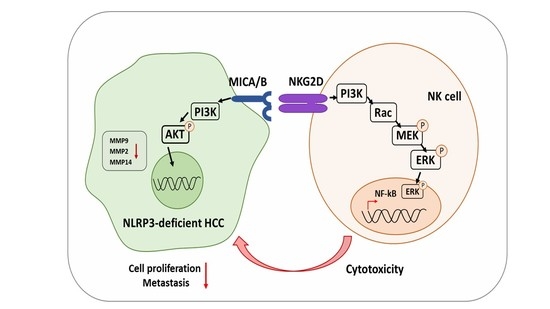
NLRP3 Deficiency in Hepatocellular Carcinoma Enhances Surveillance of NK-92 through a Modulation of MICA/B
Abstract
:1. Introduction
2. Results
2.1. High NK Cytotoxicity against Human HCC Lacking NLRP3
2.2. High Expression of NK-Activating Receptors in a Co-Culture of NLRP3 KO HCC
2.3. Increase in MICA/B Expression on the Surface of NLRP3 KO HCC
2.4. Suppression of Tumor Development and Metastases, as well as Induction of High Sensitivity to NK Cytotoxicity in Immune-Deficient Mice Developed from NLRP3 KO HCC
2.5. Increased MICA/B Expression via the Downregulation of MMP2, MMP9, and MMP14 Protein Expression in the Liver Tissue of Mice Implanted with NLRP3 KO HCC
2.6. Expression of Molecules Associated with the Epithelial-to-Mesenchymal Transition and Cancer Development in NLRP3-Lacking HCC Metastatic Liver Tissues
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture
4.2. NLRP3 Knockout in HCC Using Lentiviral CRISPR Lenti-CRISPR-NLRP3/Cas9
4.3. RT-PCR and Quantitative RT-PCR Analysis
4.4. NK Cytotoxicity to Target Cells by LDH-Release Assay
4.5. Flow Cytometry Analysis
4.6. Western Blot Analysis
4.7. In Vivo Experiment Using Xenograft Model NRG Mice
4.8. IHC Staining Analysis
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- El-Serag, H.B. Hepatocellular carcinoma. N. Engl. J. Med. 2011, 365, 1118–1127. [Google Scholar] [CrossRef]
- Marengo, A.; Rosso, C.; Bugianesi, E. Liver Cancer: Connections with Obesity, Fatty Liver, and Cirrhosis. Annu. Rev. Med. 2016, 67, 103–117. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Khemlina, G.; Ikeda, S.; Kurzrock, R. The biology of Hepatocellular carcinoma: Implications for genomic and immune therapies. Mol. Cancer 2017, 16, 149. [Google Scholar] [CrossRef]
- Zucman-Rossi, J.; Villanueva, A.; Nault, J.C.; Llovet, J.M. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015, 149, 1226–1239.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Abderrazak, A.; Syrovets, T.; Couchie, D.; El Hadri, K.; Friguet, B.; Simmet, T.; Rouis, M. NLRP3 inflammasome: From a danger signal sensor to a regulatory node of oxidative stress and inflammatory diseases. Redox Biol. 2015, 4, 296–307. [Google Scholar] [CrossRef]
- Rathinam, V.A.; Vanaja, S.K.; Fitzgerald, K.A. Regulation of inflammasome signaling. Nat. Immunol. 2012, 13, 333–342. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Hara, H.; Nunez, G. Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends Biochem. Sci. 2016, 41, 1012–1021. [Google Scholar] [CrossRef] [Green Version]
- Liston, A.; Masters, S.L. Homeostasis-altering molecular processes as mechanisms of inflammasome activation. Nat. Rev. Immunol. 2017, 17, 208–214. [Google Scholar] [CrossRef]
- Xue, Y.; Enosi Tuipulotu, D.; Tan, W.H.; Kay, C.; Man, S.M. Emerging Activators and Regulators of Inflammasomes and Pyroptosis. Trends Immunol. 2019, 40, 1035–1052. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.S. Role of inflammasomes in inflammatory autoimmune rheumatic diseases. Korean J. Physiol. Pharm. 2018, 22, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.A.; Chiang, B.L. Inflammasomes and human autoimmunity: A comprehensive review. J. Autoimmun. 2015, 61, 1–8. [Google Scholar] [CrossRef]
- Menu, P.; Vince, J.E. The NLRP3 inflammasome in health and disease: The good, the bad and the ugly. Clin. Exp. Immunol. 2011, 166, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Voet, S.; Srinivasan, S.; Lamkanfi, M.; van Loo, G. Inflammasomes in neuroinflammatory and neurodegenerative diseases. EMBO Mol. Med. 2019, 11, e10248. [Google Scholar] [CrossRef]
- Feng, Y.S.; Tan, Z.X.; Wu, L.Y.; Dong, F.; Zhang, F. The involvement of NLRP3 inflammasome in the treatment of Alzheimer’s disease. Ageing Res. Rev. 2020, 64, 101192. [Google Scholar] [CrossRef]
- Chen, Y.; Tian, Z. Innate lymphocytes: Pathogenesis and therapeutic targets of liver diseases and cancer. Cell. Mol. Immunol. 2021, 18, 57–72. [Google Scholar] [CrossRef]
- Stiglund, N.; Strand, K.; Cornillet, M.; Stål, P.; Thorell, A.; Zimmer, C.L.; Näslund, E.; Karlgren, S.; Nilsson, H.; Mellgren, G.; et al. Retained NK Cell Phenotype and Functionality in Non-alcoholic Fatty Liver Disease. Front. Immunol. 2019, 10, 1255. [Google Scholar] [CrossRef]
- Cai, L.; Zhang, Z.; Zhou, L.; Wang, H.; Fu, J.; Zhang, S.; Shi, M.; Zhang, H.; Yang, Y.; Wu, H.; et al. Functional impairment in circulating and intrahepatic NK cells and relative mechanism in hepatocellular carcinoma patients. Clin. Immunol. 2008, 129, 428–437. [Google Scholar] [CrossRef]
- Gong, J.; Fang, L.; Liu, R.; Wang, Y.; Xing, J.; Chen, Y.; Zhuang, R.; Zhang, Y.; Zhang, C.; Yang, A.; et al. UPR decreases CD226 ligand CD155 expression and sensitivity to NK cell-mediated cytotoxicity in hepatoma cells. Eur. J. Immunol. 2014, 44, 3758–3767. [Google Scholar] [CrossRef] [Green Version]
- Mantovani, S.; Oliviero, B.; Lombardi, A.; Varchetta, S.; Mele, D.; Sangiovanni, A.; Rossi, G.; Donadon, M.; Torzilli, G.; Soldani, C.; et al. Deficient Natural Killer Cell NKp30-Mediated Function and Altered NCR3 Splice Variants in Hepatocellular Carcinoma. Hepatology 2019, 69, 1165–1179. [Google Scholar] [CrossRef]
- Sun, D.; Wang, X.; Zhang, H.; Deng, L.; Zhang, Y. MMP9 mediates MICA shedding in human osteosarcomas. Cell Biol. Int. 2011, 35, 569–574. [Google Scholar] [CrossRef]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.P.; Zhang, J.J.; Liang, W.B.; Tu, M.; Lu, Z.P.; Wei, J.S.; Jiang, K.R.; Gao, W.T.; Wu, J.L.; Xu, Z.K.; et al. Elevation of MMP-9 and IDO induced by pancreatic cancer cells mediates natural killer cell dysfunction. BMC Cancer 2014, 14, 738. [Google Scholar] [CrossRef] [Green Version]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinon, F.; Burns, K.; Tschopp, J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Ghiringhelli, F.; Apetoh, L.; Tesniere, A.; Aymeric, L.; Ma, Y.; Ortiz, C.; Vermaelen, K.; Panaretakis, T.; Mignot, G.; Ullrich, E.; et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1β–dependent adaptive immunity against tumors. Nat. Med. 2009, 15, 1170–1178. [Google Scholar] [CrossRef]
- Zaki, M.H.; Boyd, K.L.; Vogel, P.; Kastan, M.B.; Lamkanfi, M.; Kanneganti, T.D. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 2010, 32, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Guo, B.; Fu, S.; Zhang, J.; Liu, B.; Li, Z. Targeting inflammasome/IL-1 pathways for cancer immunotherapy. Sci. Rep. 2016, 6, 36107. [Google Scholar] [CrossRef]
- Kantono, M.; Guo, B. Inflammasomes and Cancer: The Dynamic Role of the Inflammasome in Tumor Development. Front. Immunol. 2017, 8, 1132. [Google Scholar] [CrossRef] [Green Version]
- Wei, Q.; Mu, K.; Li, T.; Zhang, Y.; Yang, Z.; Jia, X.; Zhao, W.; Huai, W.; Guo, P.; Han, L. Deregulation of the NLRP3 inflammasome in hepatic parenchymal cells during liver cancer progression. Lab. Investig. 2014, 94, 52–62. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.E.; Lee, J.Y.; Yang, G.; Kang, H.C.; Cho, Y.-Y.; Lee, H.S.; Lee, J.Y. Inhibition of NLRP3 inflammasome in tumor microenvironment leads to suppression of metastatic potential of cancer cells. Sci. Rep. 2019, 9, 12277. [Google Scholar] [CrossRef] [Green Version]
- Mamessier, E.; Sylvain, A.; Bertucci, F.; Castellano, R.; Finetti, P.; Houvenaeghel, G.; Charaffe-Jaufret, E.; Birnbaum, D.; Moretta, A.; Olive, D. Human breast tumor cells induce self-tolerance mechanisms to avoid NKG2D-mediated and DNAM-mediated NK cell recognition. Cancer Res. 2011, 71, 6621–6632. [Google Scholar] [CrossRef] [Green Version]
- Lundholm, M.; Schröder, M.; Nagaeva, O.; Baranov, V.; Widmark, A.; Mincheva-Nilsson, L.; Wikström, P. Prostate tumor-derived exosomes down-regulate NKG2D expression on natural killer cells and CD8+ T cells: Mechanism of immune evasion. PLoS ONE 2014, 9, e108925. [Google Scholar] [CrossRef]
- Zingoni, A.; Molfetta, R.; Fionda, C.; Soriani, A.; Paolini, R.; Cippitelli, M.; Cerboni, C.; Santoni, A. NKG2D and Its Ligands: “One for All, All for One”. Front. Immunol. 2018, 9, 476. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.P.; Zhu, Y.; Zhang, J.J.; Xu, Z.K.; Qian, Z.Y.; Dai, C.C.; Jiang, K.R.; Wu, J.L.; Gao, W.T.; Li, Q.; et al. Comprehensive analysis of the percentage of surface receptors and cytotoxic granules positive natural killer cells in patients with pancreatic cancer, gastric cancer, and colorectal cancer. J. Transl. Med. 2013, 11, 262. [Google Scholar] [CrossRef] [Green Version]
- Salih, H.R.; Rammensee, H.G.; Steinle, A. Cutting edge: Down-regulation of MICA on human tumors by proteolytic shedding. J. Immunol. 2002, 169, 4098–4102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinushi, M.; Vanneman, M.; Munshi, N.C.; Tai, Y.T.; Prabhala, R.H.; Ritz, J.; Neuberg, D.; Anderson, K.C.; Carrasco, D.R.; Dranoff, G. MHC class I chain-related protein A antibodies and shedding are associated with the progression of multiple myeloma. Proc. Natl. Acad. Sci. USA 2008, 105, 1285–1290. [Google Scholar] [CrossRef] [Green Version]
- Holdenrieder, S.; Stieber, P.; Peterfi, A.; Nagel, D.; Steinle, A.; Salih, H.R. Soluble MICA in malignant diseases. Int. J. Cancer 2006, 118, 684–687. [Google Scholar] [CrossRef]
- Xing, S.; Ferrari de Andrade, L. NKG2D and MICA/B shedding: A ‘tag game’ between NK cells and malignant cells. Clin. Transl. Immunol. 2020, 9, e1230. [Google Scholar] [CrossRef]
- Yang, F.Q.; Liu, M.; Yang, F.P.; Zhang, X.L.; Yang, B.; Guo, C.C.; Huang, J.H.; Che, J.P.; Yan, Y.; Zheng, J.H. Matrix metallopeptidase 2 (MMP2) mediates MHC class I polypeptide-related sequence A (MICA) shedding in renal cell carcinoma. Actas Urol. Esp. 2014, 38, 172–178. [Google Scholar] [CrossRef]
- Liu, G.; Atteridge, C.L.; Wang, X.; Lundgren, A.D.; Wu, J.D. The membrane type matrix metalloproteinase MMP14 mediates constitutive shedding of MHC class I chain-related molecule a independent of A disintegrin and metalloproteinases. J. Immunol. 2010, 184, 3346–3350. [Google Scholar] [CrossRef] [Green Version]
- Robert, S.; Gicquel, T.; Victoni, T.; Valença, S.; Barreto, E.; Bailly-Maître, B.; Boichot, E.; Lagente, V. Involvement of matrix metalloproteinases (MMPs) and inflammasome pathway in molecular mechanisms of fibrosis. Biosci. Rep. 2016, 36, e00360. [Google Scholar] [CrossRef] [Green Version]
- Ershaid, N.; Sharon, Y.; Doron, H.; Raz, Y.; Shani, O.; Cohen, N.; Monteran, L.; Leider-Trejo, L.; Ben-Shmuel, A.; Yassin, M.; et al. NLRP3 inflammasome in fibroblasts links tissue damage with inflammation in breast cancer progression and metastasis. Nat. Commun. 2019, 10, 4375. [Google Scholar] [CrossRef] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.H.; Kim, D.; Jung, J.; Kang, H.; Cho, H. NLRP3 Deficiency in Hepatocellular Carcinoma Enhances Surveillance of NK-92 through a Modulation of MICA/B. Int. J. Mol. Sci. 2021, 22, 9285. https://doi.org/10.3390/ijms22179285
Lee HH, Kim D, Jung J, Kang H, Cho H. NLRP3 Deficiency in Hepatocellular Carcinoma Enhances Surveillance of NK-92 through a Modulation of MICA/B. International Journal of Molecular Sciences. 2021; 22(17):9285. https://doi.org/10.3390/ijms22179285
Chicago/Turabian StyleLee, Hwan Hee, Dongoh Kim, Joohee Jung, Hyojeung Kang, and Hyosun Cho. 2021. "NLRP3 Deficiency in Hepatocellular Carcinoma Enhances Surveillance of NK-92 through a Modulation of MICA/B" International Journal of Molecular Sciences 22, no. 17: 9285. https://doi.org/10.3390/ijms22179285
APA StyleLee, H. H., Kim, D., Jung, J., Kang, H., & Cho, H. (2021). NLRP3 Deficiency in Hepatocellular Carcinoma Enhances Surveillance of NK-92 through a Modulation of MICA/B. International Journal of Molecular Sciences, 22(17), 9285. https://doi.org/10.3390/ijms22179285