pathway and expression analysis was analyzed [321]. 2. Chloroplast chaperones, Rubisco large subunit, cytochrome b6f, oxygen evolving complexes and ascorbate peroxidase expression variation during drought stress were studied [322]. 3. Lutein and beta carotene detection [298]. |
| 10. | Sesuvium portulacastrum (L.) L. (Shoreline purslane, Aizoaceae) | Locally consumed in various regions of India, South East Asia, Philippines [323]. | Salt, drought and oxidative stress tolerance. Salty taste and fleshy nature of leaves [324]. | Rich source of sodium [323]. | 1. Identified Late embryogenesis abundant 2 as the gene for salt and drought tolerance [324]. Fructose-1,6-bisphosphate aldolase gene (FBA) for abiotic stress tolerance was isolated [325]. 2. Copper (Cu), iron (Fe), manganese (Mn) and zinc (Zn) accumulation during salinity tolerance was reported [326]. |
| 11. | Ipomoea batatas (L.) Lam. (Sweet potato, Convolvulaceae) | Consumed throughout the world. Asia and Pacific islands produce 92 % of the world’s sweet potato supply [327]. | It is pest and disease tolerant and adapted to high moisture conditions. Rich in complex carbohydrates, vitamin A, vitamin C, Fe and K. Orange-fleshed sweet potatoes are one of the storehouses of beta-carotene. It is a highly resistant crop [327]. | Rich source of beta carotene [327]. | 1. APX, manganese-dependent superoxide dismutase (MnSOD), LEA, early responsive to dehydration (ERD), sodium/hydrogen antiporter (NHX), aquaporin (AQP), vacuolar calcium ion transporter (CAX), metallothionein (MT), betaine aldehyde dehydrogenase (BADH), pyrophosphatase (PPase), catalase (CAT), polyphenol oxidases (PPO), ABRE-binding protein (AREB) during abiotic stress tolerance reported [328]. 2. Amino acids, carbohydrates and flavonoids were detected using metabolomics [329]. Βeta-carotene content [330]. |
| 12. | Ipomoea imperati (Vahl) Griseb. (Beach morning glory, Convolvulaceae) | Distributed in coastline all over the world [331]. Consumed by local communities for the underground tuber. | Salinity tolerant and grows well in poor nutrient soil [331]. | Rich in sodium [331]. | 1. Expression profiling of AP2/EREBP, bHLH, HD-ZIP and MYB transcription factors during salinity tolerance reported [331]. |
| 13. | Dioscorea spp. (Yam, Dioscoreaceae) | Tropical and subtropical Countries. Major food in Africa [309]. | Great source of fiber, potassium, manganese, copper and antioxidants. They also exhibit abiotic stress tolerance [332]. | Vitamin C and potassium rich [333]. | 1. Metabolite profiling revealed amino acid content, malic acid content, fatty acids and phosphate content [332]. 2. Genome sequencing revealed the hybrid origin of Dioscorea rotundata from D. prehensilis (wild rainforest plant) and Dioscorea abyssinica (Savannah adapted plant) [334]. |
| 14. | Portulaca oleracea L. (Common purslane, Portulacaceae) | Distributed all around the world such as New Zealand, Canada, America, temperate countries of Europe, Australia and is highly abundant in India [335]. | It contains high amounts of α-linolenic acid and oxalic acid in their leaves which are highly health beneficial [336]. It is also rich in carbohydrates, protein, minerals (calcium, magnesium, sodium and potassium), vitamin C, carotene, riboflavin, thiamine and nicotinic acid. It is well adapted to dry and salinity conditions, therefore ideal for arid areas [337]. | High amount of alpha-linolenic acid and oxalic acid in the leaves [336]. | Metabolomics study reported 6 amino acids, 22 phenolic compounds, 16 alkaloids and 11 fatty acids [338]. α-linolenic acid accounted for about 40 % to 60 % of total fatty acid [339]. |
| 15. | Physalis peruviana L. (Wild tomatillos, Solanaceae) | A cultural staple of Mexico, Central America, South Africa, North America and Europe [340]. | They have carotenoids, minerals and vitamin-rich fruits and seeds and show adaptability towards various environmental conditions [341,342]. | Carotenoid and vitamin-rich fruits and seeds [341]. | Metabolomic profiling reported lutein as the most abundant carotenoid (64.61 µg/g at the half-ripe stage) and the presence of gamma carotenoid (rare in fruits) [343]. |
| 16. | Rumex vesicarius L. (Ruby dock, Polygonaceae) | Cultivated in North Indian states as a vegetable [344]. | Rich in phenols, ascorbic acid, α-tocopherol and β-carotene [345]. | Vitamin rich [345]. | Metabolomic study reported 13 Phenolic compounds, ascorbic acid, α-tocopherol and β-carotene content and 6-C-glucosyl-naringenin identified as the key phenolic compound which have high antioxidant capacity [345]. |
| 17. | Corylus avellana L. (Hazelnuts, Betulaceae) | Consumed by human civilizations from Mesolithic time onwards and cultivated worldwide especially in Spain, Turkey and Italy, United States and Canada [346,347]. | Rich source of starch, protein, lipids, vitamin E and C, potassium, phosphorus, magnesium and calcium [348]. | Rich in malic acid and unsaturated fatty acids [349]. | Reported higher concentration of palmitic acid which prevents metabolic syndromes such as diabetes [350]. |
| 18. | Avena sativa L. (Oats, Poaceae) | Consumed in developing as well as developed countries [351]. | Nutritionally rich, traditionally used cereal crops as a major protein diet in cold climate countries including Northern Europe [352]. Better adapted to acid soils and variable soil types than other grain cereal crops [351]. | High dietary fiber content and 78–81.5% unsaturated fatty acids out of 5–9 % lipids [353]. | 1. Barley yellow dwarf virus tolerance QTL on chromosome 3C using genome wide association study was reported [354]. 2. Presence of polyamines detected during osmotic stress detected [355]. |
| 19. | Bacopa monnieri (L.) Pennell. (Brahmi, Plantaginaceae) | Sri Lanka, India, Nepal, China, Taiwan, Vietnam and Pakistan. Traditionally used as a medicinal plant from ancient times onwards [356]. | Rich in Fe, Mg and Zn. Studies have proven the ability of Brahmi to enhance memory. They grow well in Marshy areas [356]. | Rich source of microelements [356]. | 1. De novo assembly of transcriptome and draft chloroplast genome from RNAseq data [357]. 2. Proline content elevation during osmotic stress [358]. |
| 20 | Elaeagnus umbellata Thunb. (Autumn olive, Elaeagnaceae) | Berries consumed in tropical and temperate Asia. Nowadays it is available in European countries also [65]. | The berries are a rich source of lycopene and possess 10 times higher quantity of lycopene in their fruits than tomatoes [359]. They are rich in β-cryptoxanthin, α-cryptoxanthin, lutein, β-carotene, phytofluene and phytoene and vitamins. Exhibit drought tolerance, temperature tolerance and high tolerance to pruning. Can grow in high-saline soils [65]. | Ten times higher quantity of lycopene in their fruit than tomato [65]. | 1. Phytoene Synthase (EutPSY) gene expression correlation with lycopene [360]. 2. Sugar metabolism-related enzymes (R-amylase, UGPase, phosphoglucomutase, acid invertase and triose-phosphate isomerase) and carotenoid biosynthesis-related proteins (Acetyl-CoA C-acetyltransferase, IPP isomerase and dimethylallyl diphosphate) reported [361]. |
| 21. | Porteresia coarctata (Roxb.) Tateoka(Wild rice, Poaceae) | India, Sri Lanka, Bangladesh and Myanmar [362]. | Grows in saline estuaries and is adapted to salinity [362]. | With increase in salinity stress, carbohydrate and ash content increases [363]. | Elevation of proteins related to photosynthesis such as Rubisco large subunit, Rubisco small subunit and light harvesting complex-chlorophyll a, b reported during salinity [362]. |
| 22. | Atriplex lentiformis (Torr.) S.Watson(Quail Bush, Chenopodiaceae) | South western United States and northern Mexico [290]. | Good salinity adaptation capacity [290]. | Rich source of sodium [364]. | 1. Studied the H+-ATPase activity of plasma membranes during salinity stress, which leads the plant for K+ retention and Na+ exclusion for better salt tolerance [290]. |
| 23 | Fagopyrum esculentum Moench (Buckwheat, Polygonaceae) | Worldwide distribution [365]. | Grows in hilly areas and marginal ecosystems [365]. Rich in sulfur containing amino acids such as cysteine and methionine than any cereal. Fatless, gluten-free grains that are rich in starch and minerals such as Ca, Mo, S and vitamins [352,366]. | Excellent quality of protein with a high amount of essential amino acid lysine [98]. | 1. Draft genome of buckwheat was developed and the same study identified expression of three granule bound starch synthase (GBSS) genes [287]. 2. Differential expression of sugar biosynthesis and metabolism-related genes in F. esculentum and F. tataricum [367]. |
| 24 | Panicum miliaceum L. (Proso millet, Poaceae) | It is cultivated widely in Asian countries, some African countries and the Middle East [368]. | More efficient in water usage, because it shows the C4 pathway, hence suitable for cultivation in dry areas. High productivity in low input soil and marginal lands [263]. Rich in both major nutrients and minor nutrients such as phenolics, minerals and vitamins. Gluten-free grain [197]. | Richer in essential amino acids than wheat [197]. | 1. Genes related to C4 mechanisms such as carbonic anhydrase (CA), NAD dependent malic enzyme (NAD-ME) and NADP- malic enzyme (NADP-ME) [369]. 2. Protein related to metabolisms such as polysaccharide and starch [370]. 3. Nearly 48 metabolites including several primary metabolites and five phenolic acids were detected [371]. |
| 25 | Sclerocarya birrea (A.Rich.) Hochst. (Marula, Anacardiaceae) | Popular African tree [140]. | Ascorbic acid-, arginine- and glutamine-rich fruits [140]. | Highest level of arginine and ascorbic acid [140]. | 1. Draft genome reported and identified genes involved in starch biosynthesis pathway [265]. |
| 26 | Ziziphus jujuba Mill. (Chinese jujube, Rhamnaceae) | Mainly cultivated in Asian countries [372]. | Salt tolerant and drought tolerant [373]. Good source of phenolics, vitamin C, triterpenic acids, flavonoids and polysaccharides [374]. | Rich in unsaturated fatty acid, especially omega-6 fatty acid [375]. | 1. Expression of 5269 differentially expressed genes during salinity were recognized and among them, 2540 were downregulated and 2729 were upregulated [373]. Expression profiling of genes during heat stress led to identification of heat responsive factors [374].Expression profiling of three UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT), responsible for anthocyanin accumulation in fruit peel [376]. |
| 27 | Dacryodes edulis (G.Don.) H.J.Lam (African pear, bush pear, Burseraceae) | Cultivated in tropical countries of Africa [113]. | Rich source of protein, vitamins and lipids [113]. | Selenium content is high compared to other crops reported with selenium. Beta-carotene is higher than papaya, avocado and amaranth. They are rich in potassium [114]. | NA |
| 28 | Basella alba L. (Vine spinach, Basellaceae) | Tropical Asian countries [89]. | Heat- and drought-tolerant plants, high quantities of vitamin A, C, iron and calcium are present [89]. | Leaves are rich in calcium [89]. | NA |
| 29 | Solanum quitoense Lam. (Lulo, Solanaceae) | South American countries and nowadays found in European countries also [377]. | Adapted to shady areas and rich in vitamins [377]. | Rich in carotenoids [143]. | NA |
| 30 | Chenopodium pallidicaule Aellen (Cañiwa, Amaranthaceae) | Mainly cultivated in Bolivia and Peru [378]. | Disease and pest resistant. Adapted to salinity, heat and drought tolerance. Rich in protein [378]. | Exceptional protein content and quality, equivalent to that of milk proteins. Balanced amino acid composition [135]. | NA |
| 31 | Adansonia digitata L. (Baobab, Malvaceae) | Tropical African countries [120]. | Adapted to arid and semi-arid conditions and rich source of vitamin A and C [120]. | Fruit pulp vitamin C is almost ten times that of oranges [121]. | Performed profiling of proteins, amino acids and minerals [121]. |
| 32 | Strychnos cocculoides Baker (Monkey orange, Loganiaceae) | America, African and South tropic Asian regions [379]. | Adapted to warm climate conditions [379]. Rich in iron, zinc and vitamin C [212]. | Essential source of iron [212]. | N/A |
| 33 | Panicum sumatrense Roth(Little millet, Poaceae) | Tropical region of Asia and Africa [368]. | Grow with minimal requirements and adapted to harsh environmental conditions and rich in micronutrients [368]. | Grains are a good source of iron and calcium [196]. | 1. Complete chloroplast genome was sequenced [380]. 2. RNa sequences were performed and differential gene expression at the time of drought and salinity stress also studied. At the time of drought stress, 241 DGEs were upregulated and 134 DGEs were downregulated [381]. |
| Domesticated Crop|Related Traditional Plant (S) | Gene | Wild Trait | Domestication Trait | Function of the Gene | Reference (s) |
|---|---|---|---|---|---|
| Fragaria vesca|Pyrus pyrifolia, Rubus fruticosus, R. spectabilis, R. occidentalis [459]. | TERMINAL FLOWER 1 Homologue KSN (TFL1) | Non-frequent flowering. | Continuous flowering. | Flowering repression. Establishment of a continuous flowering habit. | [437,460,461] |
| Hordeum vulgare|H. murinum [462], H. brachyantherum, H. jubatum [463]. | nud (nud) | Palea and lemma hulls are tightly adhered to the caryopsis which results in hulled seeds. | Reduced organ adhesion between the caryopsis and the hull. | Controls caryopsis and is involved in the lipid biosynthesis pathway. | [437,464] |
| SIX-ROWED SPIKE1 (VRS1) | Two-rowed inflorescences. | Change in inflorescence architecture from two-rowed to six-rowed spikelet. | Loss of function of Vrs1 results in the conversion of the rudimentary lateral two-rowed spikelet in barley into a fully developed six-rowed fertile spikelet. | [437,465] | |
| Photoperiod-H1 (Ppd-H1) | Early flowering. | Delayed flowering time. | Candidate gene for leaf size and flowering time in the barley population. | [437,466] | |
| RESISTANT TO RALSTONIA SOLANACEARUM 2 (RRS2) | Low leaf scald resistance. | Increased leaf scald resistance. | Resistance gene to fungal pathogen Rhynchosporium secalis which causes leaf scald disease. | [437,467] | |
| EARLY FLOWERING3 (ELF3) | Late flowering. | Earlier flowering time. | Part of a circadian clock input pathway. Can regulate the initiation of flowering independently of phyB. | [437,468] | |
| INTERMEDIUM-C (INT-C) | Tillering and sterile lateral spikelets. | Increased expression causes suppression of tillering and male fertility in lateral spikelets. | Regulation of shoot system development. Mutation of the gene is correlated with lateral spikelet fertility phenotypes. | [437,469].. | |
| Oryza sativa|O. latifolia, O. glumaepatula [470]. | PROSTRATE GROWTH1 (PROG1) | Prostrate growth. | Asymmetrical growth of the tiller base leading to erect growth. | Inactive prog1 results in the conversion of prostrate to erect growth habit in domesticated rice. | [437,471] |
| SHATTERING4-1 (SH4-1) | Easily shatters seeds. | Lack of an abscission layer leads to seed non-shattering. | Responsible for rice grain shattering. | [437,472,473] | |
| BLACK HULL4 (BH4) | Black hull. | White hull. | Controls black hull color. | [437,474] | |
| Red pericarp (Rc) | Red pericarp. | White pericarp (absence of anthocyanin). | Required for red pericarp in rice- proanthocyanin synthesis-related gene. | [437,475] | |
| AMMONIUM TRANSPORTER1;1 (AMT1;1) | Poor nitrogen uptake mechanism. | Modified nitrogen uptake and response. | It is a high affinity ammonium transporter which may be involved in ammonium uptake from the soil. | [437,476] | |
| LIGULELESS1 (LG1) | Open the panicle and easily shatter seeds. | Altered panicle growth results in closed panicles and reduced shattering. | Controls laminar joint formation between leaf blade and leaf sheath and controls ligule and auricle development. | [437,477] | |
| BETAINE ALDEHYDE DEHYDROGENASE2 (BADH2) | Non-fragrant grains. | Fragrant grains. | Plays a key role in the accumulation of a fragrant compound, 2-acetyl-1-pyrroline (2AP). An inactive BADH2 promotes fragrance in rice. | [437,478] | |
| GRAIN WIDTH5 (GW5/SW5)) | Small sized seeds. | Increase seed size by increasing the cell number of the outer glume layer. | Controls rice grain width and weight. | [437,479] | |
| GRANULE BOUND STARCH SYNTHASE I (Waxy; GBSSI) | Non-glutinous grains. | Glutinous grains. | It controls amylose synthesis in the endosperm. | [437,480,481] | |
| GRAIN SIZE3 (GS3) | Short grain. | Long grain phenotype. | Contributes to seed or grain size. | [437,482] | |
| SHATTERING1 (Sh1) | Shattering. | Reduction in shattering. | Controls shattering. | [437,472] | |
| HEADINGDATE1 (HD1) | Early flowering. | Delayed flowering time. | A regulator of the florigen gene Hd3a. | [437,483] | |
| Quantitative trait locus of seed shattering on chromosome 1 (qSH1) | Shattering seeds. | Loss of seed shattering because of the absence of an abscission layer. | Regulates seed shattering. | [472,484] | |
| Zea mays|Setaria italica, Lolium perenne, Digitaria exilis, Avena sativa, Secale cereale [485]. | teosinte glume architecture 1 (Tga1) | Hard glume. | Softer glume. | Represses branching. | [437,486,487,488,489] |
| zea agamous-like1 (Zagl1) | Small female ear. | Increase in female ear length. | Role in flowering time and ear size. | [437] | |
| ramosa1 (ra1) | Many branches with multiple ears on each branch and tassel at the tip of the branch. | Affects kernel organization, altered inflorescence architecture. | Regulate the inflorescence branching systems. | [437,490] | |
| PROLAMIN BINDING FACTOR (PBF) | Less protein storage. | Altered prolamin protein levels in seeds. | Controls the expression of seed storage protein (zein) genes. | [437] | |
| teosinte branched 1 (TB1) | Many branches with multiple ears on each branch and tassel at the tip of the branch. | Increased expression causes short, ear-tipped branches. | It is involved in apical dominance. It has a significant role in repression of axillary organs. | [437,487,489,491]. | |
| SHATTERING 1-5.1, SHATTERING1-5.2 (Sh1-5.1-Sh1-5.2) | Easily shattering. | Non-shattering phenotype because of lack of abscission layer. | It plays a key role in establishment of the abscission layer and is responsible for grain shattering. | [437,472] | |
| BARREN STALK1 (BA1) | Presence of axillary meristem. | Prevents axillary meristem development. | Modulates maize inflorescence. Regulates vegetative lateral meristem. | [437,492] | |
| CO, CO-LIKE and TIMING OF CAB1 (CCT) | Late flowering. | Lower expression leads to earlier flowering. | CO, CO-like and TIMING OF CAB1 modulate flowering time. | [437,493,494] | |
| MADS19 (zmm19) | Kernels without glume covering. | Ectopic expression in inflorescences leads to kernels covered by glumes. | Loss of the MADS19 results in larger glumes. | [437,495] | |
| SUGARY1 (Su1) | Non-sweet taste. | Altered starch biosynthesis, sugary sweet taste. | Key role in starch biosynthetic process | [437,496,497] | |
| SHATTERING1 (Sh1) | Shattering phenotype. | Non-shattering phenotype because of lack of abscission layer. | Promotes grain shattering through an abscission layer. | [437,472] | |
| Glycine max|Canavalia ensiformis, C. gladiata, Lupinus mutabilis, Cajanus cajan, Phaseolus mungo, P. vulgaris, P. aconitifolius, P. calcaratus, P. lunatus, Vigna unguiculata, Lens culinaris, Vicia faba, Lathyrus sativus, Cyamopsis tetragonolobus, Dolichos lablab, Arachis hypogaea [498,499]. | TERMINAL FLOWER1b (TFL1b) | Indeterminate shoots. | Determinate shoots end with terminal inflorescence. | Maintains indeterminate growth of cells in the shoot apical meristem. | [437] |
| Setaria italica|S. faberi, S. viridis, S. pumila, Panicum glaucum, P. miliaceum [500]. | GRANULE BOUND STARCH SYNTHASE I (GBSSI) | Non-glutinous grains. | Glutinous grains. | The gene is involved in starch biosynthesis. | [437,501,502] |
| Solanum lycopersicum|S. quitoense, S. macrocarpon, Physalis prunisa, P. minima [446,503]. | FASCIATED (FAS) | Small fruit size. | Increased cell proliferation leads to larger fruit. | Promotes cell size growth. | [437,504] |
| fruit weight 2.2 (FW2.2) | Lower number of locules. | Increase in locule number in fruit. | Regulates fruit size. | [437,505,506] | |
| OVATE (OVATE) | Non-expansive fruit neck region. | Expansion of the fruit and fruit shape determination. | Key regulator of fruit shape. | [437,507] | |
| SUN (SUN) | Fruit is not elongated. | Increased growth resulting in elongated fruit. | Major gene controlling the elongated fruit shape. | [437,508] | |
| LOCULE NUMBER (LC) | Fruits have two locules. | Fruits have 3–4 locules instead of 2 locules. | Control fruit shape. | [437,504] | |
| Vitis vinifera|Cissus discolor, C. s mollissima, Cayratia pedata, Ampelocissus latifolia [509]. | myb-related transcription factor (MYBA1) | Dark colored berry. | Lack of anthocyanins lead to white berry color. | Controls the last steps in the anthocyanins biosynthesis pathway. | [437,510] |
| myb-related transcription factor (MYBA2) | Dark colored berry. | Lack of anthocyanins lead to white berry color. | Control the anthocyanin biosynthesis pathway. | [437] |
| Sl. No. | Crop Name | Method of Gene Editing | Target Gene and Effect of Mutation after Editing | References |
|---|---|---|---|---|
| 1 | Solanum lycopersicum L. | CRISPR/Cas9 system via Agrobacterium- mediated transformation and TALEN. | Anthocyanin mutant 1 (ANT1)—resulted in deep purple colored plant tissues. | [526] |
| CRISPR/Cas9 system via Agrobacterium- mediated transformation. | Mildew resistance locus O (MLO)—powdery mildew-resistant plant. | [527] | ||
| 2 | Solanum tuberosum L. | CRISPR/Cas9 system via Agrobacterium- mediated transformation. | Acetolactate synthase1 (ALS1)—resulted in reduced herbicide susceptibility. | [528] |
| CRISPR/Cas9 system PEG mediated protoplast transfection. | Granule bound starch synthase (GBSS)—resulted in absence of amylase enzyme. | [529] | ||
| 3 | Zea mays L. | CRISPR/Cas9 system via particle bombardment transformation. | ALS1, ALS2—resulted in chlorsulfuron-resistant plants. | [513] |
| CRISPR/Cas9 system via particle bombardment transformation. | Auxin-regulated gene involved in organ size (ARGOS8)—resulted in decreased ethylene response and increased grain yield under stress conditions. | [530] | ||
| CRISPR/Cas9 system via Agrobacterium- mediated transformation. | Thermosensitive genic male-sterile 5 (TMS5)-resulted in male sterility. | [531] | ||
| TALEN via PEG-mediated transformation. | Phytoene desaturase (PDS), Inositol-pentakisphosphate 2-kinase (IPK1A), Isopentenyl phosphate kinase (IPK), Multidrug resistance-associated protein 4 (MRP4)—resulted in mutation of the genes. | [532] | ||
| CRISPR/Cas9 system via PEG-mediated transformation. | Inositol phosphate kinase (IPK)—resulted in mutation. | [532] | ||
| CRISPR/Cas9 system. | G protein β subunit (Gβ)—resulted in an autoimmune response. | [533] | ||
| CRISPR/Cas9 system. | Waxy—resulted in waxy corn hybrids. | [534] | ||
| CRISPR/Cas9 system via Agrobacterium- mediated transformation. | Gibberellin-Oxidase20-3 (GA20ox3)—resulted in semi dwarf plants. | [535] | ||
| 4 | Oryza sativa L. | CRISPR/Cas9 system via particle bombardment transformation. | ALS—resulted in herbicide resistance. | [536] |
| CRISPR/Cpf1 system via particle bombardment transformation. | Chlorophyllide-a oxygenase (COA1) -resulted in precise gene insertions and indel mutations. | [537] | ||
| CRISPR/Cas9 system via particle bombardment transformation. | Nitrate transporter 1.1 (NRT1.1B)—resulted in improved nitrogen use efficiency. | [538] | ||
| CRISPR/Cas9 system via PEG mediated transformation. | Drooping leaf (DL)—resulted in a drooping leaf phenotype. | [539] | ||
| 5 | Triticum aestivum L. | CRISPR/Cas9 system via Agrobacterium- mediated transformation. | Grain width (GASR7)—resulted in mutations. | [531] |
| CRISPR/Cas9 system via particle bombardment transformation. | Grain weight (GW)—resulted in mutation of the gene. | [540] | ||
| 6 | Malus domestica Borkh. | CRISPR/Cas9 system via PEG mediated transformation. | DIPM-1, DIPM-2 and DIPM-4—resulted in mutation of the genes.. | [516] |
| 7 | Vitis vinifera L. | CRISPR/Cas9 system via PEG mediated transformation. | (MLO-7)—Resulted inmutations of the gene. | [516] |
| 8 | Brassica oleracea L. | CRISPR/Cas9 system via PEG mediated transformation. | FRIGIDA (FRI) and phytoene desaturase (PDS)—resulted in the mutations of the genes. | [541] |
| 9 | Cucumis sativus L. | CRISPR/Cas9 system via Agrobacterium- mediated transformation. | WPP domain-interacting protein 1 (WIP1)—resulted in development of gynoecious phenotype with upper node having only female flowers. | [542] |
| CRISPR/Cas9 system via Agrobacterium- mediated transformation. | Eukaryotic translation initiation factor 4E (eIF4E)—resulted in resistance against vein yellowing virus (ipomovirus), Zucchini yellow mosaic virus and Papaya ringspot mosaic virus-W (potyviruses). | [515] | ||
| 10 | Solanum nigrum L. | CRISPR/Cas9 system via Agrobacterium-mediated transformation. | Gravity response gene (Lazy1)—resulted in downward growth of the stem. | [543] |
| 11 | Brassica rapa L. | CRISPR/Cas9 system via PEG mediated transformation. | FRI and PDS genes—resulted in the mutations of the genes. | [541] |
| 12 | Musa x paradisiaca L. | CRISPR/Cas9 system via PEG mediated transformation. | PDS—resulted in mutation of the gene. | [544] |
| 13. | Nicotiana tabacum L. | CRISPR/Cas9 system. | PDS—resulted in albino phenotype. | [545] |
| 14 | Setaria viridis (L.) P. Beauv. | CRISPR/Cas9_Trex2 system via Agrobacterium-mediated transformation. | Domains rearranged methylase (Drm1) and male sterile 45 (Ms45)— resulted in the mutations of the genes. | [546] |
| CRISPR/Cas9 system. | Less Shattering1 (Les1)—reduced shattering. | [547] | ||
| 15 | Medicago truncatula Gaertn. | CRISPR/Cas9 system via Agrobacterium- mediated transformation. | Hua enhancer1 (Hen1)—results in a shrunken, shriveled seed phenotype. | [548] |
| CRISPR/Cas9 system via Agrobacterium- mediated transformation. | PDS—resulted in albino phenotypes. | [549] | ||
| 16 | Vigna unguiculata (L.) Walp. | CRISPR/Cas9 system via Agrobacterium- mediated transformation. | Meiosis gene (SPO11-1)—infertile phenotype. | [550] |
| CRISPR/Cas9 system via Agrobacterium- mediated transformation. | Symbiosis receptor-like kinase (SYMRK)—resulted in blockage of nodule formation. | [525] | ||
| 17 | Cicer arietinum L. | CRISPR/Cas9 system via PEG mediated transformation. | 4-coumarate ligase (4CL) and Reveille 7 (RVE7) genes—resulted in mutations of the genes. | [551] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, A.; Anju, T.; Kumar, S.; Chhapekar, S.S.; Sreedharan, S.; Singh, S.; Choi, S.R.; Ramchiary, N.; Lim, Y.P. Integrating Omics and Gene Editing Tools for Rapid Improvement of Traditional Food Plants for Diversified and Sustainable Food Security. Int. J. Mol. Sci. 2021, 22, 8093. https://doi.org/10.3390/ijms22158093
Kumar A, Anju T, Kumar S, Chhapekar SS, Sreedharan S, Singh S, Choi SR, Ramchiary N, Lim YP. Integrating Omics and Gene Editing Tools for Rapid Improvement of Traditional Food Plants for Diversified and Sustainable Food Security. International Journal of Molecular Sciences. 2021; 22(15):8093. https://doi.org/10.3390/ijms22158093
Chicago/Turabian StyleKumar, Ajay, Thattantavide Anju, Sushil Kumar, Sushil Satish Chhapekar, Sajana Sreedharan, Sonam Singh, Su Ryun Choi, Nirala Ramchiary, and Yong Pyo Lim. 2021. "Integrating Omics and Gene Editing Tools for Rapid Improvement of Traditional Food Plants for Diversified and Sustainable Food Security" International Journal of Molecular Sciences 22, no. 15: 8093. https://doi.org/10.3390/ijms22158093
APA StyleKumar, A., Anju, T., Kumar, S., Chhapekar, S. S., Sreedharan, S., Singh, S., Choi, S. R., Ramchiary, N., & Lim, Y. P. (2021). Integrating Omics and Gene Editing Tools for Rapid Improvement of Traditional Food Plants for Diversified and Sustainable Food Security. International Journal of Molecular Sciences, 22(15), 8093. https://doi.org/10.3390/ijms22158093