Sex Dimorphism of Nonalcoholic Fatty Liver Disease (NAFLD) in Pparg-Null Mice
Abstract
:1. Introduction
2. Results
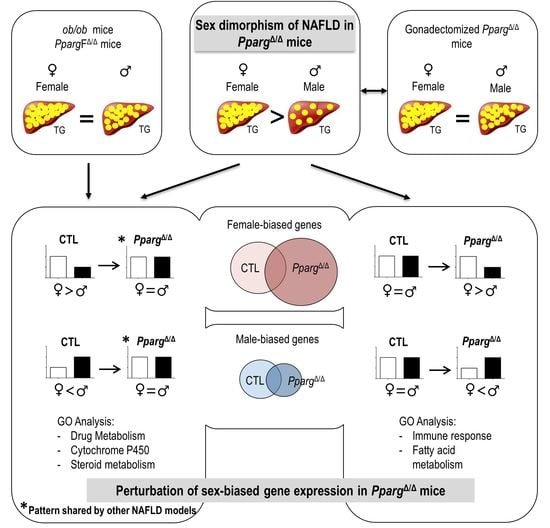
2.1. PPARγ-Null Mice Represent a New Model of NAFLD Exhibiting Sex Dimorphism
2.2. Distribution Pattern of Sex-Biased Genes in the Liver of CTL and PpargΔ/Δ Mice
2.3. Perturbation of the Physiological Sex-Biased Gene Expression by NAFLD
2.4. Modulation of Pathways Involved in Lipid Droplet Formation, Storage and Secretion in PpargΔ/Δ Mice
2.5. Analyses of Lipids and Lipid Pathways Involved in Cell Signaling and Inflammation
2.6. Relationship between Hormonal Status and Sex Dimorphism of Hepatic Lipid Accumulation
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Plasma Biochemistry
4.3. Histology and Immunohistochemistry
4.4. Gene Expression Analysis
4.5. Hepatic Lipid Content
4.6. Lipidomics Analysis
4.7. Hepatic Eicosanoids
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harrison, S.A.; Torgerson, S.; Hayashi, P.H. The natural history of nonalcoholic fatty liver disease: A clinical histopathological study. Am. J. Gastroenterol. 2003, 98, 2042–2047. [Google Scholar] [CrossRef] [PubMed]
- Day, C.P. Natural History of NAFLD: Remarkably Benign in the Absence of Cirrhosis. Gastroenterology 2005, 129, 375–378. [Google Scholar] [CrossRef]
- Farrell, G.C.; Larter, C.Z. Nonalcoholic fatty liver disease: From steatosis to cirrhosis. Hepatology 2006, 43, S99–S112. [Google Scholar] [CrossRef]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef]
- Li, Z.; Tuteja, G.; Schug, J.; Kaestner, K.H. Foxa1 and Foxa2 Are Essential for Sexual Dimorphism in Liver Cancer. Cell 2012, 148, 72–83. [Google Scholar] [CrossRef] [Green Version]
- Matsuzawa, Y.; Shimomura, I.; Nakamura, T.; Keno, Y.; Kotani, K.; Tokunaga, K. Pathophysiology and pathogenesis of visceral fat obesity. Obes Res. 1995, 3 (Suppl. 2), 187S–194S. [Google Scholar] [CrossRef] [PubMed]
- White, U.A.; Tchoukalova, Y.D. Sex dimorphism and depot differences in adipose tissue function. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 377–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beery, A.; Zucker, I. Sex bias in neuroscience and biomedical research. Neurosci. Biobehav. Rev. 2011, 35, 565–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waxman, D.J.; Holloway, M.G. Sex Differences in the Expression of Hepatic Drug Metabolizing Enzymes. Mol. Pharmacol. 2009, 76, 215–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mode, A.; Gustafsson, J.A. Sex and the liver—A journey through five decades. Drug Metab. Rev. 2006, 38, 197–207. [Google Scholar] [CrossRef]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef]
- Nadra, K.; Quignodon, L.; Sardella, C.; Joye, E.; Mucciolo, A.; Chrast, R.; Desvergne, B. PPARgamma in placental angiogenesis. Endocrinology 2010, 151, 4969–4981. [Google Scholar] [CrossRef] [PubMed]
- Gilardi, F.; Winkler, C.; Quignodon, L.; Diserens, J.G.; Toffoli, B.; Schiffrin, M.; Sardella, C.; Preitner, F.; Desvergne, B. Systemic PPARgamma deletion in mice provokes lipoatrophy, organomegaly, severe type 2 diabetes and metabolic inflexibility. Metabolism 2019, 95, 8–20. [Google Scholar] [CrossRef] [Green Version]
- Sardella, C.; Winkler, C.; Quignodon, L.; Hardman, J.A.; Toffoli, B.; Attianese, G.M.P.G.; Hundt, J.E.; Michalik, L.; Vinson, C.R.; Paus, R.; et al. Delayed Hair Follicle Morphogenesis and Hair Follicle Dystrophy in a Lipoatrophy Mouse Model of Pparg Total Deletion. J. Investig. Dermatol. 2018, 138, 500–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moitra, J.; Mason, M.M.; Olive, M.; Krylov, D.; Gavrilova, O.; Marcus-Samuels, B.; Feigenbaum, L.; Lee, E.; Aoyama, T.; Eckhaus, M.; et al. Life without white fat: A transgenic mouse. Genes Dev. 1998, 12, 3168–3181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Mullican, S.E.; DiSpirito, J.R.; Peed, L.C.; Lazar, M.A. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARgamma. Proc. Natl. Acad. Sci. USA 2013, 110, 18656–18661. [Google Scholar] [CrossRef] [Green Version]
- Conforto, T.L.; Zhang, Y.; Sherman, J.; Waxman, D.J. Impact of CUX2 on the Female Mouse Liver Transcriptome: Activation of Female-Biased Genes and Repression of Male-Biased Genes. Mol. Cell. Biol. 2012, 32, 4611–4627. [Google Scholar] [CrossRef] [Green Version]
- Abbaszade, I.G.; Clarke, T.R.; Park, C.H.; Payne, A.H. The mouse 3 beta-hydroxysteroid dehydrogenase multigene family includes two functionally distinct groups of proteins. Mol. Endocrinol. 1995, 9, 1214–1222. [Google Scholar] [CrossRef] [PubMed]
- Mattijssen, F.; Georgiadi, A.; Andasarie, T.; Szalowska, E.; Zota, A.; Krones-Herzig, A.; Heier, C.; Ratman, D.; De Bosscher, K.; Qi, L.; et al. Hypoxia-inducible Lipid Droplet-associated (HILPDA) Is a Novel Peroxisome Proliferator-activated Receptor (PPAR) Target Involved in Hepatic Triglyceride Secretion. J. Biol. Chem. 2014, 289, 19279–19293. [Google Scholar] [CrossRef] [Green Version]
- Pagadala, M.; Kasumov, T.; McCullough, A.J.; Zein, N.N.; Kirwan, J.P. Role of ceramides in nonalcoholic fatty liver disease. Trends Endocrinol. Metab. 2012, 23, 365–371. [Google Scholar] [CrossRef] [Green Version]
- Araya, J.; Rodrigo, R.; Videla, L.A.; Thielemann, L.; Orellana, M.; Pettinelli, P.; Poniachik, J. Increase in long-chain polyunsaturated fatty acid n−6/n−3 ratio in relation to hepatic steatosis in patients with non-alcoholic fatty liver disease. Clin. Sci. 2004, 106, 635–643. [Google Scholar] [CrossRef] [Green Version]
- Merino, D.M.; Johnston, H.; Clarke, S.; Roke, K.; Nielsen, D.; Badawi, A.; El-Sohemy, A.; Ma, D.; Mutch, D.M. Polymorphisms in FADS1 and FADS2 alter desaturase activity in young Caucasian and Asian adults. Mol. Genet. Metab. 2011, 103, 171–178. [Google Scholar] [CrossRef]
- Treffkorn, L.; Scheibe, R.; Maruyama, T.; Dieter, P. PGE2 exerts its effect on the LPS-induced release of TNF-alpha, ET-1, IL-1alpha, IL-6 and IL-10 via the EP2 and EP4 receptor in rat liver macrophages. Prostagland. Other Lipid. Mediat. 2004, 74, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Takeda, T.; Komiya, Y.; Koga, T.; Ishida, T.; Ishii, Y.; Kikuta, Y.; Nakaya, M.; Kurose, H.; Yokomizo, T.; Shimizu, T.; et al. Dioxin-induced increase in leukotriene B4 biosynthesis through the aryl hydrocarbon receptor and its relevance to hepatotoxicity owing to neutrophil infiltration. J. Biol. Chem. 2017, 292, 10586–10599. [Google Scholar] [CrossRef] [Green Version]
- Palmisano, B.T.; Zhu, L.; Eckel, R.H.; Stafford, J.M. Sex differences in lipid and lipoprotein metabolism. Mol. Metab. 2018, 15, 45–55. [Google Scholar] [CrossRef]
- Soares, A.F.; Paz-Montoya, J.; Lei, H.; Moniatte, M.; Gruetter, R. Sexual dimorphism in hepatic lipids is associated with the evolution of metabolic status in mice. NMR Biomed. 2017, 30, e3761. [Google Scholar] [CrossRef] [Green Version]
- Chitraju, C.; Trötzmüller, M.; Hartler, J.; Wolinski, H.; Thallinger, G.G.; Lass, A.; Zechner, R.; Zimmermann, R.; Köfeler, H.; Spener, F. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J. Lipid Res. 2012, 53, 2141–2152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, J.C.; Sasser, J.M.; Pollock, D.M.; Pollock, J.S. Sexual Dimorphism in Renal Production of Prostanoids in Spontaneously Hypertensive Rats. Hypertension 2005, 45, 406–411. [Google Scholar] [CrossRef]
- Cagen, L.M.; Baer, P.G. Effects of gonadectomy and steroid treatment on renal prostaglandin 9-ketoreductase activity in the rat. Life Sci. 1987, 40, 95–100. [Google Scholar] [CrossRef]
- Leng, S.; Winter, T.; Aukema, H.M. Dietary LA and sex effects on oxylipin profiles in rat kidney, liver, and serum differ from their effects on PUFAs. J. Lipid. Res. 2017, 58, 1702–1712. [Google Scholar] [CrossRef] [Green Version]
- Lemieux, C.; Phaneuf, D.; Labrie, F.; Giguère, V.; Richard, D.; Deshaies, Y. Estrogen receptor α-mediated adiposity-lowering and hypocholesterolemic actions of the selective estrogen receptor modulator acolbifene. Int. J. Obes. 2005, 29, 1236–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, L.K.; Jacobs, R.; Vance, D.E. Tamoxifen induces triacylglycerol accumulation in the mouse liver by activation of fatty acid synthesis. Hepatology 2010, 52, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, K.N.; Pratis, K.; Jones, M.; Simpson, E.R. Estrogen Replacement Reverses the Hepatic Steatosis Phenotype in the Male Aromatase Knockout Mouse. Endocrinology 2004, 145, 1842–1848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.-Y.; Yu, I.-C.; Wang, R.-S.; Chen, Y.-T.; Liu, N.-C.; Altuwaijri, S.; Hsu, C.-L.; Ma, W.-L.; Jokinen, J.; Sparks, J.D.; et al. Increased hepatic steatosis and insulin resistance in mice lacking hepatic androgen receptor. Hepatology 2008, 47, 1924–1935. [Google Scholar] [CrossRef] [PubMed]
- Brie, B.; Ramirez, M.C.; De Winne, C.; Vicchi, F.L.; Villarruel, L.; Sorianello, E.; Catalano, P.; Ornstein, A.M.; Becu-Villalobos, D. Brain Control of Sexually Dimorphic Liver Function and Disease: The Endocrine Connection. Cell. Mol. Neurobiol. 2019, 39, 169–180. [Google Scholar] [CrossRef]
- Gardner, C.J.; Irwin, A.J.; Daousi, C.; McFarlane, I.A.; Joseph, F.; Bell, J.D.; Thomas, E.L.; Adams, V.L.; Kemp, G.J.; Cuthbertson, D.J. Hepatic steatosis, GH deficiency and the effects of GH replacement: A Liverpool magnetic resonance spectroscopy study. Eur. J. Endocrinol. 2012, 166, 993–1002. [Google Scholar] [CrossRef] [Green Version]
- Laron, Z.; Ginsberg, S.; Webb, M. Nonalcoholic fatty liver in patients with Laron syndrome and GH gene deletion—Preliminary report. Growth Horm. IGF Res. 2008, 18, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Cordoba-Chacon, J.; Majumdar, N.; List, E.O.; Diaz-Ruiz, A.; Frank, S.J.; Manzano, A.; Bartrons, R.; Puchowicz, M.; Kopchick, J.J.; Kineman, R.D. Growth Hormone Inhibits Hepatic De Novo Lipogenesis in Adult Mice. Diabetes 2015, 64, 3093–3103. [Google Scholar] [CrossRef] [Green Version]
- Sarmento-Cabral, A.; Del Rio-Moreno, M.; Vazquez-Borrego, M.C.; Mahmood, M.; Gutierrez-Casado, E.; Pelke, N.; Guzman, G.; Subbaiah, P.V.; Cordoba-Chacon, J.; Yakar, S.; et al. GH directly inhibits steatosis and liver injury in a sex-dependent and IGF1-independent manner. J. Endocrinol. 2021, 248, 31–44. [Google Scholar] [CrossRef]
- Choi, H.K.; Waxman, D.J. Plasma Growth Hormone Pulse Activation of Hepatic JAK-STAT5 Signaling: Developmental Regulation and Role in Male-Specific Liver Gene Expression. Endocrinology 2000, 141, 3245–3255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Laz, E.V.; Waxman, D.J. Dynamic, Sex-Differential STAT5 and BCL6 Binding to Sex-Biased, Growth Hormone-Regulated Genes in Adult Mouse Liver. Mol. Cell. Biol. 2012, 32, 880–896. [Google Scholar] [CrossRef] [Green Version]
- Oshida, K.; Waxman, D.J.; Corton, J.C. Chemical and Hormonal Effects on STAT5b-Dependent Sexual Dimorphism of the Liver Transcriptome. PLoS ONE 2016, 11, e0150284. [Google Scholar]
- Smati, S.; Polizzi, A.; Fougerat, A.; Ellero-Simatos, S.; Blum, Y.; Lippi, Y.; Régnier, M.; Laroyenne, A.; Huillet, M.; Arif, M.; et al. Integrative study of diet-induced mouse models of NAFLD identifies PPARα as a sexually dimorphic drug target. Gut 2021. [Google Scholar] [CrossRef]
- Bruce, S.J.; Rey, F.; Béguin, A.; Berthod, C.; Werner, D.; Henry, H. Discrepancy between radioimmunoassay and high performance liquid chromatography tandem-mass spectrometry for the analysis of androstenedione. Anal. Biochem. 2014, 455, 20–25. [Google Scholar] [CrossRef]
- Parini, P.; Johansson, L.; Broijersen, A.; Angelin, B.; Rudling, M. Lipoprotein profiles in plasma and interstitial fluid analyzed with an automated gel-filtration system. Eur. J. Clin. Investig. 2006, 36, 98–104. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fauland, A.; Köfeler, H.; Trötzmüller, M.; Knopf, A.; Hartler, J.; Eberl, A.; Chitraju, C.; Lankmayr, E.; Spener, F. A comprehensive method for lipid profiling by liquid chromatography-ion cyclotron resonance mass spectrometry. J. Lipid Res. 2011, 52, 2314–2322. [Google Scholar] [CrossRef] [Green Version]
- Hartler, J.; Trötzmüller, M.; Chitraju, C.; Spener, F.; Köfeler, H.; Thallinger, G.G. Lipid Data Analyzer: Unattended identification and quantitation of lipids in LC-MS data. Bioinformatics 2011, 27, 572–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebisch, G.; Vizcaino, J.A.; Köfeler, H.; Trötzmüller, M.; Griffiths, W.; Schmitz, G.; Spener, F.; Wakelam, M. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Faouder, P.; Baillif, V.; Spreadbury, I.; Motta, J.-P.; Rousset, P.; Chêne, G.; Guigné, C.; Tercé, F.; Vanner, S.; Vergnolle, N.; et al. LC–MS/MS method for rapid and concomitant quantification of pro-inflammatory and pro-resolving polyunsaturated fatty acid metabolites. J. Chromatogr. B 2013, 932, 123–133. [Google Scholar] [CrossRef] [PubMed]






| Group | Gene GO Term | No. of Genes | p Value |
|---|---|---|---|
| I (208 genes) Female-biased genes in CTL mice | Microsome | 16 | 2.24 × 10−9 |
| Drug Metabolism | 11 | 1.91 × 10−8 | |
| Cytochrome P450 | 9 | 2.58 × 10−6 | |
| Arachidonic acid metabolism | 7 | 5.16 × 10−4 | |
| Endoplasmic reticulum | 19 | 6.23 × 10−4 | |
| Linoleic acid metabolism | 4 | 0.01 | |
| II (145 genes) Male-biased genes in CTL mice | ncRNA metabolic process | 7 | 3.22 × 10−3 |
| Tubulin | 4 | 3.1 × 10−4 | |
| Steroid dehydrogenase activity | 4 | 6.18 × 10−4 | |
| Metal ion binding | 38 | 0.04 | |
| Nucleolus | 8 | 5.78 × 10−3 | |
| III (292 genes) Female-biased genes in γΔ/Δ mice | Cell surface | 24 | 1.02 × 10−9 |
| Immune response | 11 | 4.06 × 10−9 | |
| Immunoglobulin domain | 28 | 4.59 × 10−9 | |
| Defense response | 26 | 1.37 × 10−8 | |
| Neutrophil-mediated immunity | 3 | 4.43 × 10−3 | |
| Fatty acid biosynthesis | 5 | 5.2 × 10−3 | |
| IV (115 genes) Male-biased genes in in γΔ/Δ mice | Cell fraction | 10 | 9.87 × 10−3 |
| Cell cycle process | 8 | 5.46 × 10−3 | |
| Cellular response to stress | 9 | 1.54 × 10−3 | |
| Oxidation reduction | 10 | 0.01 | |
| Mitochondrion | 10 | 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schiffrin, M.; Winkler, C.; Quignodon, L.; Naldi, A.; Trötzmüller, M.; Köfeler, H.; Henry, H.; Parini, P.; Desvergne, B.; Gilardi, F. Sex Dimorphism of Nonalcoholic Fatty Liver Disease (NAFLD) in Pparg-Null Mice. Int. J. Mol. Sci. 2021, 22, 9969. https://doi.org/10.3390/ijms22189969
Schiffrin M, Winkler C, Quignodon L, Naldi A, Trötzmüller M, Köfeler H, Henry H, Parini P, Desvergne B, Gilardi F. Sex Dimorphism of Nonalcoholic Fatty Liver Disease (NAFLD) in Pparg-Null Mice. International Journal of Molecular Sciences. 2021; 22(18):9969. https://doi.org/10.3390/ijms22189969
Chicago/Turabian StyleSchiffrin, Mariano, Carine Winkler, Laure Quignodon, Aurélien Naldi, Martin Trötzmüller, Harald Köfeler, Hugues Henry, Paolo Parini, Béatrice Desvergne, and Federica Gilardi. 2021. "Sex Dimorphism of Nonalcoholic Fatty Liver Disease (NAFLD) in Pparg-Null Mice" International Journal of Molecular Sciences 22, no. 18: 9969. https://doi.org/10.3390/ijms22189969
APA StyleSchiffrin, M., Winkler, C., Quignodon, L., Naldi, A., Trötzmüller, M., Köfeler, H., Henry, H., Parini, P., Desvergne, B., & Gilardi, F. (2021). Sex Dimorphism of Nonalcoholic Fatty Liver Disease (NAFLD) in Pparg-Null Mice. International Journal of Molecular Sciences, 22(18), 9969. https://doi.org/10.3390/ijms22189969