Derivation of Mouse Parthenogenetic Advanced Stem Cells
Abstract
:1. Introduction
2. Results
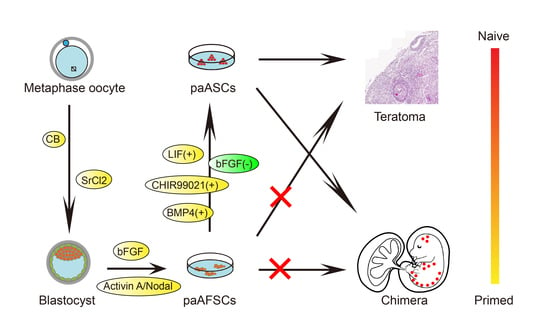
2.1. Derivation of paAFSCs
2.2. Characterization of paAFSCs
2.3. Derivation and Characterization of paASCs
2.4. The Developmental Potential of paAFSCs and paASCs
3. Discussion
4. Materials and Methods
4.1. Parthenogenetic Diploid Embryos Collection
4.2. Derivation of paAFSCs and paASCs
4.3. Alkaline Phosphatase (AP) Staining
4.4. Karyotype
4.5. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.6. Immunofluorescence
4.7. Teratomas Formation
4.8. Production of Chimaera
4.9. Cryosectioning and Immunofluorescence Staining
4.10. Bisulfite Genome Sequencing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stevens, L.C.; Varnum, D.S. The development of teratomas from parthenogenetically activated ovarian mouse eggs. Dev. Biol. 1974, 37, 369–380. [Google Scholar] [CrossRef]
- Kaufman, M.H.; Barton, S.C.; Surani, M.A.H. Normal postimplantation development of mouse parthenogenetic embryos to the forelimb bud stage. Nature 1977, 265, 53–55. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, M.H. The chromosome complement of single-pronuclear haploid mouse embryos following activation by ethanol treatment. J. Embryol. Exp. Morphol. 1982, 71, 139–154. [Google Scholar]
- Kono, T.; Obata, Y.; Wu, Q.; Niwa, K.; Ono, Y.; Yamamoto, Y.; Park, E.S.; Seo, J.S.; Ogawa, H. Birth of parthenogenetic mice that can develop to adulthood. Nature 2004, 428, 860–864. [Google Scholar] [CrossRef]
- Li, Z.; Wan, H.; Feng, G.; Wang, L.; He, Z.; Wang, Y.; Wang, X.J.; Li, W.; Zhou, Q.; Hu, B. Birth of fertile bimaternal offspring following intracytoplasmic injection of parthenogenetic haploid embryonic stem cells. Cell Res. 2016, 26, 135–138. [Google Scholar] [CrossRef] [Green Version]
- Zhong, C.; Xie, Z.; Yin, Q.; Dong, R.; Yang, S.; Wu, Y.; Yang, L.; Li, J. Parthenogenetic haploid embryonic stem cells efficiently support mouse generation by oocyte injection. Cell Res. 2016, 26, 131–134. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.T.; Choi, M.H.; Lee, E.J.; Gong, S.P.; Jang, M.; Park, S.H.; Jee, H.; Kim, D.Y.; Han, J.Y.; Lim, J.M. Establishment of autologous embryonic stem cells derived from preantral follicle culture and oocyte parthenogenesis. Fertil. Steril. 2008, 90, 1910–1920. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.K.; Wang, L.Y.; Wang, L.B.; Feng, G.H.; Yuan, X.W.; Liu, C.; Xu, K.; Li, Y.H.; Wan, H.F.; Zhang, Y.; et al. Generation of Bimaternal and Bipaternal Mice from Hypomethylated Haploid ESCs with Imprinting Region Deletions. Cell Stem Cell 2018, 23, 665–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, B.J.; Jang, H.S.; Song, H.; Park, C.; Hong, K.; Lee, J.W.; Do, J.T. Generation of Mouse Parthenogenetic Epiblast Stem Cells and Their Imprinting Patterns. Int. J. Mol. Sci. 2019, 20, 5428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, S.; Tang, F.; Li, X.; Hayashi, K.; Gillich, A.; Lao, K.; Surani, M.A. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 2009, 461, 1292–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, S.; Tang, W.W.; Wu, B.; Kim, S.; Li, J.; Li, L.; Kobayashi, T.; Lee, C.; Chen, Y.; Wei, M.; et al. Derivation of hypermethylated pluripotent embryonic stem cells with high potency. Cell Res. 2018, 28, 22–34. [Google Scholar] [CrossRef] [Green Version]
- Brons, I.G.M.; Smithers, L.E.; Trotter, M.W.; Rugg-Gunn, P.; Sun, B.; de Sousa Lopes, S.M.C.; Howlett, S.K.; Clarkson, A.; Ahrlund-Richter, L.; Pedersen, R.A.; et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007, 448, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Tesar, P.J.; Chenoweth, J.G.; Brook, F.A.; Davies, T.J.; Evans, E.P.; Mack, D.L.; Gardner, R.L.; McKay, R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007, 448, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Kline, D.; Kline, J.T. Repetitive calcium transients and the role of calcium in exocytosis and cell cycle activation in the mouse egg. Dev. Biol. 1992, 149, 80–89. [Google Scholar] [CrossRef]
- Jones, K.T.; Carroll, J.; Merriman, J.A.; Whittingham, D.G.; Kono, T. Repetitive sperm-induced Ca2+ transients in mouse oocytes are cell cycle dependent. Development 1995, 121, 3259–3266. [Google Scholar] [CrossRef]
- Liu, L.; Trimarchi, J.R.; Keefe, D.L. Haploidy but not parthenogenetic activation leads to increased incidence of apoptosis in mouse embryos. Biol. Reprod. 2002, 66, 204–210. [Google Scholar] [CrossRef] [Green Version]
- Tropepe, V.; Hitoshi, S.; Sirard, C.; Mak, T.W.; Rossant, J.; van der Kooy, D. Direct neural fate specification from embryonic stem cells: A primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron 2001, 30, 65–78. [Google Scholar] [CrossRef] [Green Version]
- Ying, Q.L.; Nichols, J.; Chambers, I.; Smith, A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 2003, 115, 281–292. [Google Scholar] [CrossRef] [Green Version]
- Ying, Q.L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.G.; Hooper, M.L. Buffalo rat liver cells produce a diffusible activity which inhibits the differentiation of murine embryonal carcinoma and embryonic stem cells. Dev. Biol. 1987, 121, 1–9. [Google Scholar] [CrossRef]
- Smith, A.G.; Heath, J.K.; Donaldson, D.D.; Wong, G.G.; Moreau, J.; Stahl, M.; Rogers, D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 1988, 336, 688–690. [Google Scholar] [CrossRef]
- Williams, R.L.; Hilton, D.J.; Pease, S.; Willson, T.A.; Stewart, C.L.; Gearing, D.P.; Wagner, E.F.; Metcalf, D.; Nicola, N.A.; Gough, N.M. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 1988, 336, 684–687. [Google Scholar] [CrossRef]
- Nichols, J.; Smith, A. Naive and primed pluripotent states. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef] [Green Version]
- Rossant, J. Mouse and human blastocyst-derived stem cells: Vive les differences. Development 2015, 142, 9–12. [Google Scholar] [CrossRef] [Green Version]
- Rossant, J.; Tam, P.P.L. New Insights into Early Human Development: Lessons for Stem Cell Derivation and Differentiation. Cell Stem Cell 2017, 20, 18–28. [Google Scholar] [CrossRef] [Green Version]
- Xing, F.; Fang, Z.; Qin, B.; Li, Y.; Hou, J.; Chen, X. Parthenogenetic embryonic stem cells derived from cryopreserved newborn mouse ovaries: A new approach to autologous stem cell therapy. Fertil. Steril. 2009, 91, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Varrault, A.; Eckardt, S.; Girard, B.; Le Digarcher, A.; Sassetti, I.; Meusnier, C.; Ripoll, C.; Badalyan, A.; Bertaso, F.; McLaughlin, K.J.; et al. Mouse Parthenogenetic Embryonic Stem Cells with Biparental-Like Expression of Imprinted Genes Generate Cortical-Like Neurons That Integrate into the Injured Adult Cerebral Cortex. Stem Cells 2018, 36, 192–205. [Google Scholar] [CrossRef] [Green Version]
- Leeb, M.; Wutz, A. Derivation of haploid embryonic stem cells from mouse embryos. Nature 2011, 479, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shuai, L.; Wan, H.; Dong, M.; Wang, M.; Sang, L.; Feng, C.; Luo, G.Z.; Li, T.; Li, X.; et al. Androgenetic haploid embryonic stem cells produce live transgenic mice. Nature 2012, 490, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Jia, Y.; Li, T.; Su, B. SOX2 Inhibition Promotes Promoter Demethylation of CDX2 to Facilitate Gastric Intestinal Metaplasia. Dig. Dis. Sci. 2017, 62, 124–132. [Google Scholar] [CrossRef]
- Wan, H.; He, Z.; Dong, M.; Gu, T.; Luo, G.Z.; Teng, F.; Xia, B.; Li, W.; Feng, C.; Li, X.; et al. Parthenogenetic haploid embryonic stem cells produce fertile mice. Cell Res. 2013, 23, 1330–1333. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.; Solter, D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 1984, 37, 179–183. [Google Scholar] [CrossRef]




| No. of MII-Stage Oocytes | No. of Parthenogenetic Blastocysts | No. of paAFSCs Cell Lines |
|---|---|---|
| 77 | 45 (58.44%) | 11 (24.44%) |
| Stage of Embryo (Collected) | No. of Transferred Embryos | No. of Collected Embryos | No. of Chimaeras |
|---|---|---|---|
| E6.5 | 65 | 24 (36.9%) | 1 (4.2%) |
| Stage of Embryo (Collected) | No. of Transferred Embryos | No. of Collected Embryos | No. of Chimaeras |
|---|---|---|---|
| E10.5 | 18 | 6 (33.3%) | 2 (33.3%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, M.; Zhang, J.; Liu, J.; Zhao, C.; Cao, S.; Yan, X.; Wu, B.; Bao, S. Derivation of Mouse Parthenogenetic Advanced Stem Cells. Int. J. Mol. Sci. 2021, 22, 8976. https://doi.org/10.3390/ijms22168976
Wei M, Zhang J, Liu J, Zhao C, Cao S, Yan X, Wu B, Bao S. Derivation of Mouse Parthenogenetic Advanced Stem Cells. International Journal of Molecular Sciences. 2021; 22(16):8976. https://doi.org/10.3390/ijms22168976
Chicago/Turabian StyleWei, Mengyi, Jindun Zhang, Jia Liu, Chaoyue Zhao, Shuo Cao, Xiaojie Yan, Baojiang Wu, and Siqin Bao. 2021. "Derivation of Mouse Parthenogenetic Advanced Stem Cells" International Journal of Molecular Sciences 22, no. 16: 8976. https://doi.org/10.3390/ijms22168976
APA StyleWei, M., Zhang, J., Liu, J., Zhao, C., Cao, S., Yan, X., Wu, B., & Bao, S. (2021). Derivation of Mouse Parthenogenetic Advanced Stem Cells. International Journal of Molecular Sciences, 22(16), 8976. https://doi.org/10.3390/ijms22168976