Directed Evolution of the Methanosarcina barkeri Pyrrolysyl tRNA/aminoacyl tRNA Synthetase Pair for Rapid Evaluation of Sense Codon Reassignment Potential
Abstract
:1. Introduction
2. Results and Discussion
2.1. Directed Evolution of M. barkeri Pyl aaRS Variants that Amionoacylate Tyrosine
2.1.1. Design and Preparation of a Library of M. barkeri Pyl aaRS Amino Acid-Binding Pocket Variants
2.1.2. High-Throughput Screening of the Targeted aaRS Library and Amplification of Apparent Tyrosine-Charging aaRS Variants
2.1.3. Characterization of M. barkeri Pyl Aminoacyl tRNA Synthetase Variants Confirms Incorporation of Tyrosine in Response to the Amber Stop Codon
2.2. System Modifications Improve the Amber Reassignment Efficiency of the Evolved aaRS
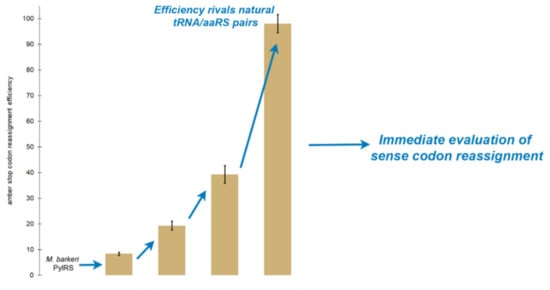
2.3. Random Mutagenesis Further Improves the Efficiency of Tyrosine Incorporation by the Evolved aaRS
2.4. Mass Spectrometry Confirms Incorporation of Tyrosine by the Evolved Aminoacyl tRNA Synthetases
2.5. Application of Evolved Tyrosine-Charging aaRS Variants for Rapid Evaluation of the Efficiency of Sense Codon Reassignment
2.5.1. The Tyrosine-Incorporating M. barkeri Pyl tRNA/aaRS Pairs Reassign Sense Codons Simply by Altering the Anticodon of the tRNA to Watson–Crick Base Pair with the Targeted Codon
2.5.2. Divergent Efficiencies of Sense Codon Reassignment by the M. barkeri and M. jannaschii Orthogonal Pairs Suggest Broad Evaluation of Sense Codon Reassignment by the M. barkeri tRNA/aaRS Pair will be Informative
2.5.3. The Tyrosine-Incorporating M. barkeri Pyl tRNA/aaRS Pairs Discriminate between Targeted and Non-Targeted Codons
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| aaRS | aminoacyl tRNA synthetase |
| EP-PCR | error-prone polymerase chain reaction |
| ESI-MS | electrospray ionization mass spectrometry |
| FACS | fluorescence-activated cell sorting |
| GFP | green fluorescent protein |
| IPTG | Isopropyl-beta-D-thiogalactoside |
| ncAA | non-canonical amino acid |
| PCR | polymerase chain reaction |
References
- Dumas, A.; Lercher, L.; Spicer, C.D.; Davis, B.G. Designing Logical Codon Reassignment—Expanding the Chemistry in Biology. Chem. Sci. 2015, 6, 50–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lajoie, M.J.; Söll, D.; Church, G.M. Overcoming Challenges in Engineering the Genetic Code. J. Mol. Biol. 2016, 428, 1004–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.C.; Schultz, P.G. Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngo, J.T.; Tirrell, D.A. Noncanonical Amino Acids in the Interrogation of Cellular Protein Synthesis. Acc. Chem. Res. 2011, 44, 677–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furter, R. Expansion of the Genetic Code: Site-Directed Pp-fluoro-phenylalanine Incorporation in Escherichia coli. Protein Sci. 1998, 7, 419–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Brock, A.; Herberich, B.; Schultz, P.G. Expanding the Genetic Code of Escherichia coli. Science 2001, 292, 498–500. [Google Scholar] [CrossRef] [Green Version]
- Melnikov, S.V.; Söll, D. Aminoacyl-tRNA Synthetases and tRNAs for an Expanded Genetic Code: What Makes them Orthogonal? Int. J. Mol. Sci. 2019, 20, 1929. [Google Scholar] [CrossRef] [Green Version]
- Polycarpo, C.R.; Herring, S.; Berube, A.; Wood, J.L.; Söll, D.; Ambrogelly, A. Pyrrolysine Analogues as Substrates for Pyrrolysyl-tRNA Synthetase. FEBS Lett. 2006, 580, 6695–6700. [Google Scholar] [CrossRef] [Green Version]
- Wan, W.; Tharp, J.M.; Liu, W.R. Pyrrolysyl-tRNA Synthetase: An Ordinary Enzyme but an Outstanding Genetic Code Expansion Tool. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 1059–1070. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.B.; Xu, J.; Shen, Z.; Takimoto, J.K.; Schultz, M.D.; Schmitz, R.J.; Xiang, Z.; Ecker, J.R.; Briggs, S.P.; Wang, L. RF1 Knockout Allows Ribosomal Incorporation of Unnatural Amino Acids at Multiple Sites. Nat. Chem. Biol. 2011, 7, 779–786. [Google Scholar] [CrossRef] [Green Version]
- Lajoie, M.J.; Rovner, A.J.; Goodman, D.B.; Aerni, H.-R.; Haimovich, A.D.; Kuznetsov, G.; Mercer, J.A.; Wang, H.H.; Carr, P.A.; Mosberg, J.A.; et al. Genomically Recoded Organisms Expand Biological Functions. Science 2013, 342, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukai, T.; Hayashi, A.; Iraha, F.; Sato, A.; Ohtake, K.; Yokoyama, S.; Sakamoto, K. Codon Reassignment in the Escherichia coli Genetic Code. Nucleic Acids Res. 2010, 38, 8188–8195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohlke, N.; Budisa, N. Sense Codon Emancipation for Proteome-Wide Incorporation of Noncanonical Amino Acids: Rare Isoleucine Codon AUA as a Target for Genetic Code Expansion. FEMS Microbiol. Lett. 2014, 351, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, I.; Kirshenbaum, K.; Tirrell, D.A. Breaking the Degeneracy of the Genetic Code. J. Am. Chem. Soc. 2003, 125, 7512–7513. [Google Scholar] [CrossRef] [Green Version]
- Ho, J.M.; Reynolds, N.M.; Rivera, K.; Connolly, M.; Guo, L.-T.; Ling, J.; Pappin, D.J.; Church, G.M.; Söll, D. Efficient Reassignment of a Frequent Serine Codon in Wild-Type Escherichia coli. ACS Synth. Biol. 2016, 5, 163–171. [Google Scholar] [CrossRef] [Green Version]
- Mukai, T.; Yamaguchi, A.; Ohtake, K.; Takahashi, M.; Hayashi, A.; Iraha, F.; Kira, S.; Yanagisawa, T.; Yokoyama, S.; Hoshi, H.; et al. Reassignment of a Rare Sense Codon to a Non-Canonical Amino Acid in Escherichia coli. Nucleic Acids Res. 2015, 43, 8111–8122. [Google Scholar] [CrossRef] [Green Version]
- Biddle, C.; Schmitt, M.A.; Fisk, J.D. Evaluating Sense Codon Reassignment with a Simple Fluorescence Screen. Biochemistry 2015, 54, 7355–7364. [Google Scholar] [CrossRef]
- Lee, B.S.; Shin, S.; Jeon, J.Y.; Jang, K.-S.; Lee, B.Y.; Choi, S.; Yoo, T.H. Incorporation of Unnatural Amino Acids in Response to the AGG codon. ACS Chem. Biol. 2015, 10, 1648–1653. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, W.W.; Liu, W.R. Towards Reassigning the Rare AGG Codon in Escherichia coli. ChemBioChem 2014, 15, 1750–1754. [Google Scholar] [CrossRef] [Green Version]
- Bacher, J.M.; Bull, J.J.; Ellington, A.D. Evolution of Phage with Chemically Ambiguous Proteomes. BMC Evol. Biol. 2003, 3, 24. [Google Scholar] [CrossRef] [Green Version]
- Thyer, R.; Ellington, A.D. The Role of tRNA in Establishing New Genetic Codes. Biochemistry 2019, 58, 1460–1463. [Google Scholar] [CrossRef] [PubMed]
- Neumann, H.; Slusarczyk, A.L.; Chin, J.W. De Novo Generation of Mutually Orthogonal Aminoacyl-tRNA Synthetase/tRNA Pairs. J. Am. Chem. Soc. 2010, 132, 2142–2144. [Google Scholar] [CrossRef]
- Reynolds, N.M.; Vargas-Rodriguez, O.; Söll, D.; Crnković, A. The Central Role of tRNA in Genetic Code Expansion. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 3001–3008. [Google Scholar] [CrossRef] [PubMed]
- Neumann, H.; Wang, K.H.; Davis, L.; Garcia-Alai, M.; Chin, J.W. Encoding Multiple Unnatural Amino Acids via Evolution of a Quadruplet-Decoding Ribosome. Nature 2010, 464, 441–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, A.; Sun, S.B.; Furman, J.L.; Xiao, H.; Schultz, P.G. A Versatile Platform for Single- and Multiple-Unnatural Amino Acid Mutagenesis in Escherichia coli. Biochemistry 2013, 52, 1828–1837. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Wang, Z.; Huang, Y.; Liu, W.R. Catalyst-Free and Site-Specific One-Pot Dual-Labeling of a Protein Directed by Two Genetically Incorporated Noncanonical Amino Acids. ChemBioChem 2012, 13, 1405–1408. [Google Scholar] [CrossRef] [Green Version]
- Fluitt, A.; Pienaar, E.; Vijoen, H. Ribosome Kinetics and aa-tRNA Competition Determine Rate and Fidelity of Peptide Synthesis. Comput. Biol. Chem. 2007, 31, 335–346. [Google Scholar] [CrossRef] [Green Version]
- Rodnina, M.V.; Wintermeyer, W. Fidelity of Aminoacyl-tRNA Selection on the Ribosome: Kinetic and Structural Mechanisms. Annu. Rev. Biochem. 2001, 70, 415–435. [Google Scholar] [CrossRef]
- Moore, P.B. How Should We Think about the Ribosome? Annu. Rev. Biophys. 2012, 41, 1–19. [Google Scholar] [CrossRef]
- Hsu, S.T.D.; Blaser, G.; Jackson, S.E. The Folding, Stability and Conformational Dynamics of Beta-Barrel Fluorescent Proteins. Chem. Soc. Rev. 2009, 38, 2951–2965. [Google Scholar] [CrossRef]
- Schmitt, M.A.; Biddle, W.; Fisk, J.D. Mapping the Plasticity of the Escherichia coli Genetic Code with Orthogonal Pair-Directed Sense Codon Reassignment. Biochemistry 2018, 57, 2762–2774. [Google Scholar] [CrossRef] [PubMed]
- Schwark, D.G.; Schmitt, M.A.; Biddle, W.; Fisk, J.D. The Influence of Competing tRNA Abundance on Translation: Quantifying the Efficiency of Sense Codon Reassignment at Rarely Used Codons. ChemBioChem 2020, 21, 2274–2286. [Google Scholar] [CrossRef] [PubMed]
- Biddle, W.; Schmitt, M.A.; Fisk, J.D. Modification of Orthogonal tRNAs: Unexpected Consequences for Sense Codon Reassignment. Nucleic Acids Res. 2016, 44, 10042–10050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elf, J.; Nilsson, D.; Tenson, T.; Ehrenberg, M. Selective Charging of tRNA Isoacceptors Explains Patterns of Codon Usage. Science 2003, 300, 1718–1722. [Google Scholar] [CrossRef] [PubMed]
- Avcilar-Kucukgoze, I.; Bartholomaus, A.; Varela, J.A.C.; Kaml, R.F.X.; Neubauer, P.; Budisa, N.; Ignatova, Z. Discharging tRNAs: A Tug of War between Translation and Detoxification in Escherichia coli. Nucleic Acids Res. 2016, 44, 8324–8334. [Google Scholar] [CrossRef] [Green Version]
- Fechter, P.; Rudinger-Thirion, J.; Tukalo, M.; Giege, R. Major Tyrosine Identity Determinants in Methanococcus jannaschii and Saccharomyces cerevisiae tRNA(Tyr) Conserved but Expressed Differently. Eur. J. Biochem. 2001, 268, 761–767. [Google Scholar] [CrossRef]
- Giege, R.; Sissler, M.; Florentz, C. Universal Rules and Idiosyncratic Features in tRNA Identity. Nucleic Acids Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [Green Version]
- Ambrogelly, A.; Gundllapalli, S.; Herring, S.; Polycarpo, C.; Frauer, C.; Söll, D. Pyrrolysine is Not Hardwired for Cotranslational Insertion at UAG Codons. Proc. Natl. Acad. Sci. USA 2007, 104, 3141–3146. [Google Scholar] [CrossRef] [Green Version]
- Krzycki, J.A. The Direct Genetic Encoding of Pyrrolysine. Curr. Opin. Microbiol. 2005, 8, 706–712. [Google Scholar] [CrossRef]
- Borrel, G.; Gaci, N.; Peyret, P.; O’Toole, P.W.; Gribaldo, S.; Brugere, J.F. Unique Characteristics of the Pyrrolysine System in the 7th Order of Methanogens: Implications for the Evolution of a Genetic Code Expansion Cassette. Archaea 2014, 2014, 374146. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, K.; O’Donoghue, P.; Gundllapalli, S.; Araiso, Y.; Ishitani, R.; Umehara, T.; Söll, D.; Nureki, O. Pyrrolysyl-tRNA Synthetase-tRNA(Pyl) Structure Reveals the Molecular Basis of Orthogonality. Nature 2009, 457, 1163–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanagisawa, T.; Ishii, R.; Fukunaga, R.; Kobayashi, T.; Sakamoto, K.; Yokoyama, S. Crystallographic Studies on Multiple Conformational States of Active-Site Loops in Pyrrolysyl-tRNA Synthetase. J. Mol. Biol. 2008, 378, 634–652. [Google Scholar] [CrossRef]
- Guo, L.-T.; Wang, Y.-S.; Nakamura, A.; Eiler, D.; Kavran, J.M.; Wong, M.; Kiessling, L.L.; Steitz, T.A.; O’Donoghue, P.; Söll, D. Polyspecific Pyrrolysyl-tRNA Synthetases from Directed Evolution. Proc. Natl. Acad. Sci. USA 2014, 111, 16724–16729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavran, J.M.; Gundllapalli, S.; O’Donoghue, P.; Englert, M.; Söll, D.; Steitz, T.A. Structure of Pyrrolysyl-tRNA Synthetase, an Archaeal Enzyme for Genetic Code Innovation. Proc. Natl. Acad. Sci. USA 2007, 104, 11268–11273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takimoto, J.K.; Dellas, N.; Noel, J.P.; Wang, L. Stereochemical Basis for Engineered Pyrrolysyl-tRNA Synthetase and the Efficient in vivo Incorporation of Structurally Divergent Non-Native Amino Acids. ACS Chem. Biol. 2011, 6, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, T.; Kuratani, M.; Seki, E.; Hino, N.; Sakamoto, K.; Yokoyama, S. Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase. Cell Chem. Biol. 2019, 26, 936–949. [Google Scholar] [CrossRef]
- Sakamoto, K.; Hayashi, A. Synthetic Tyrosine tRNA Molecules with Noncanonical Secondary Structures. Int. J. Mol. Sci. 2018, 20, 92. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Miller, C.; Guo, L.-T.; Ho, J.M.L.; Bryson, D.I.; Wang, Y.-S.; Liu, D.R.; Söll, D. Crystal Structures Reveal an Elusive Functional Domain of Pyrrolysyl-tRNA Synthetase. Nat. Chem. Biol. 2017, 13, 1261–1266. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.S.; Fang, X.Q.; Wallace, A.L.; Wu, B.; Liu, W.S.R. A Rationally Designed Pyrrolysyl-tRNA Synthetase Mutant with a Broad Substrate Spectrum. J. Am. Chem. Soc. 2012, 134, 2950–2953. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.S.; Russell, W.K.; Wang, Z.Y.; Wan, W.; Dodd, L.E.; Pai, P.J.; Russell, D.H.; Liu, W.S.R. The de novo Engineering of Pyrrolysyl-tRNA Synthetase for Genetic Incorporation of L-phenylalanine and Its Derivatives. Mol. Bio. Syst. 2011, 7, 714–717. [Google Scholar] [CrossRef]
- Ko, J.H.; Wang, Y.S.; Nakamura, A.; Guo, L.T.; Söll, D.; Umehara, T. Pyrrolysyl-tRNA Synthetase Variants Reveal Ancestral Aminoacylation Function. FEBS Lett. 2013, 587, 3243–3248. [Google Scholar] [CrossRef] [Green Version]
- Lacey, V.K.; Louie, G.V.; Noel, J.P.; Wang, L. Expanding the Library and Substrate Diversity of the Pyrrolysyl-tRNA Synthetase to Incorporate Unnatural Amino Acids Containing Conjugated Rings. ChemBioChem 2013, 14, 2100–2105. [Google Scholar] [CrossRef] [Green Version]
- Kunkel, T.A. Rapid and Efficient Site-Specific Mutagenesis without Phenotypic Selection. Proc. Natl. Acad. Sci. USA 1985, 82, 488–492. [Google Scholar] [CrossRef] [Green Version]
- Sidhu, S.S.; Weiss, G.A. Constructing Phage Display Libraries by Oligonucleotide-Directed Mutagenesis. In Phage Display a Practical Approach; Clackson, T., Lowman, H.B., Eds.; Oxford University Press: Oxford, UK, 2004; Volume 266, pp. 27–41. [Google Scholar]
- Neumann, H.; Peak-Chew, S.Y.; Chin, J.W. Genetically Encoding N-Epsilon-Acetyllysine in Recombinant Proteins. Nat. Chem. Biol. 2008, 4, 232–234. [Google Scholar] [CrossRef]
- Santoro, S.W.; Wang, L.; Herberich, B.; King, D.S.; Schultz, P.G. An Efficient System for the Evolution of Aminoacyl-tRNA Synthetase Specificity. Nat. Biotechnol. 2002, 20, 1044–1048. [Google Scholar] [CrossRef]
- Link, A.J.; Vink, M.K.S.; Agard, N.J.; Prescher, J.A.; Bertozzi, C.R.; Tirrell, D.A. Discovery of Aminoacyl-tRNA Synthetase Activity Through Cell-Surface Display of Noncanonical Amino Acids. Proc. Natl. Acad. Sci. USA 2006, 103, 10180–10185. [Google Scholar] [CrossRef] [Green Version]
- Owens, A.E.; Grasso, K.T.; Ziegler, C.A.; Fasan, R. Two-Tier Screening Platform for Directed Evolution of Aminoacyl-tRNA Synthetases with Enhanced Stop Codon Suppression Efficiency. ChemBioChem 2017, 18, 1109–1116. [Google Scholar] [CrossRef]
- Lin, A.E.; Lin, Q. Rapid Identification of Functional Pyrrolysyl-tRNA Synthetases via Fluorescence-Activated Cell Sorting. Int. J. Mol. Sci. 2019, 20, 29. [Google Scholar] [CrossRef] [Green Version]
- Kwok, H.S.; Vargas-Rodriguez, O.; Melnikov, S.V.; Söll, D. Engineered Aminoacyl-tRNA Synthetases with Improved Selectivity toward Noncanonical Amino Acids. ACS Chem. Biol. 2019, 14, 603–612. [Google Scholar] [CrossRef]
- Pott, M.; Schmidt, M.J.; Summerer, D. Evolved Sequence Contexts for Highly Efficient Amber Suppression with Noncanonical Amino Acids. ACS Chem. Biol. 2014, 9, 2815–2822. [Google Scholar] [CrossRef]
- Potts, K.A.; Stieglitz, J.T.; Lei, M.; Van Deventer, J.A. Reporter System Architecture Affects Measurements of Noncanonical Amino Acid Incorporation Efficiency and Fidelity. Mol. Syst. Des. Eng. 2020, 5, 573–588. [Google Scholar]
- Normanly, J.; Kleina, L.G.; Masson, J.M.; Abelson, J.; Miller, J.H. Construction of Escherichia-coli Amber Suppressor Transfer-RNA Genes. 3. Determination of Transfer-RNA Specificity. J. Mol. Biol. 1990, 213, 719–726. [Google Scholar] [CrossRef]
- Hao, B.; Gong, W.; Ferguson, T.K.; James, C.M.; Krzycki, J.A.; Chan, M.K. A New UAG-Encoded Residue in the Structure of a Methanogen Methyltransferase. Science 2002, 296, 1462–1466. [Google Scholar] [CrossRef]
- Young, T.S.; Ahmad, I.; Yin, J.A.; Schultz, P.G. An Enhanced System for Unnatural Amino Acid Mutagenesis in E. coli. J. Mol. Biol. 2010, 395, 361–374. [Google Scholar] [CrossRef]
- Liu, D.R.; Schultz, P.G. Progress toward the Evolution of an Organism with an Expanded Genetic Code. Proc. Natl. Acad. Sci. USA 1999, 96, 4780–4785. [Google Scholar] [CrossRef] [Green Version]
- Eggertsson, G.; Söll, D. Transfer Ribonucleic Acid-Mediated Suppression of Termination Codons in Escherichia-coli. Microbiol. Rev. 1988, 52, 354–374. [Google Scholar] [CrossRef]
- Fan, C.; Xiong, H.; Reynolds, N.M.; Söll, D. Rationally Evolving tRNA(Pyl) for Efficient Incorporation of Noncanonical Amino Acids. Nucleic Acids Res. 2015, 43, e156. [Google Scholar] [CrossRef] [Green Version]
- LaRiviere, F.J.; Wolfson, A.D.; Uhlenbeck, O.C. Uniform Binding of Aminoacyl-tRNAs to Elongation Factor Tu by Thermodynamic Compensation. Science 2001, 294, 165–168. [Google Scholar] [CrossRef] [Green Version]
- Olejniczak, M.; Dale, T.; Fahlman, R.P.; Uhlenbeck, O.C. Idiosyncratic Tuning of tRNAs to Achieve Uniform Ribosome Binding. Nat. Struct. Mol. Biol. 2005, 12, 788–793. [Google Scholar] [CrossRef]
- Mittelstaet, J.; Konevega, A.; Rodnina, M.V. A Kinetic Safety Gate Controlling the Delivery of Unnatural Amino Acids to the Ribosome. J. Am. Chem. Soc. 2013, 135, 17031–17038. [Google Scholar] [CrossRef] [Green Version]
- Dale, T.; Sanderson, L.E.; Uhlenbeck, O.C. The Affinity of Elongation Factor Tu for an Aminoacyl-tRNA is Modulated by the Esterified Amino Acid. Biochemistry 2004, 43, 6159–6166. [Google Scholar] [CrossRef]
- Hamano-Takaku, F.; Iwama, T.; Saito-Yano, S.; Takaku, K.; Monden, Y.; Kitabatake, M.; Söll, D.; Nishimura, S. A Mutant Escherichia coli Tyrosyl-tRNA Synthetase Utilizes the Unnatural Amino Acid Azatyrosine More Efficiently than Tyro-Sine. J. Biol. Chem. 2000, 275, 40324–40328. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.W.; Smith, B.A.C.; Wang, L.; Brock, A.; Cho, C.; Schultz, P.G. A New Strategy for the Site-Specific Modification of Proteins in vivo. Biochemistry 2003, 42, 6735–6746. [Google Scholar] [CrossRef]
- Ibba, M.; Söll, D. Aminoacyl-tRNA Synthesis. Annu. Rev. Biochem. 2000, 69, 617–650. [Google Scholar] [CrossRef]
- Miyake-Stoner, S.J.; Refakis, C.A.; Hammill, J.T.; Lusic, H.; Hazen, J.L.; Deiters, A.; Mehl, R.A. Generating Permissive Site-Specific Unnatural Aminoacyl-tRNA Synthetases. Biochemistry 2010, 49, 1667–1677. [Google Scholar] [CrossRef]
- Braisted, A.C.; Wells, J.A. Minimizing a Binding Domain from Protein A. Proc. Natl. Acad. Sci. USA 1996, 93, 5688–5692. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Wang, L.; Brock, A.; Wong, C.-H.; Schultz, P.G. A Method for the Generation of Glycoprotein Mimetics. J. Am. Chem. Soc. 2003, 125, 1702–1703. [Google Scholar] [CrossRef]
- Apostol, I.; Aitken, J.; Levine, J.; Lippincott, J.; Davidson, J.S.; Abbottbrown, D. Recombinant Protein Sequences Can Trigger Methylation of N-Terminal Amino-Acids in Escherichia-coli. Protein Sci. 1995, 4, 2616–2618. [Google Scholar] [CrossRef] [Green Version]
- Krishnakumar, R.; Prat, L.; Aerni, H.R.; Ling, J.Q.; Merryman, C.; Glass, J.I.; Rinehart, J.; Söll, D. Transfer RNA Misidentification Scrambles Sense Codon Recoding. ChemBioChem 2013, 14, 1967–1972. [Google Scholar] [CrossRef] [Green Version]
- Ruan, B.F.; Palioura, S.; Sabina, J.; Marvin-Guy, L.; Kochhar, S.; LaRossa, R.A.; Söll, D. Quality Control despite Mistranslation Caused by an Ambiguous Genetic Code. Proc. Natl. Acad. Sci. USA 2008, 105, 16502–16507. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Yo, P.; Keith, A.; Ragan, T.J.; Harris, T.K. Thermodynamically Balanced Inside-Out (TBIO) PCR-Based Gene Synthesis: A Novel Method of Primer Design for High-Fidelity Assembly of Longer Gene Sequences. Nucleic Acids Res. 2003, 31, 143e. [Google Scholar] [CrossRef]
- Miyazaki, K.; Wintrode, P.L.; Grayling, R.A.; Rubingh, D.N.; Arnold, F.H. Directed Evolution Study of Temperature Adaptation in a Psychrophilic Enzyme. J. Mol. Biol. 2000, 297, 1015–1026. [Google Scholar] [CrossRef] [Green Version]
- Copp, J.N.; Hanson-Manful, P.; Ackerley, D.F.; Patrick, W.M. Error-Prone PCR and Effective Generation of Gene Variant Libraries for Directed Evolution. In Directed Evolution Library Creation: Methods and Protocols, 2nd ed.; Gillam, E.M.J., Copp, J.N., Ackerley, D.F., Eds.; Springer: New York, NY, USA, 2014; pp. 3–22. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]




| Amino Acid Charged by aaRS and Organism of Origin | Amino Acid Residue Position, M. barkeri Pyl aaRS 1 | |||||
|---|---|---|---|---|---|---|
| 267 (302) | 271 (306) | 274 (309) | 311 (346) | 313 (348) | 367 (401) | |
| pyrrolysine (wild type), M. barkeri [55] | Ala | Tyr | Leu | Asn | Cys | Val |
| phenylalanine, M. mazei [51] | Leu | Met | Leu | Ser | Leu | Val |
| phenylalanine, M. mazei [50] | Ala | Tyr | Leu | Ala | Leu | Val |
| O-methyl-L-tyrosine, M. mazei [45] | Thr | Tyr | Leu | Val | Trp | Leu |
| 4-bromo-L-phenylalanine/4-iodo-L-phenylalanine, M. mazei [50] | Ala | Leu | Ser | Ser | Met | Leu |
| tyrosine, TyrGen1 M. barkeri, [this paper] | Lys | Tyr | Met | Ala | Glu | Val |
| Codon Reassigned | Instantaneous Doubling Time (minutes) | Relative Cellular Fitness 1 |
|---|---|---|
| Lys AAG | 31.0 ± 0.7 | 0.926 ± 0.032 |
| Asn AAU | 30.2 ± 1.3 | 0.951 ± 0.049 |
| Glu GAG | 33.3 ± 1.7 | 0.861 ± 0.050 |
| Arg AGG | 29.9 ± 0.9 | 0.959 ± 0.037 |
| Reference system | 28.7 ± 0.7 | 1.00 ± 0.035 |
| tRNA Anticodon | Targeted Codon | Reassignment Efficiency, TyrGen2 aaRS | Non-Targeted Codon | Reassignment Efficiency, TyrGen2 aaRS | Discrimination Ratio |
|---|---|---|---|---|---|
| CUU | AAG | 10.9 ± 0.6% 1 | AAA | B.D. 2 | 99:1 |
| AUU | AAU | 2.6 ± 0.3% | AAC | B.D. | ≥95:5 |
| CUC | GAG | 11.8 ± 0.5% | GAA | 0.2 ± 0.04% | 98:1 |
| CCU | AGG | 65.0 ± 2.7% | AGA | B.D. | >99:1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwark, D.G.; Schmitt, M.A.; Fisk, J.D. Directed Evolution of the Methanosarcina barkeri Pyrrolysyl tRNA/aminoacyl tRNA Synthetase Pair for Rapid Evaluation of Sense Codon Reassignment Potential. Int. J. Mol. Sci. 2021, 22, 895. https://doi.org/10.3390/ijms22020895
Schwark DG, Schmitt MA, Fisk JD. Directed Evolution of the Methanosarcina barkeri Pyrrolysyl tRNA/aminoacyl tRNA Synthetase Pair for Rapid Evaluation of Sense Codon Reassignment Potential. International Journal of Molecular Sciences. 2021; 22(2):895. https://doi.org/10.3390/ijms22020895
Chicago/Turabian StyleSchwark, David G., Margaret A. Schmitt, and John D. Fisk. 2021. "Directed Evolution of the Methanosarcina barkeri Pyrrolysyl tRNA/aminoacyl tRNA Synthetase Pair for Rapid Evaluation of Sense Codon Reassignment Potential" International Journal of Molecular Sciences 22, no. 2: 895. https://doi.org/10.3390/ijms22020895
APA StyleSchwark, D. G., Schmitt, M. A., & Fisk, J. D. (2021). Directed Evolution of the Methanosarcina barkeri Pyrrolysyl tRNA/aminoacyl tRNA Synthetase Pair for Rapid Evaluation of Sense Codon Reassignment Potential. International Journal of Molecular Sciences, 22(2), 895. https://doi.org/10.3390/ijms22020895