Homodimeric and Heterodimeric Interactions among Vertebrate Basic Helix–Loop–Helix Transcription Factors
Abstract
:1. Introduction
2. The bHLH TF Family
3. Classifications of the bHLH TFs
4. Dynamic Nature of the bHLH TFs
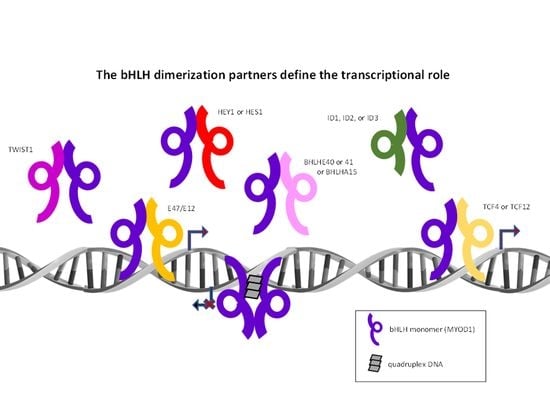
5. The Current Functional bHLH Model
6. bHLH Dimeric Interactions: The Importance of the Experimental Approach
7. Heterodimeric Interactions among bHLH TFs of Classes I, II, V and VI
8. bHLH TF Homodimers
9. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A. Methods for Detecting Protein–Protein Interactions of bHLH TFs
Appendix A.1. In Vitro Interactions
- Cell/nuclear extracts. Detection of a specific protein in the mixture can be achieved by incubation with antibodies that produce a super-shift or slower electrophoretic mobility in comparison to the protein–nucleic acid complex by itself.
- Recombinant purified proteins
- In vitro transcribed/translated proteins.
- Single short DNA probes containing one or multiple protein-binding sequences (e.g., E-boxes, N-boxes, or ESE-boxes)
Appendix A.2. In Vivo Interactions
References
- Fulton, D.L.; Sundararajan, S.; Badis, G.; Hughes, T.R.; Wasserman, W.W.; Roach, J.C.; Sladek, R. TFCat: The curated catalog of mouse and human transcription factors. Genome Biol. 2009, 10, R29. [Google Scholar] [CrossRef] [Green Version]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 175, 598–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemon, B.; Tjian, R. Orchestrated response: A symphony of transcription factors for gene control. Genes Dev. 2000, 14, 2551–2569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luscombe, N.M.; Austin, S.E.; Berman, H.M.; Thornton, J.M. An overview of the structures of protein-DNA complexes. Genome Biol. 2000, 1, 1–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaquerizas, J.M.; Kummerfeld, S.K.; Teichmann, S.A.; Luscombe, N.M. A census of human transcription factors: Function, expression and evolution. Nat. Rev. Genet. 2009, 10, 252–263. [Google Scholar] [CrossRef]
- Murre, C. Helix-loop-helix proteins and the advent of cellular diversity: 30 years of discovery. Genes Dev. 2019, 33, 6–25. [Google Scholar] [CrossRef] [Green Version]
- Murre, C.; Page-McCaw, P.; Vaessin, H.; Caudy, M.; Jan, L.; Jan, Y.N.; Cabrera, C.V.; Buskin, J.N.; Hauschka, S.D.; Lassar, A.B.; et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 1989, 58, 537–544. [Google Scholar] [CrossRef]
- Murre, C.; McCaw, P.S.; Baltimore, D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 1989, 56, 777–783. [Google Scholar] [CrossRef]
- Davis, R.L.; Cheng, P.-F.; Lassar, A.B.; Weintraub, H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell 1990, 60, 733–746. [Google Scholar] [CrossRef]
- Voronova, A.; Baltimore, D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc. Natl. Acad. Sci. USA 1990, 87, 4722–4726. [Google Scholar] [CrossRef] [Green Version]
- Ephrussi, A.; Church, G.M.; Tonegawa, S.; Gilbert, W. B Lineage—Specific Interactions of an Immunoglobulin Enhancer with Cellular Factors in Vivo. Science 1985, 227, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, M.; Sasai, Y.; Nakanishi, S.; Kageyama, R. Molecular characterization of HES-2, a mammalian helix-loop-helix factor structurally related to Drosophila hairy and Enhancer of split. Eur. J. Biochem. 1993, 215, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Belanger-Jasmin, S.; Llamosas, E.; Tang, Y.; Joachim, K.; Osiceanu, A.-M.; Jhas, S.; Stifani, S. Inhibition of cortical astrocyte differentiation by Hes6 requires amino- and carboxy-terminal motifs important for dimerization and phosphorylation. J. Neurochem. 2007, 103, 2022–2034. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-H.; Baker, N.E. E Proteins and ID Proteins: Helix-Loop-Helix Partners in Development and Disease. Dev. Cell 2015, 35, 269–280. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Ghaffari, S.; Taneja, R. bHLH-Orange Transcription Factors in Development and Cancer. Transl. Oncogenom. 2007, 2, 107–120. [Google Scholar]
- Belle, I.; Zhuang, Y. E Proteins in Lymphocyte Development and Lymphoid Diseases. Curr. Top. Dev. Biol. 2014, 110, 153–187. [Google Scholar] [CrossRef]
- Slattery, C.; Ryan, M.P.; McMorrow, T. E2A proteins: Regulators of cell phenotype in normal physiology and disease. Int. J. Biochem. Cell Biol. 2008, 40, 1431–1436. [Google Scholar] [CrossRef]
- Barinaga, M. Dimers direct development. Science 1991, 251, 1176–1177. [Google Scholar] [CrossRef]
- Weintraub, H.; Davis, R.; Tapscott, S.; Thayer, M.; Krause, M.; Benezra, R.; Blackwell, T.K.; Turner, D.; Rupp, R.; Hollenberg, S.; et al. The myoD Gene Family: Nodal Point during Specification of the Muscle Cell Lineage. Science 1991, 251, 761–766. [Google Scholar] [CrossRef]
- Benezra, R.; Davis, R.L.; Lockshon, D.; Turner, D.L.; Weintraub, H. The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell 1990, 61, 49–59. [Google Scholar] [CrossRef]
- Murre, C.; Bain, G.; van Dijk, M.A.; Engel, I.; Furnari, B.A.; Massari, M.E.; Matthews, J.R.; Quong, M.W.; Rivera, R.R.; Stuiver, M.H. Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 1994, 1218, 129–135. [Google Scholar] [CrossRef]
- Massari, M.E.; Murre, C. Helix-Loop-Helix Proteins: Regulators of Transcription in Eucaryotic Organisms. Mol. Cell. Biol. 2000, 20, 429–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, V.J.; Steenbergen, R.; Murre, C. Localization of E2A mRNA expression in developing and adult rat tissues. Proc. Natl. Acad. Sci. USA 1993, 90, 7583–7587. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, Y.; Kim, C.G.; Bartelmez, S.; Cheng, P.; Groudine, M.; Weintraub, H. Helix-loop-helix transcription factors E12 and E47 are not essential for skeletal or cardiac myogenesis, erythropoiesis, chondrogenesis, or neurogenesis. Proc. Natl. Acad. Sci. USA 1992, 89, 12132–12136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravanpay, A.C.; Olson, J.M. E protein dosage influences brain development more than family member identity. J. Neurosci. Res. 2008, 86, 1472–1481. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Arnold, H.H. The four human muscle regulatory helix-loop-helix proteins Myf3-Myf6 exhibit similar hetero-dimerization and DNA binding properties. Nucleic Acids Res. 1991, 19, 5645–5651. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, A.; Cui, X.; Naumovski, L.; Cleary, M.L. Helix-loop-helix proteins LYL1 and E2a form heterodimeric complexes with distinctive DNA-binding properties in hematolymphoid cells. Mol. Cell. Biol. 1996, 16, 2394–2401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flora, A.; Garcia, J.J.; Thaller, C.; Zoghbi, H.Y. The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc. Natl. Acad. Sci. USA 2007, 104, 15382–15387. [Google Scholar] [CrossRef] [Green Version]
- Poulin, G.; Turgeon, B.; Drouin, J. NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol. Cell. Biol. 1997, 17, 6673–6682. [Google Scholar] [CrossRef] [Green Version]
- Murakami, M.; Kataoka, K.; Tominaga, J.; Nakagawa, O.; Kurihara, H. Differential cooperation between dHAND and three different E-proteins. Biochem. Biophys. Res. Commun. 2004, 323, 168–174. [Google Scholar] [CrossRef]
- Li, S.; Mattar, P.; Zinyk, D.; Singh, K.; Chaturvedi, C.-P.; Kovach, C.P.; Dixit, R.; Kurrasch, D.M.; Ma, Y.; Chan, J.A.; et al. GSK3 Temporally Regulates Neurogenin 2 Proneural Activity in the Neocortex. J. Neurosci. 2012, 32, 7791–7805. [Google Scholar] [CrossRef] [Green Version]
- Fischer, B.; Azim, K.; Hurtado-Chong, A.; Ramelli, S.; Fernández, M.; Raineteau, O. E-proteins orchestrate the progression of neural stem cell differentiation in the postnatal forebrain. Neural Dev. 2014, 9, 23. [Google Scholar] [CrossRef] [Green Version]
- Pfurr, S.; Chu, Y.-H.; Bohrer, C.; Greulich, F.; Beattie, R.; Mammadzada, K.; Hils, M.; Arnold, S.J.; Taylor, V.; Schachtrup, K.; et al. The E2A splice variant E47 regulates the differentiation of projection neurons via p57(KIP2) during cortical development. Development 2017, 144, 3917–3931. [Google Scholar] [CrossRef] [Green Version]
- Bouderlique, T.; Peña-Pérez, L.; Kharazi, S.; Hils, M.; Li, X.; Krstic, A.; De Paepe, A.; Schachtrup, C.; Gustafsson, C.; Holmberg, D.; et al. The Concerted Action of E2-2 and HEB Is Critical for Early Lymphoid Specification. Front. Immunol. 2019, 10, 455. [Google Scholar] [CrossRef] [Green Version]
- Wedel, M.; Fröb, F.; Elsesser, O.; Wittmann, M.-T.; Lie, D.C.; Reis, A.; Wegner, M. Transcription factor Tcf4 is the preferred heterodimerization partner for Olig2 in oligodendrocytes and required for differentiation. Nucleic Acids Res. 2020, 48, 4839–4857. [Google Scholar] [CrossRef] [PubMed]
- McLellan, A.S.; Langlands, K.; Kealey, T. Exhaustive identification of human class II basic helix-loop-helix proteins by virtual library screening. Mech. Dev. 2002, 119 (Suppl. 1), S285–S291. [Google Scholar] [CrossRef]
- Carroll, P.A.; Freie, B.W.; Mathsyaraja, H.; Eisenman, R.N. The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 2018, 12, 412–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Etzioni, S.; Yafe, A.; Khateb, S.; Weisman-Shomer, P.; Bengal, E.; Fry, M. Homodimeric MyoD Preferentially Binds Tetraplex Structures of Regulatory Sequences of Muscle-specific Genes. J. Biol. Chem. 2005, 280, 26805–26812. [Google Scholar] [CrossRef] [Green Version]
- Bersten, D.C.; Sullivan, A.E.; Peet, D.J.; Whitelaw, M.L. bHLH-PAS proteins in cancer. Nat. Rev. Cancer 2013, 13, 827–841. [Google Scholar] [CrossRef]
- Kolonko, M.; Greb-Markiewicz, B. bHLH-PAS Proteins: Their Structure and Intrinsic Disorder. Int. J. Mol. Sci. 2019, 20, 3653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atchley, W.R.; Fitch, W.M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledent, V.; Vervoort, M. The Basic Helix-Loop-Helix Protein Family: Comparative Genomics and Phylogenetic Analysis. Genome Res. 2001, 11, 754–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledent, V.; Paquet, O.; Vervoort, M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Crozatier, M.; Valle, D.; Dubois, L.; Ibnsouda, S.; Vincent, A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr. Biol. 1996, 6, 707–718. [Google Scholar] [CrossRef] [Green Version]
- Jones, S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004, 5, 226. [Google Scholar] [CrossRef] [Green Version]
- Stevens, J.D.; Roalson, E.; Skinner, M.K. Phylogenetic and expression analysis of the basic helix-loop-helix transcription factor gene family: Genomic approach to cellular differentiation. Differentiation 2008, 76, 1006–1042. [Google Scholar] [CrossRef] [Green Version]
- Skinner, M.K.; Rawls, A.; Wilson-Rawls, J.; Roalson, E.H. Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation 2010, 80, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, T.; Olson, E.N. Domains outside of the DNA-binding domain impart target gene specificity to myogenin and MRF4. Mol. Cell Biol. 1991, 11, 6103–6108. [Google Scholar]
- Petropoulos, H.; Skerjanc, I.S. Analysis of the Inhibition of MyoD Activity by ITF-2B and Full-length E12/E47. J. Biol. Chem. 2000, 275, 25095–25101. [Google Scholar] [CrossRef] [Green Version]
- Yutzey, K.E.; Rhodes, S.J.; Konieczny, S.F. Differential trans activation associated with the muscle regulatory factors MyoD1, myogenin, and MRF4. Mol. Cell Biol. 1990, 10, 3934–3944. [Google Scholar] [PubMed] [Green Version]
- Quong, M.W.; Massari, M.E.; Zwart, R.; Murre, C. A new transcriptional-activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol. Cell Biol. 1993, 13, 792–800. [Google Scholar]
- Massari, M.E.; Jennings, P.A.; Murre, C. The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol. Cell Biol. 1996, 16, 121–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Sloan, S.R. The basic helix-loop-helix domain of the E47 transcription factor requires other protein regions for full DNA binding activity. Biochem. Biophys. Res. Commun. 2002, 290, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Hyndman, B.D.; Thompson, P.; Denis, C.M.; Chitayat, S.; Bayly, R.; Smith, S.P.; LeBrun, D.P. Mapping acetylation sites in E2A identifies a conserved lysine residue in activation domain 1 that promotes CBP/p300 recruitment and transcriptional activation. Biochim. Biophys. Acta 2012, 1819, 375–381. [Google Scholar] [CrossRef]
- Henthorn, P.; Kiledjian, M.; Kadesch, T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science 1990, 247, 467–470. [Google Scholar] [CrossRef]
- Pagliuca, A.; Gallo, P.; De Luca, P.; Lania, L. Class A helix-loop-helix proteins are positive regulators of several cyclin-dependent kinase inhibitors’ promoter activity and negatively affect cell growth. Cancer Res. 2000, 60, 1376–1382. [Google Scholar]
- Perez-Moreno, M.A.; Locascio, A.; Rodrigo, I.; Dhondt, G.; Portillo, F.; Nieto, M.A.; Cano, A. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J. Biol. Chem. 2001, 276, 27424–27431. [Google Scholar] [CrossRef] [Green Version]
- Kondo, M.; Cubillo, E.; Tobiume, K.; Shirakihara, T.; Fukuda, N.; Suzuki, H.; Shimizu, K.; Takehara, K.; Cano, A.; Saitoh, M.; et al. A role for Id in the regulation of TGF-beta-induced epithelial-mesenchymal transdifferentiation. Cell Death Differ. 2004, 11, 1092–1101. [Google Scholar] [CrossRef]
- Kumar, M.S.; Hendrix, J.A.; Johnson, A.D.; Owens, G.K. Smooth muscle alpha-actin gene requires two E-boxes for proper expression in vivo and is a target of class I basic helix-loop-helix proteins. Circ. Res. 2003, 92, 840–847. [Google Scholar] [CrossRef] [Green Version]
- Sepp, M.; Kannike, K.; Eesmaa, A.; Urb, M.; Timmusk, T. Functional diversity of human basic helix-loop-helix transcription factor TCF4 isoforms generated by alternative 5’ exon usage and splicing. PLoS ONE 2001, 6, e22138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobrado, V.R.; Moreno-Bueno, G.; Cubillo, E.; Holt, L.; Nieto, M.A.; Portillo, F.; Cano, A. The class I bHLH factors E2-2A and E2-2B regulate EMT. J. Cell Sci. 2009, 122, 1014–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furumura, M.; Potterf, S.B.; Toyofuku, K.; Matsunaga, J.; Muller, J.; Hearing, V.J. Involvement of ITF2 in the transcriptional regulation of melanogenic genes. J. Biol. Chem. 2001, 276, 28147–28154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.S.; Olson, E.N.; Kingston, R.E. HEB, a helix-loop-helix protein related to E2A and ITF2 that can modulate the DNA-binding ability of myogenic regulatory factors. Mol. Cell Biol. 1992, 12, 1031–1042. [Google Scholar]
- Weintraub, H.; Davis, R.; Lockshon, D.; Lassar, A. MyoD binds cooperatively to two sites in a target enhancer sequence: Occupancy of two sites is required for activation. Proc. Natl. Acad. Sci. USA 1990, 87, 5623–5627. [Google Scholar] [CrossRef] [Green Version]
- Lassar, A.B.; Davis, R.L.; Wright, W.E.; Kadesch, T.; Murre, C.; Voronova, A.; Baltimore, D.; Weintraub, H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 1991, 66, 305–315. [Google Scholar] [CrossRef]
- Shirakata, M.; Paterson, B.M. The E12 inhibitory domain prevents homodimer formation and facilitates selective heterodimerization with the MyoD family of gene regulatory factors. EMBO J. 1995, 14, 1766–1772. [Google Scholar] [CrossRef]
- Johnson, S.E.; Wang, X.; Hardy, S.; Taparowsky, E.J.; Konieczny, S.F. Casein kinase II increases the transcriptional activities of MRF4 and MyoD independently of their direct phosphorylation. Mol. Cell Biol. 1996, 16, 1604–1613. [Google Scholar] [CrossRef] [Green Version]
- Yafe, A.; Shklover, J.; Weisman-Shomer, P.; Bengal, E.; Fry, M. Differential binding of quadruplex structures of muscle-specific genes regulatory sequences by MyoD, MRF4 and myogenin. Nucleic Acids Res. 2008, 36, 3916–3925. [Google Scholar] [CrossRef] [Green Version]
- Braun, T.; Bober, E.; Winter, B.; Rosenthal, N.; Arnold, H.H. Myf-6, a new member of the human gene family of myogenic determination factors: Evidence for a gene cluster on chromosome 12. EMBO J. 1990, 9, 821–831. [Google Scholar] [CrossRef]
- Braun, T.; Buschhausen-Denker, G.; Bober, E.; Tannich, E.; Arnold, H.H. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J. 1989, 8, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Winter, B.; Bober, E.; Arnold, H.H. Transcriptional activation domain of the muscle-specific gene-regulatory protein myf5. Nature 1990, 346, 663–665. [Google Scholar] [CrossRef]
- Brennan, T.J.; Olson, E.N. Myogenin resides in the nucleus and acquires high affinity for a conserved enhancer element on heterodimerization. Genes Dev. 1990, 4, 582–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millar, S.E.; Lader, E.; Liang, L.F.; Dean, J. Oocyte-specific factors bind a conserved upstream sequence required for mouse zona pellucida promoter activity. Mol. Cell Biol. 1991, 11, 6197–6204. [Google Scholar] [PubMed] [Green Version]
- Liang, L.; Soyal, S.M.; Dean, J. FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development 1997, 124, 4939–4947. [Google Scholar] [CrossRef]
- Cserjesi, P.; Brown, D.; Ligon, K.L.; Lyons, G.E.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Olson, E.N. Scleraxis: A basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development 1995, 121, 1099–1110. [Google Scholar] [CrossRef]
- Carlberg, A.L.; Tuan, R.S.; Hall, D.J. Regulation of scleraxis function by interaction with the bHLH protein E47. Mol. Cell Biol. Res. Commun. 2000, 3, 82–86. [Google Scholar] [CrossRef]
- Furumatsu, T.; Shukunami, C.; Amemiya-Kudo, M.; Shimano, H.; Ozaki, T. Scleraxis and E47 cooperatively regulate the Sox9-dependent transcription. Int. J. Biochem. Cell Biol. 2010, 42, 148–156. [Google Scholar] [CrossRef] [Green Version]
- Wilson-Rawls, J.; Rhee, J.M.; Rawls, A. Paraxis is a basic helix-loop-helix protein that positively regulates transcription through binding to specific E-box elements. J. Biol. Chem. 2004, 279, 37685–37692. [Google Scholar] [CrossRef] [Green Version]
- Spicer, D.B.; Rhee, J.; Cheung, W.L.; Lassar, A.B. Inhibition of Myogenic bHLH and MEF2 Transcription Factors by the bHLH Protein Twist. Science 1996, 272, 1476–1480. [Google Scholar] [CrossRef]
- Hamamori, Y.; Wu, H.Y.; Sartorelli, V.; Kedes, L. The basic domain of myogenic basic helix-loop-helix (bHLH) proteins is the novel target for direct inhibition by another bHLH protein, Twist. Mol. Cell. Biol. 1997, 17, 6563–6573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco, H.L.; Casasnovas, J.; Rodríguez-Medina, J.R.; Cadilla, C.L. Redundant or separate entities?—Roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res. 2011, 39, 1177–1186. [Google Scholar] [CrossRef] [Green Version]
- Gong, X.Q.; Li, L. Dermo-1, a multifunctional basic helix-loop-helix protein, represses MyoD transactivation via the HLH domain, MEF2 interaction, and chromatin deacetylation. J. Biol. Chem. 2002, 277, 12310–12317. [Google Scholar] [CrossRef] [Green Version]
- Verzi, M.P.; Anderson, J.P.; Dodou, E.; Kelly, K.K.; Greene, S.B.; North, B.J.; Cripps, R.M.; Black, B.L. N-twist, an evolutionarily conserved bHLH protein expressed in the developing CNS, functions as a transcriptional inhibitor. Dev. Biol. 2002, 249, 174–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morin, S.; Pozzulo, G.; Robitaille, L.; Cross, J.; Nemer, M. MEF2-dependent recruitment of the HAND1 transcription factor results in synergistic activation of target promoters. J. Biol. Chem. 2005, 280, 32272–32278. [Google Scholar] [CrossRef] [Green Version]
- Hollenberg, S.M.; Sternglanz, R.; Cheng, P.F.; Weintraub, H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell Biol. 1995, 15, 3813–3822. [Google Scholar] [CrossRef] [Green Version]
- Knofler, M.; Meinhardt, G.; Bauer, S.; Loregger, T.; Vasicek, R.; Bloor, D.J.; Kimber, S.J.; Husslein, P. Human Hand1 basic helix-loop-helix (bHLH) protein: Extra-embryonic expression pattern, interaction partners and identification of its transcriptional repressor domains. Biochem. J. 2002, 361, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Bounpheng, M.A.; Morrish, T.A.; Dodds, S.G.; Christy, B.A. Negative Regulation of Selected bHLH Proteins by eHAND. Exp. Cell Res. 2000, 257, 320–331. [Google Scholar] [CrossRef]
- Scott, I.C.; Anson-Cartwright, L.; Riley, P.; Reda, D.; Cross, J.C. The HAND1 Basic Helix-Loop-Helix Transcription Factor Regulates Trophoblast Differentiation via Multiple Mechanisms. Mol. Cell. Biol. 2000, 20, 530–541. [Google Scholar] [CrossRef] [Green Version]
- Firulli, B.A.; Hadzic, D.B.; McDaid, J.R.; Firulli, A.B. The basic helix-loop-helix transcription factors dHAND and eHAND exhibit dimerization characteristics that suggest complex regulation of function. J. Biol. Chem. 2000, 275, 33567–33573. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.S.; Cserjesi, P. The basic helix-loop-helix factor, HAND2, functions as a transcriptional activator by binding to E-boxes as a heterodimer. J. Biol. Chem. 2002, 277, 12604–12612. [Google Scholar] [CrossRef] [Green Version]
- Funato, N.; Chapman, S.L.; McKee, M.D.; Funato, H.; Morris, J.A.; Shelton, J.M.; Richardson, J.A.; Yanagisawa, H. Hand2 controls osteoblast differentiation in the branchial arch by inhibiting DNA binding of Runx2. Development 2009, 136, 615–625. [Google Scholar] [CrossRef] [Green Version]
- Beres, T.M.; Masui, T.; Swift, G.H.; Shi, L.; Henke, R.M.; MacDonald, R.J. PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol. Cell Biol. 2006, 26, 117–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, S.D.; Swift, G.H.; Peyton, M.J.; Hammer, R.E.; MacDonald, R.J. The role of PTF1-P48 in pancreatic acinar gene expression. J. Biol. Chem. 2001, 276, 44018–44026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutoh, H.; Fung, B.P.; Naya, F.J.; Tsai, M.J.; Nishitani, J.; Leiter, A.B. The basic helix-loop-helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc. Natl. Acad. Sci. USA 1997, 94, 3560–3564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Moore, M.; Marcora, E.; Lee, J.E.; Qiu, Y.; Samaras, S.; Stein, R. The NeuroD1/BETA2 sequences essential for insulin gene transcription colocalize with those necessary for neurogenesis and p300/CREB binding protein binding. Mol. Cell Biol. 1999, 19, 704–713. [Google Scholar] [CrossRef] [Green Version]
- Farah, M.H.; Olson, J.M.; Sucic, H.B.; Hume, R.I.; Tapscott, S.J.; Turner, D.L. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 2000, 127, 693–702. [Google Scholar] [CrossRef]
- Breslin, M.B.; Zhu, M.; Lan, M.S. NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J. Biol. Chem. 2003, 278, 38991–38997. [Google Scholar] [CrossRef] [Green Version]
- Westerman, B.A.; Chhatta, A.; Poutsma, A.; van Vegchel, T.; Oudejans, C.B. NEUROD1 acts in vitro as an upstream regulator of NEUROD2 in trophoblast cells. Biochim. Biophys. Acta 2004, 1676, 96–103. [Google Scholar] [CrossRef]
- Lynn, F.C.; Sanchez, L.; Gomis, R.; German, M.S.; Gasa, R. Identification of the bHLH Factor Math6 as a Novel Component of the Embryonic Pancreas Transcriptional Network. PLoS ONE 2008, 3, e2430. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Nadal-Vicens, M.; Misono, S.; Lin, M.; Zubiaga, A.; Hua, X.; Fan, G.; Greenberg, M.E. Neurogenin Promotes Neurogenesis and Inhibits Glial Differentiation by Independent Mechanisms. Cell 2001, 104, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Galvez, H.; Tena, J.J.; Giraldez, F.; Abello, G. The Repression of Atoh1 by Neurogenin1 during Inner Ear Development. Front. Mol. Neurosci. 2017, 10, 321. [Google Scholar] [CrossRef] [PubMed]
- Roztocil, T.; Matter-Sadzinski, L.; Alliod, C.; Ballivet, M.; Matter, J.M. NeuroM, a neural helix-loop-helix transcription factor, defines a new transition stage in neurogenesis. Development 1997, 124, 3263–3272. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, S.; Fukumura, R.; Higuchi, T.; Kobayashi, S. YB-1 transcription in the postnatal brain is regulated by a bHLH transcription factor Math2 through an E-box sequence in the 5’-UTR of the gene. Mol. Cell Biochem. 2009, 327, 267–275. [Google Scholar] [CrossRef]
- Uittenbogaard, M.; Martinka, D.L.; Chiaramello, A. The basic helix-loop-helix differentiation factor Nex1/MATH-2 functions as a key activator of the GAP-43 gene. J. Neurochem. 2003, 84, 678–688. [Google Scholar] [CrossRef] [Green Version]
- Akazawa, C.; Ishibashi, M.; Shimizu, C.; Nakanishi, S.; Kageyama, R. A Mammalian Helix-Loop-Helix Factor Structurally Related to the Product of Drosophila Proneural Gene atonal Is a Positive Transcriptional Regulator Expressed in the Developing Nervous System. J. Biol. Chem. 1995, 270, 8730–8738. [Google Scholar] [CrossRef] [Green Version]
- Henke, R.M.; Meredith, D.M.; Borromeo, M.D.; Savage, T.K.; Johnson, J.E. Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev. Biol. 2009, 328, 529–540. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.P.; Liu, M.; El-Hodiri, H.M.; Chu, K.; Jamrich, M.; Tsai, M.J. Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Mol. Cell Biol. 2000, 20, 3292–3307. [Google Scholar] [CrossRef] [Green Version]
- Vetere, A.; Li, W.C.; Paroni, F.; Juhl, K.; Guo, L.; Nishimura, W.; Dai, X.; Bonner-Weir, S.; Sharma, A. OVO homologue-like 1 (Ovol1) transcription factor: A novel target of neurogenin-3 in rodent pancreas. Diabetologia 2010, 53, 115–122. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Mo, Z.; Xiang, M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc. Natl. Acad. Sci. USA 2001, 98, 1649–1654. [Google Scholar] [CrossRef] [Green Version]
- Matter-Sadzinski, L.; Matter, J.M.; Ong, M.T.; Hernandez, J.; Ballivet, M. Specification of neurotransmitter receptor identity in developing retina: The chick ATH5 promoter integrates the positive and negative effects of several bHLH proteins. Development 2001, 128, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Varghese, J.; Masaratana, P.; Latunde-Dada, G.O.; Jacob, M.; Simpson, R.J.; McKie, A.T. The transcription factor ATOH8 is regulated by erythropoietic activity and regulates HAMP transcription and cellular pSMAD1,5,8 levels. Br. J. Haematol. 2014, 164, 586–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, F.; Wasserman, S.M.; Torres-Vazquez, J.; Weinstein, B.; Cao, F.; Li, Z.; Wilson, K.D.; Yue, W.; Wu, J.C.; Xie, X.; et al. The role of Hath6, a newly identified shear-stress-responsive transcription factor, in endothelial cell differentiation and function. J. Cell Sci. 2014, 127, 1428–1440. [Google Scholar]
- Ejarque, M.; Altirriba, J.; Gomis, R.; Gasa, R. Characterization of the transcriptional activity of the basic helix–loop–helix (bHLH) transcription factor Atoh8. Biochim. Biophys. Acta (BBA)—Bioenerg. 2013, 1829, 1175–1183. [Google Scholar] [CrossRef]
- Tran, T.; Jia, D.; Sun, Y.; Konieczny, S.F. The bHLH domain of Mistl is sufficient to activate gene transcription. Gene Expr. 2007, 13, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Lemercier, C.; To, R.Q.; Carrasco, R.A.; Konieczny, S.F. The basic helix-loop-helix transcription factor Mist1 functions as a transcriptional repressor of myoD. EMBO J. 1998, 17, 1412–1422. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.E.; Birren, S.J.; Saito, T.; Anderson, D.J. DNA binding and transcriptional regulatory activity of mammalian achaete-scute homologous (MASH) proteins revealed by interaction with a muscle-specific enhancer. Proc. Natl. Acad. Sci. USA 1992, 89, 3596–3600. [Google Scholar] [CrossRef] [Green Version]
- Jiang, B.; Kamat, A.; Mendelson, C.R. Hypoxia prevents induction of aromatase expression in human trophoblast cells in culture: Potential inhibitory role of the hypoxia-inducible transcription factor Mash-2 (mammalian achaete-scute homologous protein-2). Mol. Endocrinol. 2000, 14, 1661–1673. [Google Scholar] [CrossRef]
- Yoshida, S.; Ohbo, K.; Takakura, A.; Takebayashi, H.; Okada, T.; Abe, K.; Nabeshima, Y. Sgn1, a basic helix-loop-helix transcription factor delineates the salivary gland duct cell lineage in mice. Dev. Biol. 2001, 240, 517–530. [Google Scholar] [CrossRef] [Green Version]
- Hsu, H.L.; Wadman, I.; Tsan, J.T.; Baer, R. Positive and negative transcriptional control by the TAL1 helix-loop-helix protein. Proc. Natl. Acad. Sci. USA 1994, 91, 5947–5951. [Google Scholar] [CrossRef] [Green Version]
- Park, S.T.; Sun, X.H. The Tal1 oncoprotein inhibits E47-mediated transcription. Mechanism of inhibition. J. Biol. Chem. 1998, 273, 7030–7037. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Qiu, Y.; Stein, R.W.; Brandt, S.J. p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene 1999, 18, 4958–4967. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Brandt, S.J. mSin3A Regulates Murine Erythroleukemia Cell Differentiation through Association with the TAL1 (or SCL) Transcription Factor. Mol. Cell. Biol. 2000, 20, 2248–2259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Li, X.; Valverde, K.; Fu, X.; Noguchi, C.; Qiu, Y.; Huang, S. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc. Natl. Acad. Sci. USA 2009, 106, 10141–10146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldfarb, A.N.; Lewandowska, K. Inhibition of cellular differentiation by the SCL/tal oncoprotein: Transcriptional repression by an Id-like mechanism. Blood 1995, 85, 465–471. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Brown, L.; Yang, C.Y.; Tsan, J.T.; Siciliano, M.J.; Espinosa, R., III; Le Beau, M.M.; Baer, R.J. TAL2, a helix-loop-helix gene activated by the (7;9)(q34;q32) translocation in human T-cell leukemia. Proc. Natl. Acad. Sci. USA 1991, 88, 11416–11420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deleuze, V.; El-Hajj, R.; Chalhoub, E.; Dohet, C.; Pinet, V.; Couttet, P.; Mathieu, D. Angiopoietin-2 is a direct transcriptional target of TAL1, LYL1 and LMO2 in endothelial cells. PLoS ONE 2012, 7, e40484. [Google Scholar] [CrossRef] [Green Version]
- San-Marina, S.; Han, Y.; Suarez Saiz, F.; Trus, M.R.; Minden, M.D. Lyl1 interacts with CREB1 and alters expression of CREB1 target genes. Biochim. Biophys. Acta 2008, 1783, 503–517. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Y.; Jiang, L.; Hiai, H.; Toyokuni, S.; Yamada, Y. Overexpression of a transcription factor LYL1 induces T- and B-cell lymphoma in mice. Oncogene 2007, 26, 6937–6947. [Google Scholar] [CrossRef] [Green Version]
- Manetopoulos, C.; Hansson, A.; Karlsson, J.; Jonsson, J.I.; Axelson, H. The LIM-only protein LMO4 modulates the transcriptional activity of HEN1. Biochem. Biophys. Res. Commun. 2003, 307, 891–899. [Google Scholar] [CrossRef]
- Fox, D.L.; Good, D.J. Nescient helix-loop-helix 2 interacts with signal transducer and activator of transcription 3 to regulate transcription of prohormone convertase 1/3. Mol. Endocrinol. 2008, 22, 1438–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isogai, E.; Ohira, M.; Ozaki, T.; Oba, S.; Nakamura, Y.; Nakagawara, A. Oncogenic LMO3 collaborates with HEN2 to enhance neuroblastoma cell growth through transactivation of Mash1. PLoS ONE 2011, 6, e19297. [Google Scholar]
- Massari, M.E.; Rivera, R.R.; Voland, J.R.; Quong, M.W.; Breit, T.M.; van Dongen, J.J.; de Smit, O.; Murre, C. Characterization of ABF-1, a novel basic helix-loop-helix transcription factor expressed in activated B lymphocytes. Mol. Cell Biol. 1998, 18, 3130–3139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Webb, R.; Richardson, J.A.; Olson, E.N. MyoR: A muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proc. Natl. Acad. Sci. USA 1999, 96, 552–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; MacQuarrie, K.L.; Analau, E.; Tyler, A.E.; Dilworth, F.J.; Cao, Y.; Diede, S.J.; Tapscott, S.J. MyoD and E-protein heterodimers switch rhabdomyosarcoma cells from an arrested myoblast phase to a differentiated state. Genes Dev. 2009, 23, 694–707. [Google Scholar] [CrossRef] [Green Version]
- Hidai, H.; Bardales, R.; Goodwin, R.; Quertermous, T.; Quertermous, E.E. Cloning of capsulin, a basic helix-loop-helix factor expressed in progenitor cells of the pericardium and the coronary arteries. Mech. Dev. 1998, 73, 33–43. [Google Scholar] [CrossRef]
- Miyagishi, M.; Hatta, M.; Ohshima, T.; Ishida, J.; Fujii, R.; Nakajima, T.; Fukamizu, A. Cell type-dependent transactivation or repression of mesoderm-restricted basic helix-loop-helix protein, POD-1/Capsulin. Mol. Cell Biochem. 2000, 205, 141–147. [Google Scholar] [CrossRef]
- Funato, N.; Ohyama, K.; Kuroda, T.; Nakamura, M. Basic Helix-Loop-Helix Transcription Factor Epicardin/Capsulin/Pod-1 Suppresses Differentiation by Negative Regulation of Transcription. J. Biol. Chem. 2003, 278, 7486–7493. [Google Scholar] [CrossRef] [Green Version]
- Franca, M.M.; Ferraz-de-Souza, B.; Santos, M.G.; Lerario, A.M.; Fragoso, M.C.; Latronico, A.C.; Kuick, R.D.; Hammer, G.D.; Lotfi, C.F. POD-1 binding to the E-box sequence inhibits SF-1 and StAR expression in human adrenocortical tumor cells. Mol. Cell Endocrinol. 2013, 371, 140–147. [Google Scholar] [CrossRef] [Green Version]
- Narumi, O.; Mori, S.; Boku, S.; Tsuji, Y.; Hashimoto, N.; Nishikawa, S.; Yokota, Y. OUT, a novel basic helix-loop-helix transcription factor with an Id-like inhibitory activity. J. Biol. Chem. 2000, 275, 3510–3521. [Google Scholar] [CrossRef] [Green Version]
- Malik, S.; Percin, F.E.; Bornholdt, D.; Albrecht, B.; Percesepe, A.; Koch, M.C.; Landi, A.; Fritz, B.; Khan, R.; Mumtaz, S.; et al. Mutations affecting the BHLHA9 DNA-binding domain cause MSSD, mesoaxial synostotic syndactyly with phalangeal reduction, Malik-Percin type. Am. J. Hum. Genet. 2014, 95, 649–659. [Google Scholar] [CrossRef] [Green Version]
- Ross, S.E.; McCord, A.E.; Jung, C.; Atan, D.; Mok, S.I.; Hemberg, M.; Kim, T.K.; Salogiannis, J.; Hu, L.; Cohen, S.; et al. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron 2012, 73, 292–303. [Google Scholar] [CrossRef] [Green Version]
- Peyton, M.; Stellrecht, C.M.; Naya, F.J.; Huang, H.P.; Samora, P.J.; Tsai, M.J. BETA3, a novel helix-loop-helix protein, can act as a negative regulator of BETA2 and MyoD-responsive genes. Mol. Cell Biol. 1996, 16, 626–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.P.; Dutra, A.; Stellrecht, C.M.; Wu, C.; Piatigorsky, J.; Saunders, G.F. Functional and structural characterization of the human gene BHLHB5, encoding a basic helix-loop-helix transcription factor. Genomics 2002, 80, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Bramblett, D.E.; Copeland, N.G.; Jenkins, N.A.; Tsai, M.J. BHLHB4 is a bHLH transcriptional regulator in pancreas and brain that marks the dimesencephalic boundary. Genomics 2002, 79, 402–412. [Google Scholar] [CrossRef]
- Samanta, J.; Kessler, J.A. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development 2004, 131, 4131–4142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silbereis, J.C.; Nobuta, H.; Tsai, H.H.; Heine, V.M.; McKinsey, G.L.; Meijer, D.H.; Howard, M.A.; Petryniak, M.A.; Potter, G.B.; Alberta, J.A.; et al. Olig1 function is required to repress dlx1/2 and interneuron production in Mammalian brain. Neuron 2014, 81, 574–587. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; de Faria, J.P.; Andrew, P.; Nitarska, J.; Richardson, W.D. Phosphorylation Regulates OLIG2 Cofactor Choice and the Motor Neuron-Oligodendrocyte Fate Switch. Neuron 2011, 69, 918–929. [Google Scholar] [CrossRef] [Green Version]
- Novitch, B.G.; Chen, A.I.; Jessell, T.M. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 2001, 31, 773–789. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.K.; Lee, B.; Ruiz, E.C.; Pfaff, S.L. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005, 19, 282–294. [Google Scholar] [CrossRef] [Green Version]
- Muller, T.; Anlag, K.; Wildner, H.; Britsch, S.; Treier, M.; Birchmeier, C. The bHLH factor Olig3 coordinates the specification of dorsal neurons in the spinal cord. Genes Dev. 2005, 19, 733–743. [Google Scholar] [CrossRef] [Green Version]
- Boudjelal, M.; Taneja, R.; Matsubara, S.; Bouillet, P.; Dolle, P.; Chambon, P. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev. 1997, 11, 2052–2065. [Google Scholar] [CrossRef] [Green Version]
- St-Pierre, B.; Flock, G.; Zacksenhaus, E.; Egan, S.E. Stra13 homodimers repress transcription through class B E-box elements. J. Biol. Chem. 2002, 277, 46544–46551. [Google Scholar] [CrossRef] [Green Version]
- Azmi, S.; Ozog, A.; Taneja, R. Sharp-1/DEC2 Inhibits Skeletal Muscle Differentiation through Repression of Myogenic Transcription Factors. J. Biol. Chem. 2004, 279, 52643–52652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimoto, K.; Hamaguchi, H.; Hashiba, T.; Nakamura, T.; Kawamoto, T.; Sato, F.; Noshiro, M.; Bhawal, U.K.; Suardita, K.; Kato, Y. Transcriptional repression by the basic helix-loop-helix protein Dec2: Multiple mechanisms through E-box elements. Int. J. Mol. Med. 2007, 19, 925–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jen, Y.; Weintraub, H.; Benezra, R. Overexpression of Id protein inhibits the muscle differentiation program: In vivo association of Id with E2A proteins. Genes Dev. 1992, 6, 1466–1479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jogi, A.; Persson, P.; Grynfeld, A.; Pahlman, S.; Axelson, H. Modulation of basic helix-loop-helix transcription complex formation by Id proteins during neuronal differentiation. J. Biol. Chem. 2002, 277, 9118–9126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loveys, D.A.; Streiff, M.B.; Kato, G.J. E2A basic-helix-loop-helix transcription factors are negatively regulated by serum growth factors and by the Id3 protein. Nucleic Acids Res. 1996, 24, 2813–2820. [Google Scholar] [CrossRef] [Green Version]
- Riechmann, V.; van Cruchten, I.; Sablitzky, F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 1994, 22, 749–755. [Google Scholar] [CrossRef]
- Sun, J.; Kamei, C.N.; Layne, M.D.; Jain, M.K.; Liao, J.K.; Lee, M.E.; Chin, M.T. Regulation of myogenic terminal differentiation by the hairy-related transcription factor CHF2. J. Biol. Chem. 2001, 276, 18591–18596. [Google Scholar] [CrossRef] [Green Version]
- Iso, T.; Sartorelli, V.; Poizat, C.; Iezzi, S.; Wu, H.Y.; Chung, G.; Kedes, L.; Hamamori, Y. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol. Cell. Biol. 2001, 21, 6080–6089. [Google Scholar] [CrossRef] [Green Version]
- Chin, M.T.; Maemura, K.; Fukumoto, S.; Jain, M.K.; Layne, M.D.; Watanabe, M.; Hsieh, C.M.; Lee, M.E. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. J. Biol. Chem. 2000, 275, 6381–6387. [Google Scholar] [CrossRef] [Green Version]
- Lavery, D.N.; Villaronga, M.A.; Walker, M.M.; Patel, A.; Belandia, B.; Bevan, C.L. Repression of androgen receptor activity by HEYL, a third member of the Hairy/Enhancer-of-split-related family of Notch effectors. J. Biol. Chem. 2011, 286, 17796–17808. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, Y.T.; Nakamura, M.; Hino, N.; Nogami, J.; Tsuji, S.; Sato, T.; Zhang, L.; Tsujikawa, K.; Tanaka, T.; Izawa, K.; et al. Cell-autonomous and redundant roles of Hey1 and HeyL in muscle stem cells: HeyL requires Hes1 to bind diverse DNA sites. Development 2019, 146, dev163618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, B.G.; Solum, D.; Song, E.J.; Lee, K.J.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell 2004, 119, 815–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasai, Y.; Kageyama, R.; Tagawa, Y.; Shigemoto, R.; Nakanishi, S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992, 6, 2620–2634. [Google Scholar] [CrossRef] [Green Version]
- Ross, D.A.; Hannenhalli, S.; Tobias, J.W.; Cooch, N.; Shiekhattar, R.; Kadesch, T. Functional analysis of Hes-1 in preadipocytes. Mol. Endocrinol. 2006, 20, 698–705. [Google Scholar] [CrossRef] [Green Version]
- Kawamata, S.; Du, C.; Li, K.; Lavau, C. Overexpression of the Notch target genes Hes in vivo induces lymphoid and myeloid alterations. Oncogene 2002, 21, 3855–3863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murato, Y.; Yamaguti, M.; Katamura, M.; Cho, K.W.; Hashimoto, C. Two modes of action by which Xenopus hairy2b establishes tissue demarcation in the Spemann-Mangold organizer. Int. J. Dev. Biol. 2006, 50, 463–471. [Google Scholar] [CrossRef] [Green Version]
- Akazawa, C.; Sasai, Y.; Nakanishi, S.; Kageyama, R. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J. Biol. Chem. 1992, 267, 21879–21885. [Google Scholar] [CrossRef]
- Gao, X.; Chandra, T.; Gratton, M.O.; Quelo, I.; Prud’homme, J.; Stifani, S.; St-Arnaud, R. HES6 acts as a transcriptional repressor in myoblasts and can induce the myogenic differentiation program. J. Cell. Biol. 2001, 154, 1161–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cossins, J.; Vernon, A.E.; Zhang, Y.; Philpott, A.; Jones, P.H. Hes6 regulates myogenic differentiation. Development 2002, 129, 2195–2207. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Bessho, Y.; Hojo, M.; Kageyama, R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development 2000, 127, 2933–2943. [Google Scholar] [CrossRef]
- Gratton, M.O.; Torban, E.; Jasmin, S.B.; Theriault, F.M.; German, M.S.; Stifani, S. Hes6 promotes cortical neurogenesis and inhibits Hes1 transcription repression activity by multiple mechanisms. Mol. Cell Biol. 2003, 23, 6922–6935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bessho, Y.; Miyoshi, G.; Sakata, R.; Kageyama, R. Hes7: A bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells 2001, 6, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, T.; Mizuhara, E.; Minaki, Y.; Sakamoto, Y.; Ono, Y. Helt, a novel basic-helix-loop-helix transcriptional repressor expressed in the developing central nervous system. J. Biol. Chem. 2004, 279, 16356–16367. [Google Scholar] [CrossRef] [Green Version]
- Ryan, D.P.; Duncan, J.L.; Lee, C.; Kuchel, P.W.; Matthews, J.M. Assembly of the oncogenic DNA-binding complex LMO2-Ldb1-TAL1-E12. Proteins: Struct. Funct. Bioinform. 2007, 70, 1461–1474. [Google Scholar] [CrossRef]
- Fairman, R.; Beran-Steed, R.K.; Anthony-Cahill, S.J.; Lear, J.D.; Stafford, W.F., III; DeGrado, W.F.; Benfield, P.A.; Brenner, S.L. Multiple oligomeric states regulate the DNA binding of helix-loop-helix peptides. Proc. Natl. Acad. Sci. USA 1993, 90, 10429–10433. [Google Scholar] [CrossRef] [Green Version]
- Jolma, A.; Yin, Y.; Nitta, K.; Dave, K.; Popov, A.; Taipale, M.; Enge, M.; Kivioja, T.; Morgunova, E.; Taipale, J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 2015, 527, 384–388. [Google Scholar] [CrossRef]
- Slattery, M.; Riley, T.; Liu, P.; Abe, N.; Gomez-Alcala, P.; Dror, I.; Zhou, T.; Rohs, R.; Honig, B.; Bussemaker, H.J.; et al. Cofactor Binding Evokes Latent Differences in DNA Binding Specificity between Hox Proteins. Cell 2011, 147, 1270–1282. [Google Scholar] [CrossRef] [Green Version]
- El Omari, K.; Hoosdally, S.J.; Tuladhar, K.; Karia, D.; Hall-Ponselé, E.; Platonova, O.; Vyas, P.; Patient, R.; Porcher, C.; Mancini, E.J. Structural Basis for LMO2-Driven Recruitment of the SCL:E47bHLH Heterodimer to Hematopoietic-Specific Transcriptional Targets. Cell Rep. 2013, 4, 135–147. [Google Scholar] [CrossRef] [Green Version]
- Matthews, J.M.; Lester, K.; Joseph, S.; Curtis, D.J. LIM-domain-only proteins in cancer. Nat. Rev. Cancer 2013, 13, 111–122. [Google Scholar] [CrossRef]
- Langlands, K.; Yin, X.; Anand, G.; Prochownik, E.V. Differential Interactions of Id Proteins with Basic-Helix-Loop-Helix Transcription Factors. J. Biol. Chem. 1997, 272, 19785–19793. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Cserjesi, P.; Olson, E.N. Dermo-1: A novel twist-related bHLH protein expressed in the developing dermis. Dev. Biol. 1995, 172, 280–292. [Google Scholar] [CrossRef] [Green Version]
- Spinner, D.S.; Liu, S.; Wang, S.W.; Schmidt, J. Interaction of the myogenic determination factor myogenin with E12 and a DNA target: Mechanism and kinetics. J. Mol. Biol. 2002, 317, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Takagi, A.; Hiraoka, S.; Koseki, H.; Kanno, J.; Rawls, A.; Saga, Y. Transcription factors Mesp2 and Paraxis have critical roles in axial musculoskeletal formation. Dev. Dyn. 2007, 236, 1484–1494. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Z.J.; Qin, Y.; Shi, Y.; Wang, S.; Choi, Y.; Simpson, J.L.; Rajkovic, A. Transcription factor FIGLA is mutated in patients with premature ovarian failure. Am. J. Hum. Genet. 2008, 82, 1342–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dear, T.N.; Hainzl, T.; Follo, M.; Nehls, M.; Wilmore, H.; Matena, K.; Boehm, T. Identification of interaction partners for the basic-helix-loop-helix protein E47. Oncogene 1997, 14, 891–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muir, T.; Sadler-Riggleman, I.; Skinner, M.K. Role of the basic helix-loop-helix transcription factor, scleraxis, in the regulation of Sertoli cell function and differentiation. Mol. Endocrinol. 2005, 19, 2164–2174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muir, T.; Wilson-Rawls, J.; Stevens, J.D.; Rawls, A.; Schweitzer, R.; Kang, C.; Skinner, M.K. Integration of CREB and bHLH transcriptional signaling pathways through direct heterodimerization of the proteins: Role in muscle and testis development. Mol. Reprod. Dev. 2008, 75, 1637–1652. [Google Scholar] [CrossRef] [Green Version]
- Blanar, M.A.; Crossley, P.H.; Peters, K.G.; Steingrimsson, E.; Copeland, N.G.; Jenkins, N.A.; Martin, G.R.; Rutter, W.J. Meso1, a basic-helix-loop-helix protein involved in mammalian presomitic mesoderm development. Proc. Natl. Acad. Sci. USA 1995, 92, 5870–5874. [Google Scholar] [CrossRef] [Green Version]
- Palumbo-Zerr, K.; Soare, A.; Zerr, P.; Liebl, A.; Mancuso, R.; Tomcik, M.; Sumova, B.; Dees, C.; Chen, C.-W.; Wohlfahrt, T.; et al. Composition of TWIST1 dimers regulates fibroblast activation and tissue fibrosis. Ann. Rheum. Dis. 2016, 76, 244–251. [Google Scholar] [CrossRef]
- Firulli, B.A.; Krawchuk, D.; Centonze, V.E.; Vargesson, N.; Virshup, D.M.; Conway, S.J.; Cserjesi, P.; Laufer, E.; Firulli, A.B. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat. Genet. 2005, 37, 373–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teachenor, R.; Beck, K.; Wright, L.Y.T.; Shen, Z.; Briggs, S.P.; Murre, C. Biochemical and Phosphoproteomic Analysis of the Helix-Loop-Helix Protein E47. Mol. Cell. Biol. 2012, 32, 1671–1682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kophengnavong, T.; Michnowicz, J.E.; Blackwell, T.K. Establishment of distinct MyoD, E2A, and twist DNA binding specificities by different basic region-DNA conformations. Mol. Cell Biol. 2000, 20, 261–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connerney, J.; Andreeva, V.; Leshem, Y.; Muentener, C.; Mercado, M.A.; Spicer, D.B. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev. Dyn. 2006, 235, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- Firulli, B.A.; Redick, B.A.; Conway, S.J.; Firulli, A.B. Mutations within Helix I of Twist1 Result in Distinct Limb Defects and Variation of DNA Binding Affinities. J. Biol. Chem. 2007, 282, 27536–27546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Firulli, B.A.; Howard, M.J.; McDaid, J.R.; McIlreavey, L.; Dionne, K.M.; Centonze, V.E.; Cserjesi, P.; Virshup, D.M.; Firulli, A.B. PKA, PKC, and the Protein Phosphatase 2A Influence HAND Factor Function: A Mechanism for Tissue-Specific Transcriptional Regulation. Mol. Cell 2003, 12, 1225–1237. [Google Scholar] [CrossRef]
- Hill, A.A.; Riley, P.R. Differential regulation of Hand1 homodimer and Hand1-E12 heterodimer activity by the cofactor FHL2. Mol. Cell Biol. 2004, 24, 9835–9847. [Google Scholar] [CrossRef] [Green Version]
- Topno, N.S.; Kannan, M.; Krishna, R. Mechanistic insights into the activity of Ptf1-p48 (pancreas transcription factor 1a): Probing the interactions levels of Ptf1-p48 with E2A-E47 (transcription factor E2-alpha) and ID3 (inhibitor of DNA binding 3). J. Biomol. Struct. Dyn. 2017, 36, 1834–1852. [Google Scholar] [CrossRef]
- Meredith, D.M.; Masui, T.; Swift, G.H.; Macdonald, R.J.; Johnson, J.E. Multiple Transcriptional Mechanisms Control Ptf1a Levels during Neural Development Including Autoregulation by the PTF1-J Complex. J. Neurosci. 2009, 29, 11139–11148. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Chu, K.; Kim, H.J.; Seong, H.A.; Park, K.C.; Sanyal, S.; Takeda, J.; Ha, H.; Shong, M.; Tsai, M.J.; et al. Orphan nuclear receptor small heterodimer partner, a novel corepressor for a basic helix-loop-helix transcription factor BETA2/neuroD. Mol. Endocrinol. 2004, 18, 776–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, A.; Guanga, G.P.; Rose, R.B. Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: Heterodimer selectivity and DNA recognition. Biochemistry 2008, 47, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, R.; Yasuhara, N.; Oe, S.; Nagai, M.; Yoneda, Y. Synergistic nuclear import of NeuroD1 and its partner transcription factor, E47, via heterodimerization. Exp. Cell Res. 2009, 315, 1639–1652. [Google Scholar] [CrossRef]
- Ray, S.K.; Leiter, A.B. The Basic Helix-Loop-Helix Transcription Factor NeuroD1 Facilitates Interaction of Sp1 with the Secretin Gene Enhancer. Mol. Cell. Biol. 2007, 27, 7839–7847. [Google Scholar] [CrossRef] [Green Version]
- Gradwohl, G.; Fode, C.; Guillemot, F. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev. Biol. 1996, 180, 227–241. [Google Scholar] [CrossRef]
- Qiu, Y.; Guo, M.; Huang, S.; Stein, R. Acetylation of the BETA2 Transcription Factor by p300-associated Factor Is Important in Insulin Gene Expression. J. Biol. Chem. 2004, 279, 9796–9802. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-K.; Pfaff, S.L. Synchronization of Neurogenesis and Motor Neuron Specification by Direct Coupling of bHLH and Homeodomain Transcription Factors. Neuron 2003, 38, 731–745. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, C.; Akazawa, C.; Nakanishi, S.; Kageyama, R. MATH-2, a mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal, is specifically expressed in the nervous system. Eur. J. Biochem. 1995, 229, 239–248. [Google Scholar]
- Cheng, Y.-F.; Tong, M.; Edge, A.S.B. Destabilization of Atoh1 by E3 Ubiquitin Ligase Huwe1 and Casein Kinase 1 Is Essential for Normal Sensory Hair Cell Development. J. Biol. Chem. 2016, 291, 21096–21109. [Google Scholar] [CrossRef] [Green Version]
- Roark, R.; Itzhaki, L.; Philpott, A. Complex regulation controls Neurogenin3 proteolysis. Biol. Open 2012, 1, 1264–1272. [Google Scholar] [CrossRef] [Green Version]
- Atac, D.G.; Koller, S.; Hanson, J.V.M.; Feil, S.; Tiwari, A.; Bahr, A.; Baehr, L.; Magyar, I.; Kottke, R.; Gerth-Kahlert, C.; et al. Atonal homolog 7 (ATOH7) loss-of-function mutations in predominant bilateral optic nerve hypoplasia. Hum. Mol. Genet. 2019, 29, 132–148. [Google Scholar] [CrossRef] [PubMed]
- Lemercier, C.; To, R.Q.; Swansonbc, B.J.; Lyons, G.E.; Konieczny, S.F. Mist1: A Novel Basic Helix-Loop-Helix Transcription Factor Exhibits a Developmentally Regulated Expression Pattern. Dev. Biol. 1997, 182, 101–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meierhans, D.; El-Ariss, C.; Neuenschwander, M.; Sieber, M.; Stackhouse, J.F.; Allemann, R.K. DNA Binding Specificity of the Basic-Helix-Loop-Helix Protein MASH-1. Biochemistry 1995, 34, 11026–11036. [Google Scholar] [CrossRef] [PubMed]
- Sriuranpong, V.; Borges, M.W.; Strock, C.L.; Nakakura, E.K.; Watkins, D.N.; Blaumueller, C.M.; Nelkin, B.D.; Ball, D.W. Notch Signaling Induces Rapid Degradation of Achaete-Scute Homolog. Mol. Cell Biol. 2002, 22, 3129–3139. [Google Scholar] [CrossRef] [Green Version]
- Hsu, H.L.; Cheng, J.T.; Chen, Q.; Baer, R. Enhancer-binding activity of the tal-1 oncoprotein in association with the E47/E12 helix-loop-helix proteins. Mol. Cell Biol. 1991, 11, 3037–3042. [Google Scholar]
- Xia, Y.; Hwang, L.Y.; Cobb, M.; Baer, R. Products of the TAL2 oncogene in leukemic T cells: bHLH phosphoproteins with DNA-binding activity. Oncogene 1994, 9, 1437–1446. [Google Scholar] [PubMed]
- Brown, L.; Baer, R. HEN1 encodes a 20-kilodalton phosphoprotein that binds an extended E-box motif as a homodimer. Mol. Cell Biol. 1994, 14, 1245–1255. [Google Scholar]
- Lu, J.; Richardson, J.A.; Olson, E.N. Capsulin: A novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mech. Dev. 1998, 73, 23–32. [Google Scholar] [CrossRef]
- Miyagishi, M.; Nakajima, T.; Fukamizu, A. Molecular characterization of mesoderm-restricted basic helix-loop-helix protein, POD-1/Capsulin. Int. J. Mol. Med. 2000, 5, 27–31. [Google Scholar] [CrossRef]
- Azmi, S.; Sun, H.; Ozog, A.; Taneja, R. mSharp-1/DEC2, a Basic Helix-Loop-Helix Protein Functions as a Transcriptional Repressor of E Box Activity and Stra13 Expression. J. Biol. Chem. 2003, 278, 20098–20109. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, W.; Wang, J.; Malovannaya, A.; Xi, Y.; Li, W.; Guerra, R.; Hawke, D.H.; Qin, J.; Chen, J. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol. Syst. Biol. 2015, 11, 775. [Google Scholar] [CrossRef] [PubMed]
- Kotlyar, M.; Pastrello, C.; Pivetta, F.; Sardo, A.L.; Cumbaa, C.; Li, H.; Naranian, T.; Niu, Y.; Ding, Z.; Vafaee, F.; et al. In silico prediction of physical protein interactions and characterization of interactome orphans. Nat. Methods 2014, 12, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Sepp, M.; Pruunsild, P.; Timmusk, T. Pitt–Hopkins syndrome-associated mutations in TCF4 lead to variable impairment of the transcription factor function ranging from hypomorphic to dominant-negative effects. Hum. Mol. Genet. 2012, 21, 2873–2888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, H.L.; Huang, L.; Tsan, J.T.; Funk, W.; Wright, W.E.; Hu, J.S.; Kingston, R.E.; Baer, R. Preferred sequences for DNA recognition by the TAL1 helix-loop-helix proteins. Mol. Cell Biol. 1994, 14, 1256–1265. [Google Scholar]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- Sun, X.H.; Baltimore, D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell 1991, 64, 459–470. [Google Scholar] [CrossRef]
- Neuhold, L.A.; Wold, B. HLH forced dimers: Tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell 1993, 74, 1033–1042. [Google Scholar] [CrossRef]
- Thayer, M.J.; Weintraub, H. A cellular factor stimulates the DNA-binding activity of MyoD and E47. Proc. Natl. Acad. Sci. USA 1993, 90, 6483–6487. [Google Scholar] [CrossRef] [Green Version]
- Blackwell, T.K.; Weintraub, H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 1990, 250, 1104–1110. [Google Scholar] [CrossRef] [Green Version]
- Wendt, H.; Thomas, R.M.; Ellenberger, T. DNA-mediated folding and assembly of MyoD-E47 heterodimers. J. Biol. Chem. 1998, 273, 5735–5743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lingbeck, J.M.; Trausch-Azar, J.S.; Ciechanover, A.; Schwartz, A.L. E12 and E47 modulate cellular localization and proteasome-mediated degradation of MyoD and Id1. Oncogene 2005, 24, 6376–6384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molkentin, J.D.; Black, B.L.; Martin, J.F.; Olson, E.N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 1995, 83, 1125–1136. [Google Scholar] [CrossRef] [Green Version]
- Hsu, H.L.; Wadman, I.; Baer, R. Formation of in vivo complexes between the TAL1 and E2A polypeptides of leukemic T cells. Proc. Natl. Acad. Sci. USA 1994, 91, 3181–3185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wadman, I.; Li, J.; Bash, R.; Forster, A.; Osada, H.; Rabbitts, T.; Baer, R. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J. 1994, 13, 4831–4839. [Google Scholar] [CrossRef]
- Fu, J.; Qin, L.; He, T.; Qin, J.; Hong, J.; Wong, J.; Liao, L.; Xu, J. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011, 21, 275–289. [Google Scholar] [CrossRef] [Green Version]
- Sato, F.; Kawamoto, T.; Fujimoto, K.; Noshiro, M.; Honda, K.K.; Honma, S.; Honma, K.; Kato, Y. Functional analysis of the basic helix-loop-helix transcription factor DEC1 in circadian regulation. Interaction with BMAL1. Eur. J. Biochem. 2004, 271, 4409–4419. [Google Scholar] [CrossRef]
- Teo, Z.; Chan, J.S.K.; Chong, H.C.; Sng, M.K.; Choo, C.C.; Phua, G.Z.M.; Teo, D.J.R.; Zhu, P.; Choong, C.; Wong, M.T.C.; et al. Angiopoietin-like 4 induces a beta-catenin-mediated upregulation of ID3 in fibroblasts to reduce scar collagen expression. Sci. Rep. 2017, 7, 6303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cakouros, D.; Isenmann, S.; Hemming, S.E.; Menicanin, D.; Camp, E.; Zannetinno, A.C.W.; Gronthos, S.; Zannettino, A.C.W. Novel Basic Helix–Loop–Helix Transcription Factor Hes4 Antagonizes the Function of Twist-1 to Regulate Lineage Commitment of Bone Marrow Stromal/Stem Cells. Stem Cells Dev. 2015, 24, 1297–1308. [Google Scholar] [CrossRef]
- Ghosh, B.; Leach, S.D. Interactions between Hairy/Enhancer of Split-related proteins and the pancreatic transcription factor Ptf1-p48 modulate function of the PTF1 transcriptional complex. Biochem. J. 2006, 393, 679–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taelman, V.; Van Wayenbergh, R.; Sölter, M.; Pichon, B.; Pieler, T.; Christophe, D.; Bellefroid, E.J. Sequences downstream of the bHLH domain of the Xenopus hairy-related transcription factor-1 act as an extended dimerization domain that contributes to the selection of the partners. Dev. Biol. 2004, 276, 47–63. [Google Scholar] [CrossRef]
- Solter, M.; Locker, M.; Boy, S.; Taelman, V.; Bellefroid, E.J.; Perron, M.; Pieler, T. Characterization and function of the bHLH-O protein XHes2: Insight into the mechanisms controlling retinal cell fate decision. Development 2006, 133, 4097–4108. [Google Scholar] [CrossRef] [Green Version]
- Liu, A.; Li, J.; Marin-Husstege, M.; Kageyama, R.; Fan, Y.; Gelinas, C.; Casaccia-Bonnefil, P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: Old partners and new players. EMBO J. 2006, 25, 4833–4842. [Google Scholar] [CrossRef] [PubMed]
- Wende, C.-Z.; Zoubaa, S.; Blak, A.; Echevarría, D.; Martinez, S.; Guillemot, F.; Wurst, W.; Guimera, J. Hairy/Enhancer-of-Split MEGANE and Proneural MASH1 Factors Cooperate Synergistically in Midbrain GABAergic Neurogenesis. PLoS ONE 2015, 10, e0127681. [Google Scholar] [CrossRef] [PubMed]
- Sotoca, A.M.; Prange, K.H.; Reijnders, B.; Mandoli, A.; Nguyen, L.N.; Stunnenberg, H.G.; Martens, J.H. The oncofusion protein FUS–ERG targets key hematopoietic regulators and modulates the all-trans retinoic acid signaling pathway in t(16;21) acute myeloid leukemia. Oncogene 2015, 35, 1965–1976. [Google Scholar] [CrossRef] [Green Version]
- Springhorn, J.P.; Singh, K.; Kelly, R.A.; Smith, T.W. Posttranscriptional regulation of Id1 activity in cardiac muscle. Alternative splicing of novel Id1 transcript permits homodimerization. J. Biol. Chem. 1994, 269, 5132–5136. [Google Scholar] [CrossRef]
- Guo, S.-J.; Hu, J.-G.; Zhao, B.-M.; Shen, L.; Wang, R.; Zhou, J.-S.; Lü, H.-Z. Olig1 and ID4 interactions in living cells visualized by bimolecular fluorescence complementation technique. Mol. Biol. Rep. 2010, 38, 4637–4642. [Google Scholar] [CrossRef]
- Ellenberger, T.; Fass, D.; Arnaud, M.; Harrison, S.C. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994, 8, 970–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Horton, J.R.; Li, J.; Huang, Y.; Zhang, X.; Blumenthal, R.M.; Cheng, X. Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine. Nucleic Acids Res. 2019, 47, 8375–8387. [Google Scholar] [CrossRef]
- Ma, P.C.; Rould, M.A.; Weintraub, H.; Pabo, C.O. Crystal structure of MyoD bHLH domain-DNA complex: Perspectives on DNA recognition and implications for transcriptional activation. Cell 1994, 77, 451–459. [Google Scholar] [CrossRef]
- Liu, Y.; Watanabe, H.; Nifuji, A.; Yamada, Y.; Olson, E.N.; Noda, M. Overexpression of a single helix-loop-helix-type transcription factor, scleraxis, enhances aggrecan gene expression in osteoblastic osteosarcoma ROS17/2.8 cells. J. Biol. Chem. 1997, 272, 29880–29885. [Google Scholar] [CrossRef] [Green Version]
- Centonze, V.E.; Firulli, B.A.; Firulli, A.B. Fluorescence resonance energy transfer (FRET) as a method to calculate the dimerization strength of basic Helix-Loop-Helix (bHLH) proteins. Biol. Proced. Online 2004, 6, 78–82. [Google Scholar] [CrossRef] [Green Version]
- Aguado-Llera, D.; Goormaghtigh, E.; de Geest, N.; Quan, X.J.; Prieto, A.; Hassan, B.A.; Gomez, J.; Neira, J.L. The basic helix-loop-helix region of human neurogenin 1 is a monomeric natively unfolded protein which forms a “fuzzy” complex upon DNA binding. Biochemistry 2010, 49, 1577–1589. [Google Scholar] [CrossRef]
- Zhu, L.; Tran, T.; Rukstalis, J.M.; Sun, P.; Damsz, B.; Konieczny, S.F. Inhibition of Mist1 homodimer formation induces pancreatic acinar-to-ductal metaplasia. Mol. Cell Biol. 2004, 24, 2673–2681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Künne, A.G.E.; Sieber, M.; Meierhans, A.D.; Allemann, R.K. Thermodynamics of the DNA Binding Reaction of Transcription Factor MASH-1. Biochemistry 1998, 37, 4217–4223. [Google Scholar] [CrossRef]
- Oasa, S.; Vukojevic, V.; Rigler, R.; Tsigelny, I.F.; Changeux, J.-P.; Terenius, L. A strategy for designing allosteric modulators of transcription factor dimerization. Proc. Natl. Acad. Sci. USA 2020, 117, 2683–2686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Choi, G.; Anderson, D.J. The bHLH Transcription Factor Olig2 Promotes Oligodendrocyte Differentiation in Collaboration with Nkx2. Neuron 2001, 31, 791–807. [Google Scholar] [CrossRef] [Green Version]
- Wright, W.E.; Binder, M.; Funk, W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol. Cell Biol. 1991, 11, 4104–4110. [Google Scholar]
- Xing, S.; Wallmeroth, N.; Berendzen, K.; Grefen, C. Techniques for the analysis of protein-protein interactions in vivo. Plant Physiol. 2016, 171, 727–758. [Google Scholar] [CrossRef] [Green Version]
- Podobnik, M.; Krasevec, N.; Bedina Zavec, A.; Naneh, O.; Flasker, A.; Caserman, S.; Hodnik, V.; Anderluh, G. How to Study Protein-protein Interactions. Acta Chim. Slov. 2016, 63, 424–439. [Google Scholar] [CrossRef] [Green Version]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Huttlin, E.L.; Bruckner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 2021, 184, 3022–3040.e28. [Google Scholar] [CrossRef] [PubMed]
- Rolland, T.; Taşan, M.; Charloteaux, B.; Pevzner, S.J.; Zhong, Q.; Sahni, N.; Yi, S.; Lemmens, I.; Fontanillo, C.; Mosca, R.; et al. A Proteome-Scale Map of the Human Interactome Network. Cell 2014, 159, 1212–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kageyama, R.; Ohtsuka, T.; Kobayashi, T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development 2007, 134, 1243–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calkhoven, C.; Ab, G. Multiple steps in the regulation of transcription-factor level and activity. Biochem. J. 1996, 317, 329–342. [Google Scholar] [CrossRef]
- Ma, Q.; Kintner, C.; Anderson, D.J. Identification of neurogenin, a Vertebrate Neuronal Determination Gene. Cell 1996, 87, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Bramblett, D.E.; Pennesi, M.E.; Wu, S.M.; Tsai, M.-J. The Transcription Factor Bhlhb4 Is Required for Rod Bipolar Cell Maturation. Neuron 2004, 43, 779–793. [Google Scholar] [CrossRef] [Green Version]
- Yamada, M.; Shida, Y.; Takahashi, K.; Tanioka, T.; Nakano, Y.; Tobe, T.; Yamada, M. Prg1 is regulated by the basic helix-loop-helix transcription factor Math2. J. Neurochem. 2008, 106, 2375–2384. [Google Scholar] [CrossRef] [PubMed]
- Fong, A.P.; Yao, Z.; Zhong, J.W.; Johnson, N.M.; Farr, G.H., III; Maves, L.; Tapscott, S.J. Conversion of MyoD to a neurogenic factor: Binding site specificity determines lineage. Cell Rep. 2015, 10, 1937–1946. [Google Scholar] [CrossRef] [Green Version]
- Weng, P.-L.; Vinjamuri, M.; Ovitt, M.V.C.E. Ascl3 transcription factor marks a distinct progenitor lineage for non-neuronal support cells in the olfactory epithelium. Sci. Rep. 2016, 6, 38199. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Shahi, P.; Huang, J.T.W.; Phan, N.N.; Sun, Z.; Lin, Y.-C.; Lai, M.-D.; Werb, Z. Systematic analysis of the achaete-scute complex-like gene signature in clinical cancer patients. Mol. Clin. Oncol. 2016, 6, 7–18. [Google Scholar] [CrossRef] [Green Version]
- Al-Hattab, D.S.; Safi, H.A.; Nagalingam, R.S.; Bagchi, R.A.; Stecy, M.T.; Czubryt, M.P. Scleraxis regulates Twist1 and Snai1 expression in the epithelial-to-mesenchymal transition. Am. J. Physiol. Circ. Physiol. 2018, 315, H658–H668. [Google Scholar] [CrossRef]
- Ramirez-Aragon, M.; Hernandez-Sanchez, F.; Rodriguez-Reyna, T.S.; Buendia-Roldan, I.; Guitron-Castillo, G.; Nunez-Alvarez, C.A.; Hernandez-Ramirez, D.F.; Benavides-Suarez, S.A.; Esquinca-Gonzalez, A.; Torres-Machorro, A.L.; et al. The Transcription Factor SCX is a Potential Serum Biomarker of Fibrotic Diseases. Int. J. Mol. Sci. 2020, 21, 5012. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, C.; Alonso, M. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. EMBO J. 1991, 10, 2965–2973. [Google Scholar] [CrossRef] [PubMed]
- Van Doren, M.; Bailey, A.M.; Esnayra, J.; Ede, K.; Posakony, J.W. Negative regulation of proneural gene activity: Hairy is a direct transcriptional repressor of achaete. Genes Dev. 1994, 8, 2729–2742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [Green Version]
- Iso, T.; Kedes, L.; Hamamori, Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J. Cell Physiol. 2003, 194, 237–255. [Google Scholar] [CrossRef]
- Atchley, W.R.; Wollenberg, K.R.; Fitch, W.M.; Terhalle, W.; Dress, A.W. Correlations among Amino Acid Sites in bHLH Protein Domains: An Information Theoretic Analysis. Mol. Biol. Evol. 2000, 17, 164–178. [Google Scholar] [CrossRef] [Green Version]
- Maia, A.M.; da Silva, J.H.; Mencalha, A.L.; Caffarena, E.R.; Abdelhay, E. Computational modeling of the bHLH domain of the transcription factor TWIST1 and R118C, S144R and K145E mutants. BMC Bioinform. 2012, 13, 184. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Schafer, N.; Davtyan, A.; Papoian, G.; Wolynes, P.G. Predictive energy landscapes for protein-protein association. Proc. Natl. Acad. Sci. USA 2012, 109, 19244–19249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgoulia, P.S.; Bjelic, S. Prediction of Protein–Protein Binding Interactions in Dimeric Coiled Coils by Information Contained in Folding Energy Landscapes. Int. J. Mol. Sci. 2021, 22, 1368. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr.; Pallas, D.C.; DeCaprio, J.A.; Kaye, F.J.; Livingston, D.M. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell 1991, 64, 521–532. [Google Scholar] [CrossRef]
- Wang, P.; Wilson, S.R. Mass spectrometry-based protein identification by integrating de novo sequencing with database searching. BMC Bioinform. 2013, 14, S24. [Google Scholar] [CrossRef] [Green Version]
- Pfaff, S.J.; Chimenti, M.S.; Kelly, M.J.S.; Arkin, M.R. Biophysical Methods for Identifying Fragment-Based Inhibitors of Protein-Protein Interactions. Protein-Protein Interact. 2015, 1278, 587–613. [Google Scholar] [CrossRef]
- Zhou, M.; Li, Q.; Wang, R. Current Experimental Methods for Characterizing Protein-Protein Interactions. ChemMedChem 2016, 11, 738–756. [Google Scholar] [CrossRef]
- Dey, B.; Thukral, S.; Krishnan, S.; Chakrobarty, M.; Gupta, S.; Manghani, C.; Rani, V. DNA-protein interactions: Methods for detection and analysis. Mol. Cell Biochem. 2012, 365, 279–299. [Google Scholar] [CrossRef]
- Hall, R.A. Studying Protein-Protein Interactions via Blot Overlay/Far Western Blot. Protein-Protein Interact. 2015, 1278, 371–379. [Google Scholar] [CrossRef]
- Takahashi, Y. Co-immunoprecipitation from Transfected Cells. Protein-Protein Interact. 2015, 1278, 381–389. [Google Scholar] [CrossRef]
- Fields, S.; Song, O. A novel genetic system to detect protein-protein interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef]
- Stynen, B.; Tournu, H.; Tavernier, J.; Van Dijck, P. Diversity in Genetic In Vivo Methods for Protein-Protein Interaction Studies: From the Yeast Two-Hybrid System to the Mammalian Split-Luciferase System. Microbiol. Mol. Biol. Rev. 2012, 76, 331–382. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.-D.; Chinenov, Y.; Kerppola, T.K. Visualization of Interactions among bZIP and Rel Family Proteins in Living Cells Using Bimolecular Fluorescence Complementation. Mol. Cell 2002, 9, 789–798. [Google Scholar] [CrossRef]


| bHLH Class | Characteristic | Homodimerization | Heterodimerization | Examples | Activity | PDB ID + |
|---|---|---|---|---|---|---|
| I | E proteins | Yes | Classes I, II, V, and VI TFs | TCF3, TCF4 | A | 6OD4 |
| II | Tissue specific | Yes | Classes I, II, V, and VI TFs | NEUROD1, TWIST1 | A or R | 2QL2 |
| III | LZ domain | * | Classes III and IV | MYC, SRBEF1 | A or R | 2A93 |
| IV | LZ domain | * | Classes III and IV | MAD, MAX | A or R | 1R05 |
| V | No basic domain | No | Classes I, II, V, and VI | ID1, ID4 | R | 6MGN |
| VI | Proline in the basic domain | Yes | Classes I, II, V, and VI | HES1, HEY1 | R | 2MH3 |
| VII | PAS domain | No | Class VII | ARNT, HIF1A | A or R | 5SY7 |
| HGNC Gene Symbol and Aliases (a) | Classification | Function | Organism (b) | ||
|---|---|---|---|---|---|
| [42,44] | [21,22,36] | [47,48] | As homodimers and/or heterodimers | ||
| Class I | |||||
| TCF3/E47 (E2-5)/ITF1 | Group A | Class I | BHLH2/B | E2A: A [52,53,54,55], E2-5 [56], E47 [57]. CR [33,50,58,59] | Hs, Mm, Rn |
| TCF3/E12 | Group A | Class I | BHLH2/B | A [33,50,57,60] | Hs, Mm, Rn, Gg |
| TCF4/E2-2A/ITF2 | Group A | Class I | BHLH2/B | A [52,53,56,57,60,61]. CR [62,63] | Hs, Mm |
| TCF4/E2-2B | Group A | Class I | BHLH2/B | CR [62,63] | Hs, Mm |
| HEB/TCF12 | Group A | Class I | BHLH2/B | A [52,57,60,64] | Hs, Mm |
| Class II | |||||
| MYOD1/MYOD/MYF3 | Group A | Class II | BHLH3/C | A [65,66,67,68]. TI [69] | Hs, Mm, Rn, Gg |
| MYOG/MYF4/Myogenin | Group A | Class II | BHLH3/C | A [49,70]. TI [69] | Hs, Mm, Rn |
| MYF5 | Group A | Class II | BHLH3/C | A [71,72] | Hs, Mm |
| MYF6/MRF4/Herculin | Group A | Class II | BHLH3/C | A [49,68,70,73]. TI [69] | Hs, Mm, Rn, Gg |
| MESP1/BHLHC5 | Group A | Class II | BHLH3/C | Mm | |
| MESP2/BHLHC6 | Group A | Class II | BHLH3/C | Mm | |
| FIGLA/FIGα/BHLHC8 | Group A | Class II | BHLH3/C | A [74,75] | Hs, Mm |
| SCX/Scleraxis | Group A | Class II | BHLH1/A | A [76,77,78] | Hs, Mm, Rn, M |
| TCF15/Paraxis/Meso1 | Group A | Class II | BHLH1/A | A [79] | Hs, Mm |
| TWIST1 | Group A | Class II | BHLH1/A | R [80,81,82] | Hs, Mm, Gg |
| TWIST2/DERMO1 | Group A | Class II | BHLH1/A | R [82,83] | Hs, Mm |
| FERD3L/NTWIST | Group A | Class II | BHLH1/A | R [84] | Hs, Mm, Dm |
| HAND1/EHAND/Thing1 | Group A | Class II | BHLH1/A | A [85,86]. R [86,87,88,89,90] | Mm |
| HAND2/DHAND/Thing2 | Group A | Class II | BHLH1/A | A [91]. CR [90,92] | Mm |
| PTF1A/P48 | Group A | Class II | BHLH1/A | A [93,94] | Hs, Mm, Rn, M |
| NEUROD1/BETA2/NEUROD | Group A | Class II | BHLH1/A | A [29,95,96,97,98,99] | Hs, Mm, Ma, Rn, Xl |
| NEUROD2 | Group A | Class II | BHLH1/A | A [97,100] | Hs, Mm |
| NEUROG1/NGN1/NEUROD3/Neurogenin1 | Group A | Class II | BHLH1/A | A [97]. R [101,102] | Hs, Mm, Rn, Xl, Gg (c) |
| NEUROD4/ATOH3/MATH3/NeuroM | Group A | Class II | BHLH1/A | A [97,103] | Mm, Gg, Xl |
| NEUROD6/ATOH2/MATH2/NEX1 | Group A | Class II | BHLH1/A | A [97,104]. R [105] | Hs, Mm, Rn |
| ATOH1/MATH1 | Group A | Class II | BHLH1/A | A [28,97,106] | Mm |
| NEUROG2/ATOH4/MATH4A/Neurogenin2 | Group A | Class II | BHLH1/A | A [97,107]. R [31] | Mm, Gg |
| NEUROG3/ATOH5/MATH4B/Neurogenin3 | Group A | Class II | BHLH1/A | A [97,108]. CR [109] | Mm, Hs |
| ATOH7/MATH5 | Group A | Class II | BHLH1/A | A [110,111] | Hs, Mm, Gg |
| ATOH8/MATH6 | Group A | Class II | BHLH1/A | wA, wR [100,112,113,114] | Hs, Mm |
| BHLHA15/MIST1 | Group A | Class II | BHLH1/A | A [115]. R [116] | Hs, Rn, Mm |
| ASCL1/MASH1 | Group A | Class II | BHLH1/A | A [100,107,117]. R [111] | Mm, Rn, Gg |
| ASCL2/MASH2 | Group A | Class II | BHLH1/A | A [117]. CR [118] | Mm, Rn, Hs |
| ASCL3/SGN1 | Group A | Class II | BHLH1/A | R [119] | Hs, Mm |
| ASCL4/HASH4 | Group A | Class II | BHLH1/A | Hs | |
| ASCL5 | Group A | Class II | BHLH1/A | Hs | |
| TAL1/SCL | Group A | Class II | BHLH1/A | CA, CR [120,121,122,123,124,125] | Hs, Mm |
| TAL2 | Group A | Class II | BHLH1/A | Predicted similar to TAL1 [126] | Hs, Mm |
| LYL1 | Group A | Class II | BHLH1/A | A [127,128]. R [129] | Hs, Mm |
| NHLH1/HEN1/NSCL | Group A | Class II | BHLH1/A | A, R [130] | Hs, Mm |
| NHLH2/HEN2/NSCL2 | Group A | Class II | BHLH1/A | A [131]. R [132] | Hs, Mm |
| MSC/Musculin/ABF-1/MyoR | Group A | Class II | BHLH1/A | R [133,134,135] | Hs, Mm |
| TCF21/Capsulin/POD1 | Group A | Class II | BHLH1/A | A, R [136,137,138,139] | Hs, Mm |
| TCF23/OUT | Group A | Class II | BHLH1/A | R [140] | Mm |
| TCF24/OUT2 | Group A | Class II | BHLH1/A | Hs | |
| BHLHA9/Fingerin/BHLHF42 | Group A | Class II | BHLH1/A | R [141] | Hs, Mm |
| BHLHE22/BHLHB5/BETA3 | Group A | Class II | BHLH5/E | R [142,143,144] | Hs, Mm, Ma |
| BHLHE23/BHLHB4/BETA4 | Group A | Class II | BHLH5/E | R [145] | Mm |
| OLIG1 | Group A | Class II | BHLH5/E | A [146]. R [147] | Hs, Mm |
| OLIG2 | Group A | Class II | BHLH5/E | A [148]. R [149,150] | Mm, Gg, Rn |
| OLIG3 | Group A | Class II | BHLH5/E | R [151] | Mm |
| BHLHE40/SHARP2/STRA13/DEC1 | Group A | Class II | BHLH5/E | R [152,153] | Hs, Mm, Rn |
| BHLHE41/SHARP1/DEC2 | Group A | Class II | BHLH5/E | R [154,155] | Hs, Mm, Rn |
| Class V | |||||
| ID1 | Group D | Class V | BHLH2/B | R [20,156] | Mm, Hs, Rn |
| ID2 | Group D | Class V | BHLH2/B | R [157] | Mm, Hs |
| ID3 | Group D | Class V | BHLH2/B | R [158] | Mm, Hs |
| ID4 | Group D | Class V | BHLH2/B | R [159] | Mm |
| Class VI | |||||
| HEY1/HRT1/CHF2/HERP2/Hesr1 | Group E | Class VI | BHLH2/B | R [160,161] | Hs, Mm |
| HEY2/HRT2/CHF1 gridlock/HERP1 | Group E | Class VI | BHLH2/B | R [161,162] | Hs, Mm, Rn, Gg |
| HEYL/HERP3/HRT3 | Group E | Class VI | BHLH2/B | R [163,164] | Hs, Mm |
| HES1/HRY/Xhairy1 | Group E | Class VI | BHLH2/B | A [165]. R [106,166,167,168] | Hs, Mm, Rn, M |
| HES2 | Group E | Class VI | BHLH2/B | R [12] | Rn, Xl |
| HES3 | Group E | Class VI | BHLH3/C | R [166] | Mm, Hs |
| HES4/Xhairy2 | Group E | Class VI | BHLH2/B | R [169] | Xl, Hs (d) |
| HES5/ESR9 | Group E | Class VI | BHLH2/B | R [168,170] | Rn, Mm, Xl, Gg |
| HES6 | Group E | Class VI | BHLH3/C | R [171,172]. Inhibits Hes1 [173,174] | Hs, Mm, Xl |
| HES7 | Group E | Class VI | BHLH2/B | R [175] | Hs, Mm |
| HELT/MGN/HESL/MEGANE | Group E | Class VI | BHLH2/B | R [176] | Mm, Hs, Rn |
| Color key: | |||||
| Binds DNA as homodimer | Transactivator and repressor | ||||
| Titrates E-proteins | Transactivator (A) | ||||
| Titrates E-proteins and binds DNA as homodimer | Repressor (R)/Context dependent repressor (CR) Transcriptionally inactive (TI) |
| Part A. | Heterodimers with E47 or E12 | |
|---|---|---|
| Class II TFs/Eprot | E47 | E12 |
| MYOD1 | E2A: MS. Er(4), Ek(2), Ei(4), Ee(2), C(2), cIP(2), FS, qY2H, Sd, GST, Y2H, NI | Ei (5), Er, Ek, Ee, C(3), MIF, cIP(2), qY2H, Y2H, MS, NI, ChIP |
| MYOG | Er, Ek, Ei, cIP, qY2H [26,69,183] | Ei(3), Er, ChIP MIF, cIP, Y2H, MS [26,64,73,134,135,184,185] |
| MYF5 | Ei, cIP, ChIP, qY2H [26,183,185] | Ei(2), cIP, MIF, ChIP, qY2H, MS [26,72,135,183,185] |
| MYF6 | Er, Ek, Ei, cIP, qY2H [26,69,183] | Ei, MIF, cIP, ChIP, qY2H [26,183,185] |
| MESP1 | Y2H [186] | |
| MESP2 | Y2H [186] | |
| FIGLA | E2A: Ee(2) [74,75]. Y2H [187] | Ee [74] |
| SCX | cIP(2), Ee(2), Y2H(2), ChIP, Sd [77,78,188,189] | Er, Ei(2), Y2H(2), MS [76,135,184,190] |
| TCF15 | Ei, Y2H, Sd [186,188,191] | Ei (2), cIP [79,190] |
| TWIST1 | E2A: cIP [192], F, MS [193,194] | GST, cIP, Ei(3), cE, C [80,81,195,196,197] |
| TWIST2 | Sd, GST [157,188,193] | Ei, Y2H [76,184] |
| FERD3L | Ei, M2H [84] | |
| HAND1 | GST, Er, Ei(Dbox)(2), cE, C, cIP(2), F [85,86,87,89,198] | Ei (2)(Dbox), cIP(3), F, cE [87,88,198,199] |
| HAND2 | E2A: Y2H, GST, Ee [30]. Y2H, Ei, cIP, F, M2H [91,157,193] | GST, Y2H, M2H, Ei(2), C [91,197] |
| PTF1A | MDS, Ei [93,200] | Ee, Ei(3) [93,94,201] |
| NEUROD1 | Ee(2), Ei, Er, Cr, NI [29,95,98,143,202,203,204] | Ei (3), Ee (2), Er, GST [29,95,98,205,206,207] |
| NEUROD2 | cIP, Ei [25] | |
| NEUROG1 | Ei [101] | |
| NEUROD4 | Y2H, Er [208] | |
| NEUROD6 | ChIP, Er [104,209] | |
| ATOH1 | TCF3:MS [106,210] | |
| NEUROG2 | cIP, ChIP, Y2H, GST [31,150] | GST, Ei(2), cIP [107,206] |
| NEUROG3 | Ei [108] | Ei [211] |
| ATOH7 | ELISA [212] | |
| ATOH8 | cIP, MS [114,194] | |
| BHLHA15 | Ei, Er, cE, GST, MS [115,116,194] | Ei [213] |
| ASCL1 | TCF3: cIP [35] | Ei(2), Er, Y2H, CD, Ek, cIP [107,184,206,214,215] |
| ASCL2 | cIP, Ei [89] | Ei [117] |
| ASCL3 | Y2H, GST, Ei, C [119] | Y2H, GST [119] |
| ASCL4 | ||
| ASCL5 | ||
| TAL1 | E2A: Ei, GST, Y2H, cIP, MS. Ee, Ei(2),C, cIP, ChIP, Cr, Y2H. | Er, GST(2), SEC/MALLS [177,216] |
| TAL2 | Ei [217] | Y2H [184] |
| LYL1 | E2A: cIP(2), Ee, GST [27,129]. cIP, ChIP [127] | Ei [26] |
| NHLH1 | GST, M2H, Ee [130,218] | Ei(2), GST [107,218] |
| NHLH2 | * | |
| MSC | Y2H, Ei [133] | Ei(3), Y2H, MS, GST, cIP [133,134,135] |
| TCF21 | Ei, Y2H(2), M2H, IF [138,219,220] | |
| TCF23 | cIP, cE [140] | |
| TCF24 | ||
| BHLHA9 | E2A: Y2H [141] | |
| BHLHE22 | cIP, cE [143] | cIP, cE [143] |
| BHLHE23 | (b) | |
| OLIG1 | TCF3: cIP (2) [35,146] | cIP [146] |
| OLIG2 | Y2H, GST, Ei, cIP(2) [35,146,150] | cIP [146] |
| OLIG3 | ||
| BHLHE40 | Sd, Y2H, cIP [188] | w: GST [152] |
| BHLHE41 | cIP, cE, GST [154,221] | |
| Part B. Heterodimers with TCF4 or TCF12 | ||
| Class II TFs/Eprot | TCF4 | TCF12 |
| MYOD1 | Ee, Ei, cIP, MS, Fw [26,66,125,135] | Ei, MS [64,135] |
| MYOG | Ei(2), cIP [26,64] | Ei [64] |
| MYF5 | Ei, cIP, ChIP [26,185] | |
| MYF6 | Ei, cIP, ChIP [26,185] | |
| MESP1 | ||
| MESP2 | ||
| FIGLA | Ee [75] | Ee [75] |
| SCX | Y2H [189] | |
| TCF15 | ||
| TWIST1 | MS, GST [222,223] | |
| TWIST2 | ||
| FERD3L | ||
| HAND1 | MS [222] | Ei, Y2H, cIP (Dbox) [87,88] |
| HAND2 | Y2H(2), GST, Ee, Ei, M2H [30,91,157] | Y2H, GST, Ee(nDB) [30] |
| PTF1A | Ei (2) [93,94] | |
| NEUROD1 | Ee [29] | Ee [29] |
| NEUROD2 | Ei(2), NI, cIP [25,224] | cIP, Ei [25] |
| NEUROG1 | ||
| NEUROD4 | ||
| NEUROD6 | ||
| ATOH1 | cIP, Y2H, Ee [28] | MS [210] |
| NEUROG2 | (a) | (a) |
| NEUROG3 | ||
| ATOH7 | ||
| ATOH8 | ||
| BHLHA15 | MS [222] | |
| ASCL1 | Ei, NI, cIP, M2H [35,157,224] | cIP [35] |
| ASCL2 | Ei [89] | Ei [89] |
| ASCL3 | Y2H, GST [119] | Y2H [119] |
| ASCL4 | M2H [36] | |
| ASCL5 | ||
| TAL1 | Ei, C, GST, Fw, qY2H [125,183,225] | Ei, C, MS [124,225] |
| TAL2 | qY2H [183] | |
| LYL1 | qY2H [183] | |
| NHLH1 | ||
| NHLH2 | ||
| MSC | Y2H [133] | Y2H [133] |
| TCF21 | Y2H(2), GST, cIP [219,220] | Y2H(2) [219,220] |
| TCF23 | M2H [36] | |
| TCF24 | MS (2) [222,226] | |
| BHLHA9 | Y2H [141] | Y2H [141] |
| BHLHE22 | ||
| BHLHE23 | ||
| OLIG1 | cIP [35] | cIP [35] |
| OLIG2 | cIP [35] | |
| OLIG3 | w:M2H [36] | |
| BHLHE40 | ||
| BHLHE41 | ||
| Color key: | ||
| Both in vitro and in vivo | (Independent experiments) | |
| Only in vitro assays | ||
| Only EMSA | ||
| Only in vivo assays | ||
| Part A. Class II-Class II Interactions | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MYOD1 | MYOG | MYF5 | MYF6 | TWIST1 | HAND1 | HAND2 | NEUROD1 | NEUROG2 | NEUROG3 | ASCL1 | TAL1 | OLIG1 | BHLHE40 | |
| MYOD1 | ||||||||||||||
| MYOG | ||||||||||||||
| MYF5 | ||||||||||||||
| MYF6 | GST (71) | |||||||||||||
| MESP1 | ||||||||||||||
| MESP2 | ||||||||||||||
| FIGLA | ||||||||||||||
| SCX | ||||||||||||||
| TCF15 | ||||||||||||||
| TWIST1 | cIP, GST [81] | GST [81] | GST [81] | GST [81] | x | |||||||||
| TWIST2 | MS [236] | |||||||||||||
| FERD3L | ||||||||||||||
| HAND1 | M2H [90] | x | cIP [193] | |||||||||||
| HAND2 | cIP, F [193] | * | x | x | ||||||||||
| PTF1A | ||||||||||||||
| NEUROD1 | ||||||||||||||
| NEUROD2 | ||||||||||||||
| NEUROG1 | ||||||||||||||
| NEUROD4 | ||||||||||||||
| NEUROD6 | ||||||||||||||
| ATOH1 | ||||||||||||||
| NEUROG2 | ||||||||||||||
| NEUROG3 | ||||||||||||||
| ATOH7 | ||||||||||||||
| ATOH8 | cIP [100] | x | cIP [100] | |||||||||||
| BHLHA15 | Ei (nDB), GST [116] | x | ||||||||||||
| ASCL1 | GST [90] | x | ** | x | ||||||||||
| ASCL2 | cIP [88] | x | ||||||||||||
| ASCL3 | GST [119] | x | ||||||||||||
| ASCL4 | ||||||||||||||
| ASCL5 | ||||||||||||||
| TAL1 | ||||||||||||||
| TAL2 | ||||||||||||||
| LYL1 | **** | |||||||||||||
| NHLH1 | ||||||||||||||
| NHLH2 | ||||||||||||||
| MSC | ||||||||||||||
| TCF21 | ||||||||||||||
| TCF23 | ||||||||||||||
| TCF24 | ||||||||||||||
| BHLHA9 | w: Y2H [141] | Y2H [141] | x | |||||||||||
| BHLHE22 | ||||||||||||||
| BHLHE23 | ||||||||||||||
| OLIG1 | x | |||||||||||||
| OLIG2 | *** | x | cIP, M2H [148] | |||||||||||
| OLIG3 | ||||||||||||||
| BHLHE40 | cIP [155] | x | GST (nDB) [152] | x | ||||||||||
| BHLHE41 | cIP(2), cE, GST(2) | x | GST [237] | |||||||||||
| Part B. Class II-Class V or VI interactions | ||||||||||||||
| ID1 | ID2 | ID3 | ID4 | HEY1 | HEY2 | HEYL | HES1 | HES2 | HES4 | HES5 | HELT | |||
| MYOD1 | ***** | qY2H, cIP, M2H [183] | cIP, Y2H, cEMSA [158] | cIP, cE [160] | x | w: SSPC [164] | x | |||||||
| MYOG | ||||||||||||||
| MYF5 | qY2H, M2H [183] | qY2H, M2H [183] | M2H [183] | x | ||||||||||
| MYF6 | cIP [183] | cIP [183] | cIP [183] | x | ||||||||||
| MESP1 | ||||||||||||||
| MESP2 | ||||||||||||||
| FIGLA | ||||||||||||||
| SCX | cIP [238] | x | ||||||||||||
| TCF15 | ||||||||||||||
| TWIST1 | cIP [192] | x | cIP [192] | x | cIP [239] | x | ||||||||
| TWIST2 | ||||||||||||||
| FERD3L | ||||||||||||||
| HAND1 | GST [90] | GST [90] | GST [90] | x | ||||||||||
| HAND2 | GST [90] | GST [90] | GST [90] | x | ||||||||||
| PTF1A | MDS [200] | x | cIP [240] | cIP [240] | x | cIP, Y2H, GST [240] | x | |||||||
| NEUROD1 | w: cIP [241] | x | cIP [242] | x | cIP [241] | x | ||||||||
| NEUROD2 | ||||||||||||||
| NEUROG1 | ||||||||||||||
| NEUROD4 | w: cIP [241] | x | cIP [242] | x | cIP [241] | x | ||||||||
| NEUROD6 | ||||||||||||||
| ATOH1 | ||||||||||||||
| NEUROG2 | cIP [241] | x | ||||||||||||
| NEUROG3 | ||||||||||||||
| ATOH7 | ||||||||||||||
| ATOH8 | ||||||||||||||
| BHLHA15 | ||||||||||||||
| ASCL1 | x | cIP(2) [170,243] | +++ | |||||||||||
| ASCL2 | ||||||||||||||
| ASCL3 | ||||||||||||||
| ASCL4 | ||||||||||||||
| ASCL5 | ||||||||||||||
| TAL1 | ||||||||||||||
| TAL2 | ||||||||||||||
| LYL1 | ||||||||||||||
| NHLH1 | ||||||||||||||
| NHLH2 | GST, cIP [132] | x | ||||||||||||
| MSC | ||||||||||||||
| TCF21 | ||||||||||||||
| TCF23 | ||||||||||||||
| TCF24 | ||||||||||||||
| BHLHA9 | ||||||||||||||
| BHLHE22 | ||||||||||||||
| BHLHE23 | ||||||||||||||
| OLIG1 | cIP, b2H, IF [146] | x | + | x | ||||||||||
| OLIG2 | cIP, b2H, IF [146] | x | ++ | x | ||||||||||
| OLIG3 | ||||||||||||||
| BHLHE40 | x | x | ||||||||||||
| BHLHE41 | ||||||||||||||
| Factor | Homodimer | EMSA | 2H | GST | cIP | MS | Biophysical | Other | Function (a) |
|---|---|---|---|---|---|---|---|---|---|
| Class I | Biochemical | ||||||||
| TCF3/E47 | Y | Y: Ei [7,8], Er [227], Ee (2) [30,54] | Y: [194] | Y: Cr [248], FS, CD [231] | Y: C, Ek [227]; MIF [8] | A [52,53,54,55,56,57] | |||
| TCF3/E12 | Y | * | Y: Y2H [184] | Y: [135] | Y: CD [177] | Y: C, Ek [227] (b) | A [52,57,60] | ||
| TCF4 | Y | Y: Er [249], Ei [224] | Y: [222] | Y: Cr, (f) [249] | A [52,56,57,60,61] | ||||
| TCF12 | Y | Y: Ei [64] | A [52,57,60] | ||||||
| Class II | |||||||||
| MYOD1 (e) | Y | ** | w: qY2H [183] | Y: [135] | Y: Cr [250], CD, FS [231] | Y: MIF [70] | TI [69] | ||
| MYOG (e) | Y | Y: Er (2) [72,185], wq: Er [69] | w: qY2H [183] | Y: MIF [70] | TI [69] | ||||
| MYF5 (e) | Y | Y: Er [72] | w: qY2H [183] | Y: MIF [70] | |||||
| MYF6 (e) | Y | w: Er [70], q: Er [69] | N: qY2H [183] | TI [69] | |||||
| MESP1 | ? | N: Y2H [186] | |||||||
| MESP2 | ? | N: Y2H [186] | |||||||
| FIGLA | ? | ||||||||
| SCX | ? | N: Ei, Er [76]; Y: Ei [251] | A [251] | ||||||
| TCF15 | ? | N: Ei (3) [79,190,191] | N: Y2H [186] | ||||||
| TWIST1 | Y | Y: Ei (2) [196,197] | Y: [81] | Y: [236] | Y: [236] | Y: FRET [193], (g) [196] | |||
| TWIST2 | ? | N: Ei [184] | |||||||
| FERD3L | ? | N: Ei [84] | |||||||
| HAND1 | Y | N: Er [86], Ei [87] | Y: M2H, Y2H [90] | Y: [90] | Y: [89,90] | Y: FRET [252], C(nDB) [90] | A? [85,199] | ||
| HAND2 | Y | N: Ei [91], Ee [30] | Y: Y2H w: M2H [91] | Y: [90,91] | Y: [92] | N: C [91] | TI? [91] | ||
| PTF1A | ? | N: Ei [94,201], Ee[93]. | |||||||
| NEUROD1 | ? | ||||||||
| NEUROD2 | ? | ||||||||
| NEUROG1 | ? | Y fuzzy E-box: CD [253] | |||||||
| NEUROD4 | ? | N: Ei [208] | A? [103] | ||||||
| NEUROD6 | Y? | Y: Er [105], Ee [104] | A [105] | ||||||
| ATOH1 | Y | N: Er [106]. Y: Ee [28] | Y: [210] | ||||||
| NEUROG2 | Y | N: Ei [206] | N: Y2H [150] | Y: [31] | Y: ChIP [31](c) | A [31] (i) | |||
| NEUROG3 | N? | N: Ei [108] | N: CD (nDB) [253] | ||||||
| ATOH7 | ? | Y: Er [111] | N: ELISA (h) [212] | A [111] | |||||
| ATOH8 | ? | ||||||||
| BHLHA15 | Y | Y: Ei [115,116,213]; Ee [254] | Y: [116] | Y: [115,254] | Y: C [115], BMFCS [254] | A [115,254]. R [116] | |||
| ASCL1 | Y | Y: Ei [107] | Y: Y2H [244] | Y: [244] | Y: CD [255] | Y: Ek [214] | A [107]. R [111] | ||
| ASCL2 | ? | N: Ei [89] | |||||||
| ASCL3 | Y | N: Ei [119] | Y: Y2H [119] | Y: [119] | N: C [119] | R [119] | |||
| ASCL4 | ? | ||||||||
| ASCL5 | ? | ||||||||
| TAL1 | Y | N: Ei [216], Er [177] | N: qY2H [183] | N: [125,127] | Y: [124] | Y: CD [177] | |||
| TAL2 | ? | N: [217] | N: qY2H [183] | ||||||
| LYL1 | Y | N: qY2H [183] | Y: [127] | Y: [127] | |||||
| NHLH1 | Y | Y: Ei, Ee [130,218] | Y: M2H [130] | Y: [218] | Y: C [218] | A [130] | |||
| NHLH2 | Y | Y: Ee [131] | Y: [131] | A [131] | |||||
| MSC | Y | Y: Ei [133,134] | R [133,134] | ||||||
| TCF21 | N? | N: Ei [219] | N: Y2H [137,220] | ||||||
| TCF23 | ? | ||||||||
| TCF24 | ? | ||||||||
| BHLHA9 | ? | Y: Y2H [141] | |||||||
| BHLHE22 | Y | N: [143] | Y: [142] | Y: ChIP [142](d) | R [142,144] | ||||
| BHLHE23 (c) | ? | R [145] | |||||||
| OLIG1 | Y | Y: Er [147] | R [147] | ||||||
| OLIG2 | Y | Y: Ei [150] | Y: Y2H [150], M2H [148] | Y: [150] | Y: [148] | Y: FCCS [256] | R [149,150,257] | ||
| OLIG3 | ? | ||||||||
| BHLHE40 | Y | Y: Ei [153,155,237] | Y: [153] | R [153,155] | |||||
| BHLHE41 | Y | Y: Ei [155,221] | Y: [155] | R [155,221] | |||||
| Color key: | Experiment | DNA binding | Function | ||||||
| only EMSA | No DNA binding | (Independent experiments) | Transactivator (A) | ||||||
| only in vitro assays | DNA binding | Repressor (R) | |||||||
| only in vivo assays | Opposite DNA binding results | A and R | |||||||
| In vitro and in vivo | (j) | ||||||||
| Untested | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Machorro, A.L. Homodimeric and Heterodimeric Interactions among Vertebrate Basic Helix–Loop–Helix Transcription Factors. Int. J. Mol. Sci. 2021, 22, 12855. https://doi.org/10.3390/ijms222312855
Torres-Machorro AL. Homodimeric and Heterodimeric Interactions among Vertebrate Basic Helix–Loop–Helix Transcription Factors. International Journal of Molecular Sciences. 2021; 22(23):12855. https://doi.org/10.3390/ijms222312855
Chicago/Turabian StyleTorres-Machorro, Ana Lilia. 2021. "Homodimeric and Heterodimeric Interactions among Vertebrate Basic Helix–Loop–Helix Transcription Factors" International Journal of Molecular Sciences 22, no. 23: 12855. https://doi.org/10.3390/ijms222312855
APA StyleTorres-Machorro, A. L. (2021). Homodimeric and Heterodimeric Interactions among Vertebrate Basic Helix–Loop–Helix Transcription Factors. International Journal of Molecular Sciences, 22(23), 12855. https://doi.org/10.3390/ijms222312855