Palm Oil-Rich Diet Affects Murine Liver Proteome and S-Palmitoylome
Abstract
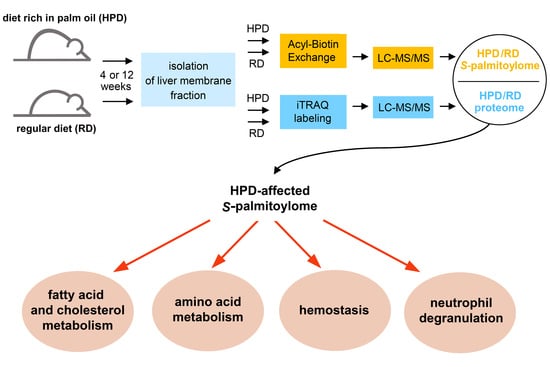
:1. Introduction
2. Results
2.1. Influence of Palm Oil-Rich Diet on Fatty Acid Composition of Liver and Peritoneal Macrophages
2.2. Metabolic Abnormalities of Mice Fed Palm Oil-Rich Diet
2.3. Protein S-Palmitoylation in HPD and RD Livers
2.4. Functional Analysis of S-Palmitoylated Proteins Affected by Palm Oil-Rich Diet
2.4.1. HPD-Affected Metabolism, Hemostasis and Neutrophil Degranulation Found by Reactome
2.4.2. Gene Ontology Biological Processes Affected by HPD Diet
2.5. HPD-Affected S-Palmitoylated Proteins Associated with NAFLD
2.6. The Effect of Palm Oil-Rich Diet on Immune Responses in the Liver
3. Discussion
4. Materials and Methods
4.1. Animal Feeding, Sample Collection
4.2. Serum Analyses, Oil Red O Staining
4.3. GCMS Analysis of Fatty Acid Composition
4.4. Mass Spectrometry Analysis of Proteome and S-Palmitoylome of Liver Membranes
4.4.1. Isolation of Membrane-Enriched Liver Fraction
4.4.2. Acyl-Biotin Exchange
4.4.3. Sample Preparation for Mass Spectrometry Analysis of Liver Membrane S-Palmitoylome
4.4.4. ITRAQ Labeling and Sample Preparation for Mass Spectrometry Analysis of Liver Membrane Proteome
4.4.5. Mass Spectrometry
4.4.6. Analysis of Mass Spectrometry Data
4.5. Functional Analyses of Proteomics Data
4.6. SDS-PAGE and Immunoblotting
4.7. RNA Isolation and RT-qPCR Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Carta, G.; Murru, E.; Banni, S.; Manca, C. Palmitic Acid: Physiological role, metabolism and nutritional implications. Front. Physiol. 2017, 8, 902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobocińska, J.; Roszczenko-Jasińska, P.; Ciesielska, A.; Kwiatkowska, K. Protein palmitoylation and its role in bacterial and viral infections. Front. Immunol. 2018, 8, 2003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, A.F.; Wan, J.; Bailey, A.O.; Sun, B.; Kuchar, J.A.; Green, W.N.; Phinney, B.S.; Yates, J.R., III; Davis, N.G. Global analysis of protein palmitoylation in yeast. Cell 2006, 125, 1003–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hang, H.C.; Geutjes, E.J.; Grotenbreg, G.; Pollington, A.M.; Bijlmakers, M.J.; Ploegh, H.L. Chemical probes for the rapid detection of fatty-acylated proteins in mammalian cells. J. Am. Chem. Soc. 2007, 129, 2744–2745. [Google Scholar] [CrossRef]
- Martin, B.R.; Cravatt, B.F. Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods 2009, 6, 135–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gould, N.S.; Evans, P.; Martínez-Acedo, P.; Marino, S.M.; Gladyshev, V.N.; Carroll, K.S.; Ischiropoulos, H. Site-specific proteomic mapping identifies selectively modified regulatory cysteine residues in functionally distinct protein networks. Chem. Biol. 2015, 22, 965–975. [Google Scholar] [CrossRef] [Green Version]
- Serwa, R.A.; Abaitua, F.; Krause, E.; Tate, E.W.; O’Hare, P. Systems analysis of protein fatty acylation in herpes simplex virus-infected cells using chemical proteomics. Chem. Biol. 2015, 22, 1008–1017. [Google Scholar] [CrossRef] [Green Version]
- Sobocinska, J.; Roszczenko-Jasinska, P.; Zareba-Koziol, M.; Hromada-Judycka, A.; Matveichuk, O.V.; Traczyk, G.; Łukasiuk, K.; Kwiatkowska, K. LPS upregulates palmitoylated enzymes of the phosphatidylinositol cycle: An insight from proteomic studies. Mol. Cell. Proteom. 2018, 17, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Thinon, E.; Fernandez, J.P.; Molina, H.; Hang, H.C.S. Selective enrichment and direct analysis of protein S-palmitoylation sites. J. Proteome Res. 2018, 17, 1907–1922. [Google Scholar] [CrossRef]
- Zareba-Kozioł, M.; Bartkowiak-Kaczmarek, A.; Figiel, I.; Krzystyniak, A.; Wojtowicz, T.; Bijata, M.; Włodarczyk, J. Stress-induced changes in the S-palmitoylation and S-nitrosylation of synaptic proteins. Mol. Cell. Proteom. 2019, 18, 1916–1938. [Google Scholar] [CrossRef]
- Ji, B.; Skup, M. Roles of palmitoylation in structural long-term synaptic plasticity. Mol. Brain 2021, 14, 8. [Google Scholar] [CrossRef]
- Sutton, L.M.; Sanders, S.S.; Butland, S.L.; Singaraja, R.R.; Franciosi, S.; Southwell, A.L.; Dot, C.N.; Schmidt, M.E.; Mui, K.K.; Kovalik, V.; et al. Hip14l-deficient mice develop neuropathological and behavioural features of Huntington disease. Hum. Mol. Genet. 2013, 22, 452–465. [Google Scholar] [CrossRef] [Green Version]
- Kouskou, M.; Thomson, D.M.; Brett, R.R.; Wheeler, L.; Tate, R.J.; Pratt, J.A.; Chamberlain, L.H. Disruption of the Zdhhc9 intellectual disability gene leads to behavioural abnormalities in a mouse model. Exp. Neurol. 2018, 308, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.F.; Chen, Y.J.; Liu, K.M.; Haddad, A.N.S.; Song, I.W.; Roan, H.Y.; Chen, L.Y.; Yen, J.J.Y.; Chen, Y.J.; Wu, J.Y.; et al. Role of S-palmitoylation by ZDHHC13 in mitochondrial function and metabolism in liver. Sci. Rep. 2017, 7, 2182. [Google Scholar] [CrossRef] [Green Version]
- Marra, F.; Svegliati-Baroni, G. Lipotoxicity and the gut-liver axis in NASH pathogenesis. J. Hepatol. 2018, 68, 280–295. [Google Scholar] [CrossRef] [PubMed]
- Palomer, X.; Pizarro-Delgado, J.; Barroso, E.; Vázquez-Carrera, M. Palmitic and oleic acid: The Yin and Yang of tatty acids in type 2 diabetes mellitus. Trends Endocrinol. Metab. 2018, 29, 178–190. [Google Scholar] [CrossRef]
- Fatima, S.; Hu, X.; Gong, R.H.; Huang, C.; Chen, M.; Wong, H.L.X.; Bian, Z.; Kwan, H.Y. Palmitic acid is an intracellular signaling molecule involved in disease development. Cell. Mol. Life Sci. 2019, 76, 2547–2557. [Google Scholar] [CrossRef] [PubMed]
- Coll, T.; Eyre, E.; Rodriguez-Calvo, R.; Palomer, X.; Sanchez, R.M.; Merlos, M.; Laguna, J.C.; Vázquez-Carrera, M. Oleate reverses palmitate-induced insulin resistance and inflammation in skeletal muscle cells. J. Biol. Chem. 2008, 283, 11107–11116. [Google Scholar] [CrossRef] [Green Version]
- Schilling, J.D.; Machkovech, H.M.; He, L.; Sidhu, R.; Fujiwara, H.; Weber, K.; Ory, D.S.; Schaffer, J.E. Palmitate and lipopolysaccharide trigger synergistic ceramide production in primary macrophages. J. Biol. Chem. 2013, 288, 2923–2932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leamy, A.K.; Egnatchik, R.A.; Shiota, M.; Ivanova, P.T.; Myers, D.S.; Brown, H.A.; Young, J.D. Enhanced synthesis of saturated phospholipids is associated with ER stress and lipotoxicity in palmitate-treated hepatic cells. J. Lipid Res. 2014, 55, 1478–1488. [Google Scholar] [CrossRef] [Green Version]
- Wong, K.L.; Wu, Y.R.; Cheng, K.S.; Chan, P.; Cheung, C.W.; Lu, D.Y.; Su, T.H.; Liu, Z.M.; Leung, Y.M. Palmitic acid-induced lipotoxicity and protection by (+)-catechin in rat cortical astrocytes. Pharmacol. Rep. 2014, 66, 1106–1113. [Google Scholar] [CrossRef]
- Ciesielska, A.; Kwiatkowska, K. Modification of pro-inflammatory signaling by dietary components: The plasma membrane as a target. Bioessays 2015, 37, 789–801. [Google Scholar] [CrossRef]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Qian, Y.; Fang, Q.; Zhong, P.; Li, W.; Wang, L.; Fu, W.; Zhang, Y.; Xu, Z.; Li, X.; et al. Saturated palmitic acid induces myocardial inflammatory injuries through direct binding to TLR4 accessory protein MD2. Nat. Comm. 2017, 8, 13997. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhang, X.; Lu, Z.; Perry, D.M.; Li, Y.; Russo, S.B.; Cowart, L.A.; Hannun, Y.A.; Huang, Y. Acid sphingomyelinase plays a key role in palmitic acid-amplified inflammatory signaling triggered by lipopolysaccharide at low concentrations in macrophages. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E853–E867. [Google Scholar] [CrossRef]
- Lancaster, G.I.; Langley, K.G.; Berglund, N.A.; Kammoun, H.L.; Reibe, S.; Estevez, E.; Weir, J.; Mellett, N.A.; Pernes, G.; Conway, J.R.W.; et al. Evidence that TLR4 is not a receptor for saturated fatty acids but mediates lipid-induced inflammation by reprogramming macrophage metabolism. Cell. Metab. 2018, 27, 1096–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korbecki, J.; Bajdak-Rusinek, K. The effect of palmitic acid on inflammatory response in macrophages: An overview of molecular mechanisms. Inflamm. Res. 2019, 68, 915–932. [Google Scholar] [CrossRef] [Green Version]
- Burgoyne, J.R.; Haeussler, D.J.; Kumar, V.; Ji, Y.; Pimental, D.R.; Zee, R.S.; Costello, C.E.; Lin, C.; McComb, M.E.; Cohen, R.A.; et al. Oxidation of HRas cysteine thiols by metabolic stress prevents palmitoylation in vivo and contributes to endothelial cell apoptosis. FASEB J. 2012, 26, 832–841. [Google Scholar] [CrossRef] [Green Version]
- Pandey, N.R.; Zhou, X.; Qin, Z.; Zaman, T.; Gomez-Smith, M.; Keyhanian, K.; Anisman, H.; Brunel, J.M.; Stewart, A.F.; Chen, H.H. The LIM domain only 4 protein is a metabolic responsive inhibitor of protein tyrosine phosphatase 1B that controls hypothalamic leptin signaling. J. Neurosci. 2013, 33, 12647–12655. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, M.; Fusco, S.; Mainardi, M.; Scala, F.; Natale, F.; Lapenta, R.; Mattera, A.; Rinaudo, M.; Li Puma, D.D.; Ripoli, C.; et al. Brain insulin resistance impairs hippocampal synaptic plasticity and memory by increasing GluA1 palmitoylation through FoxO3a. Nat. Commun. 2017, 8, 2009. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, C.; Luo, X.; Wang, P.; Zhou, W.; Zhong, S.; Xie, Y.; Jiang, Y.; Yang, P.; Tang, R.; et al. CD36 palmitoylation disrupts free fatty acid metabolism and promotes tissue inflammation in non-alcoholic steatohepatitis. J. Hepatol. 2018, 69, 705–717. [Google Scholar] [CrossRef]
- Senyilmaz, D.; Virtue, S.; Xu, X.; Tan, C.Y.; Griffin, J.L.; Miller, A.K.; Vidal-Puig, A.; Teleman, A.A. Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature 2015, 525, 124–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fattore, E.; Bosetti, C.; Brighenti, F.; Agostoni, C.; Fattore, G. Palm oil and blood lipid-related markers of cardiovascular disease: A systematic review and meta-analysis of dietary intervention trials. Am. J. Clin. Nutr. 2014, 99, 1331–1350. [Google Scholar] [CrossRef] [Green Version]
- May, C.Y.; Nesaretnam, K. Research advancements in palm oil nutrition. Eur. J. Lipid Sci. Technol. 2014, 116, 1301–1315. [Google Scholar] [CrossRef] [Green Version]
- Mancini, A.; Imperlini, E.; Nigro, E.; Montagnese, C.; Daniele, A.; Orru, S.; Buono, P. Biological and nutritional properties of palm oil and palmitic acid: Effects on health. Molecules 2015, 20, 17339–17361. [Google Scholar] [CrossRef] [PubMed]
- Gesteiro, E.; Guijarro, L.; Sanchez-Muniz, F.J.; Vidal-Carou, M.D.C.; Troncoso, A.; Venanci, L.; Jimeno, V.; Quilez, J.; Anadón, A.; González-Gross, M. Palm oil on the edge. Nutrients 2019, 11, 2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blasbalg, T.L.; Hibbeln, J.R.; Ramsden, C.E.; Majchrzak, S.F.; Rawlings, R.R. Changes in consumption of omega-3 and omega-6 fatty acids in the United States during the 20th century. Am. J. Clin. Nutr. 2011, 93, 950–962. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Hao, J.W.; Wang, X.; Guo, H.; Sun, H.H.; Lai, X.Y.; Liu, L.Y.; Zhu, M.; Wang, H.Y.; Li, Y.F.; et al. DHHC4 and DHHC5 facilitate fatty acid uptake by palmitoylating and targeting CD36 to the plasma membrane. Cell Rep. 2019, 26, 209–221. [Google Scholar] [CrossRef] [Green Version]
- Hao, J.W.; Wang, J.; Guo, H.; Zhao, Y.Y.; Sun, H.H.; Li, Y.F.; Lai, X.Y.; Zhao, N.; Wang, X.; Xie, C.; et al. CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nat. Commun. 2020, 11, 4765. [Google Scholar] [CrossRef]
- Ruiz, M.; Bodhicharla, R.; Svensk, E.; Devkota, R.; Busayavalasa, K.; Palmgren, H.; Ståhlman, M.; Boren, J.; Pilon, M. Membrane fluidity is regulated by the C. elegans transmembrane protein FLD-1 and its human homologs TLCD1/2. Elife 2018, 7, e40686. [Google Scholar] [CrossRef]
- Jeon, J.H.; Hong, C.W.; Kim, E.Y.; Lee, J.M. Current understanding on the metabolism of neutrophils. Immune Netw. 2020, 20, e46. [Google Scholar] [CrossRef] [PubMed]
- Morrison, E.; Kuropka, B.; Kliche, S.; Brügger, B.; Krause, E.; Freund, C. Quantitative analysis of the human T cell palmitome. Sci. Rep. 2015, 5, 11598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhungana, S.; Merrick, B.A.; Tomer, K.B.; Fessler, M.B. Quantitative proteomics analysis of macrophage rafts reveals compartmentalized activation of the proteasome and of proteasome-mediated ERK activation in response to lipopolysaccharide. Mol. Cell. Proteom. 2009, 8, 201–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, S.; Schaffer, M.; Beck, F.; Mosalaganti, S.; Asano, S.; Thomas, H.F.; Plitzko, J.M.; Beck, M.; Baumeister, W.; Engel, B.D. Proteasomes tether to two distinct sites at the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2017, 114, 13726–13731. [Google Scholar] [CrossRef] [Green Version]
- Geng, Y.; Faber, K.N.; de Meijer, V.E.; Blokzijl, H.; Moshage, H. How does hepatic lipid accumulation lead to lipotoxicity in non-alcoholic fatty liver disease? Hepatol Int. 2021, 15, 21–35. [Google Scholar] [CrossRef]
- Byrne, C.D.; Targher, G. Non-alcoholic fatty liver disease-related risk of cardiovascular disease and other cardiac complications. Diabetes Obes. Metab. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Estrada, L.D.; Ahumada, P.; Cabrera, D.; Arab, J.P. Liver dysfunction as a novel player in Alzheimer’s progression: Looking outside the brain. Front. Aging Neurosci. 2019, 11, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kjærgaard, K.; Mikkelsen, A.C.D.; Wernberg, C.W.; Grønkjær, L.L.; Eriksen, P.L.; Damholdt, M.F.; Mookerjee, R.P.; Vilstrup, H.; Lauridsen, M.M.; Thomsen, K.L. Cognitive dysfunction in non-alcoholic fatty liver disease-current knowledge, mechanisms and perspectives. J. Clin. Med. 2021, 10, 673. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Imajo, K.; Honda, Y.; Kessoku, T.; Tomeno, W.; Kato, S. Palmitate-induced lipotoxicity is crucial for the pathogenesis of nonalcoholic fatty liver disease in cooperation with gut-derived endotoxin. Sci. Rep. 2018, 8, 11365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gowda, S.; Yeang, C.; Wadgaonkar, S.; Anjum, F.; Grinkina, N.; Cutaia, M.; Jiang, X.-C.; Wadgaonkar, R. Sphingomyelin synthase 2 (SMS2) deficiency attenuates LPS-induced lung injury. Am. J. Physiol. Lung Cell. Molec. Physiol. 2011, 300, L430–L440. [Google Scholar] [CrossRef] [Green Version]
- Sawant, K.V.; Sepuru, K.M.; Lowry, E.; Penaranda, B.; Frevert, C.W.; Garofalo, R.P.; Rajarathnam, K. Neutrophil recruitment by chemokines Cxcl1/KC and Cxcl2/MIP2: Role of Cxcr2 activation and glycosaminoglycan interactions. J. Leukoc. Biol. 2021, 109, 777–791. [Google Scholar] [CrossRef]
- Benard, O.; Lim, J.; Apontes, P.; Jing, X.; Angeletti, R.H.; Chi, Y. Impact of high-fat diet on the proteome of mouse liver. J. Nutr. Biochem. 2016, 31, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Sang, J.; Qu, H.; Gu, R.; Chen, D.; Chen, X.; Yin, B.; Huang, Y.; Xi, W.; Wang, C.; Huang, Y. Proteomics study of the effect of high-fat diet on rat liver. Br. J. Nutr. 2019, 122, 1062–1072. [Google Scholar] [CrossRef]
- Shao, M.; Ye, Z.; Qin, Y.; Wu, T. Abnormal metabolic processes involved in the pathogenesis of non-alcoholic fatty liver disease. Exp. Ther. Med. 2020, 20, 26. [Google Scholar] [CrossRef] [PubMed]
- Karaca, M.; Martin-Levilain, J.; Grimaldi, M.; Li, L.; Dizin, E.; Emre, Y.; Maechler, P. Liver glutamate dehydrogenase controls whole-body energy partitioning through amino acid-derived gluconeogenesis and ammonia homeostasis. Diabetes 2018, 67, 1949–1961. [Google Scholar] [CrossRef] [Green Version]
- Heinrichsen, E.T.; Zhang, H.; Robinson, J.E.; Ngo, J.; Diop, S.; Bodmer, R.; Joiner, W.J.; Metallo, C.M.; Haddad, G.G. Metabolic and transcriptional response to a high-fat diet in Drosophila melanogaster. Mol. Metab. 2013, 3, 42–54. [Google Scholar] [CrossRef]
- Huang, B.; Yao, Y.; Li, Y.; Yang, H.; Liu, H.; Liu, H.; Li, D.; Shu, W.; Chen, M. Proteomics approach to investigate dynamic protein profile involved in high fat diet-induced fatty liver disease in rats. J. Toxicol. Pathol. 2019, 32, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.M.; McGillicuddy, F.C.; Harford, K.A.; Finucane, O.M.; Mills, K.H.; Roche, H.M. Dietary saturated fatty acids prime the NLRP3 inflammasome via TLR4 in dendritic cells-implications for diet-induced insulin resistance. Mol. Nutr. Food Res. 2012, 56, 1212–1222. [Google Scholar] [CrossRef]
- Liaskou, E.; Wilson, D.V.; Oo, Y.H. Innate immune cells in liver inflammation. Mediat. Inflamm. 2012, 2012, 949157. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Kubes, P.A. Reservoir of mature cavity macrophages that can rapidly invade visceral organs to affect tissue repair. Cell 2016, 1656, 668–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simoes, I.C.M.; Karkucinska-Wieckowska, A.; Janikiewicz, J.; Szymanska, S.; Pronicki, M.; Dobrzyn, P.; Dabrowski, M.; Dobrzyn, A.; Oliveira, P.J.; Zischka, H.; et al. Western diet causes obesity-induced nonalcoholic fatty liver disease development by differentially compromising the autophagic response. Antioxidants 2020, 9, 995. [Google Scholar] [CrossRef]
- Hörber, S.; Lehmann, R.; Stefan, N.; Machann, J.; Birkenfeld, A.L.; Wagner, R.; Heni, M.; Häring, H.U.; Fritsche, A.; Peter, A. Hemostatic alterations linked to body fat distribution, fatty liver, and insulin resistance. Mol. Metab. 2021, 53, 101262. [Google Scholar] [CrossRef] [PubMed]
- Grandl, G.; Wolfrum, C. Hemostasis, endothelial stress, inflammation, and the metabolic syndrome. Semin. Immunopathol. 2018, 40, 215–224. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Neelakantan, N.; Wu, Y.; Lote-Oke, R.; Pan, A.; van Dam, R.M. Palm oil consumption increases LDL cholesterol compared with vegetable oils low in saturated fat in a meta-analysis of clinical trials. J. Nutr. 2015, 145, 1549–1558. [Google Scholar] [CrossRef] [Green Version]
- Odia, O.J.; Ofori, S.; Maduka, O. Palm oil and the heart: A review. World J. Cardiol. 2015, 7, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S.R.; Maarof, S.K.; Siedar Ali, S.S.; Ali, A. Systematic review of palm oil consumption and the risk of cardiovascular disease. PLoS ONE 2018, 13, e0193533. [Google Scholar] [CrossRef] [PubMed]
- Bier, D.M. Saturated fats and cardiovascular disease: Interpretations not as simple as they once were. Crit. Rev. Food Sci. Nutr. 2016, 56, 1943–1946. [Google Scholar] [CrossRef]
- Borzecka-Solarz, J.; Dembinska, A.; Hromada-Judycka, G.; Traczyk, A.; Ciesielska, E.A.; Świątkowska, A.; Kwiatkowska, K. Association of Lyn kinase with membrane rafts determines its negative influence on LPS-induced signaling. Mol. Biol. Cell 2017, 28, 1147–1159. [Google Scholar] [CrossRef] [Green Version]
- Mehlem, A.; Hagberg, C.E.; Muhl, L.; Eriksson, U.; Falkevall, A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat. Protoc. 2013, 8, 1149–1154. [Google Scholar] [CrossRef] [Green Version]
- Folch, J.; Lees, M.; Sloane, S.G.H. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Hughes, C.S.; Moggridge, S.; Muller, T.; Sorensen, P.H.; Morin, G.B.; Krijgsveld, J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019, 14, 68–85. [Google Scholar] [CrossRef]
- McDowell, G.S.; Gaun, A.; Steen, H. iFASP: Combining isobaric mass tagging with filter-aided sample preparation. J. Proteome Res. 2013, 12, 3809–3812. [Google Scholar] [CrossRef] [Green Version]
- Malinowska, A.; Kistowski, M.; Bakun, M.; Rubel, T.; Tkaczyk, M.; Mierzejewska, J.; Dadlez, M. Diffprot—Software for non-parametric statistical analysis of differential proteomics data. J. Proteom. 2012, 75, 4062–4073. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; Jensen, L.J.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mlecnik, B.; Galon, J.; Bindea, G. Comprehensive functional analysis of large lists of genes and proteins. J. Proteom. 2018, 171, 2–10. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.P.; Murphy, C.G.; Saraceni-Richards, C.A.; Rosenstein, M.C.; Wiegers, T.C.; Mattingly, C.J. Comparative toxicogenomics database: A knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res. 2009, 37, D786–D792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]










| Protein ID | Gene ID | Protein Name | 4-Week HPDpalm/RDpalm | 4-Week HPDp/RDp | 12-Week HPDpalm/RDpalm | 12-Week HPDp/RDp |
|---|---|---|---|---|---|---|
| Fatty acid synthesis | ||||||
| Q62264 | Thrsp | Thyroid hormone-inducible hepatic protein | >100 | 0.60 | 0.62 | 0.76 |
| Q9JLJ4 * | Elovl2 | Elongation of very long chain fatty acids protein 2 | 2.26 | 0.68 | 4.51 | 0.73 |
| P19096 * | Fasn | Fatty acid synthase | 2.55 | 0.50 | 1.01 | 0.83 |
| Q8K2C9 * | Hacd3 | Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase 3 | 1.62 | 0.95 | 0.88 | 1.17 |
| Q91V92 | Acly | ATP-citrate synthase | 1.52 | 0.50 | 1.03 | 0.88 |
| Amino acid synthesis | ||||||
| Q9CQT1 | Mir1 | Methylthioribose-1-phosphate isomerase | 0.01 | 0.93 | 1.13 | 1.09 |
| Q8VBT2 | Sds | L-serine dehydratase/L-threonine deaminase | 0.29 | 0.87 | 0.97 | 0.93 |
| O35490 | Bhmt | Betaine-homocysteine S-methyltransferase 1 | 0.33 | 1.85 | 0.67 | 1.40 |
| P15105 | Glul | Glutamine synthetase | 0.56 | 1.27 | 1.09 | 1.12 |
| Amino acid degradation | ||||||
| Q9D2G2 * | Dlst | 2-oxoglutarate dehydrogenase complex component E2, mitochondrial | 0.34 | 1.23 | 1.32 | 0.95 |
| Q9D7B6 | Acad8 | Isobutyryl-CoA dehydrogenase, mitochondrial | 0.37 | 1.01 | 4.09 | 0.92 |
| Q8BH00 | Aldh8a1 | Aldehyde dehydrogenase family 8 member A1 | 0.46 | 1.26 | 0.77 | 0.99 |
| Q8BWF0 * | Aldh5a1 | Succinate-semialdehyde dehydrogenase, mitochondrial | 0.47 | 1.13 | 0.73 | 1.11 |
| O09173 | Hgd | Homogentisate 1,2-dioxygenase | 0.49 | 1.28 | 0.97 | 0.96 |
| P11725 | Otc | Ornithine carbamoyltransferase, mitochondrial | 0.49 | 1.36 | 0.94 | 0.95 |
| Q61425 | Hadh | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | 0.51 | 1.40 | 0.96 | 1.16 |
| P61922 | Abat | 4-aminobutyrate aminotransferase, mitochondrial | 0.52 | 1.31 | 1.32 | 1.03 |
| P26443 | Glud1 | Glutamate dehydrogenase 1, mitochondrial | 0.50 | 1.27 | 0.98 | 1.12 |
| Q91XD4 | Ftcd | Formimidoyltransferase-cyclodeaminase | 0.59 | 1.25 | 0.99 | 0.98 |
| Q8VC12 | Uroc1 | Urocanate hydratase | 0.60 | 1.24 | 0.84 | 1.02 |
| Q3ULD5 | Mccc2 | Methylcrotonoyl-CoA carboxylase β chain, mitochondrial | 0.63 | 1.12 | 0.84 | 1.07 |
| P49429 | Hpd | 4-Hydroxyphenylpyruvate dioxygenase | 0.64 | 1.27 | 1.28 | 1.08 |
| Q9DBA8 | Amdhd1 | Probable imidazolonepropionase | 0.79 | 1.13 | 6.28 | 0.95 |
| O88986 * | Gcat | 2-amino-3-ketobutyrate CoA ligase, mitochondria | 1.24 | 0.84 | 1.81 | 0.9 |
| Neutrophil degranulation | ||||||
| Q9Z0K8 | Vnn1 | Pantetheinase | >100 | 1.51 | 29.5 | 1.18 |
| Q8C7E7 | Stbd1 | Starch-binding domain-containing protein 1 | 6.50 | 0.87 | 0.18 | 1.10 |
| P60904 | Dnajc5 | DnaJ homolog subfamily C member 5 | 5.19 | 1.00 | >100 | 1.06 |
| Q3TDN2 | Faf2 | FAS-associated factor 2 | 3.52 | 0.94 | 0.96 | 1.02 |
| Q99JI6 | Rap1b | Ras-related protein Rap-1b | 2.74 | 0.93 | 0.16 | 1.00 |
| O08807 | Prdx4 | Peroxiredoxin-4 | 0.64 | 1.09 | 2.83 | 0.88 |
| Q08857 | Cd36 | Platelet glycoprotein 4 | 0.45 | 1.12 | 6.05 | 1.28 |
| Q91W90 | Txndc5 | Thioredoxin domain-containing protein 5 | 0.25 | 1.19 | 0.52 | 0.99 |
| P63158 | Hmgb1 | High mobility group protein B1 | 0.24 | 1.04 | 1.62 | 1.06 |
| O09044 | Snap23 | Synaptosomal-associated protein 23 | 0.18 | 1.12 | 2.16 | 1.00 |
| P18242 | Ctsd | Cathepsin D | 0.13 | 1.02 | 2.25 | 0.96 |
| Q91W53 | Golga7 | Golgin subfamily A member 7 | 0.11 | 1.00 | 0.36 | 0.86 |
| Q64310 | Surf4 | Surfeit locus protein 4 | 0.96 | 0.98 | 2.45 | 0.84 |
| P11835 | Itgb2 | Integrin β-2 | 0.94 | 0.99 | 0.13 | 0.93 |
| Hemostasis | ||||||
| O08677 | Kng1 | Kininogen-1 | >100 | 0.95 | 0.41 | 1.03 |
| P04441 * | Cd74 | H-2 class II histocompatibility antigen γ chain | >100 | 1.00 | >100 | 1.00 |
| O35678 * | Mgll | Monoglyceride lipase | 3.31 | 0.93 | 7.37 | 0.94 |
| Q01339 | Apoh | β-2-glycoprotein 1 | 2.61 | 1.00 | 1.29 | 1.06 |
| Q76MZ3 | Ppp2r1a | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A α isoform | 2.39 | 0.96 | 0.72 | 1.00 |
| P21279 | Gnaq | Guanine nucleotide-binding protein G(q) subunit α | 2.32 | 0.97 | 0.41 | 0.95 |
| Q9DC51 | Gnai3 | Guanine nucleotide-binding protein G(i) subunit α-3 | 2.10 | 1.01 | 0.71 | 0.93 |
| P14094 | Atp1b1 | Sodium/potassium-transporting ATPase subunit β-1 | 1.56 | 0.91 | 1.14 | 0.98 |
| P47757 | Capzb | F-actin-capping protein subunit β | 1.82 | 0.97 | 1.82 | 0.92 |
| O08715 | Akap1 | A-kinase anchor protein 1, mitochondrial | 0.77 | 0.96 | 0.38 | 1.11 |
| Q61411 | Hras | GTPase HRas | 0.66 | 0.98 | 3.49 | 0.96 |
| P27601 | Gna13 | Guanine nucleotide-binding protein subunit α-13 | 0.63 | 0.89 | 1.49 | 0.89 |
| P09055 | Itgb1 | Integrin β-1 | 0.53 | 0.96 | 1.08 | 1.07 |
| Q08857 | Cd36 | Platelet glycoprotein 4 | 0.45 | 1.12 | 6.05 | 1.28 |
| E9PV24 | Fga | Fibrinogen α chain | 0.43 | 1.08 | 1.94 | 0.83 |
| Q8VCM7 | Fgg | Fibrinogen γ chain | 0.43 | 1.15 | 0.87 | 0.85 |
| Q91WC9 * | Daglb | sn1-specific diacylglycerol lipase β | 0.38 | 0.93 | 1.49 | 1.00 |
| P63001 | Rac1 | Ras-related C3 botulinum toxin substrate 1 | 0.29 | 1.05 | 0.99 | 1.00 |
| Q8K0E8 | Fgb | Fibrinogen β chain | 0.28 | 1.28 | 0.58 | 0.86 |
| Q64727 | Vcl | Vinculin | 1.27 | 1.02 | 3.21 | 1.08 |
| P25911 | Lyn | Tyrosine-protein kinase Lyn | 1.43 | 0.99 | 0.63 | 0.98 |
| P21278 | Gna11 | Guanine nucleotide-binding protein subunit α-11 | 1.01 | 0.94 | 0.41 | 1.01 |
| P18572 | Bsg | Basigin | 1.07 | 0.98 | 0.32 | 1.11 |
| Q04736 | Yes1 | Tyrosine-protein kinase Yes | 0.73 | 0.93 | 2.32 | 0.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziemlińska, E.; Sobocińska, J.; Świątkowska, A.; Hromada-Judycka, A.; Traczyk, G.; Malinowska, A.; Świderska, B.; Mietelska-Porowska, A.; Ciesielska, A.; Kwiatkowska, K. Palm Oil-Rich Diet Affects Murine Liver Proteome and S-Palmitoylome. Int. J. Mol. Sci. 2021, 22, 13094. https://doi.org/10.3390/ijms222313094
Ziemlińska E, Sobocińska J, Świątkowska A, Hromada-Judycka A, Traczyk G, Malinowska A, Świderska B, Mietelska-Porowska A, Ciesielska A, Kwiatkowska K. Palm Oil-Rich Diet Affects Murine Liver Proteome and S-Palmitoylome. International Journal of Molecular Sciences. 2021; 22(23):13094. https://doi.org/10.3390/ijms222313094
Chicago/Turabian StyleZiemlińska, Ewelina, Justyna Sobocińska, Anna Świątkowska, Aneta Hromada-Judycka, Gabriela Traczyk, Agata Malinowska, Bianka Świderska, Anna Mietelska-Porowska, Anna Ciesielska, and Katarzyna Kwiatkowska. 2021. "Palm Oil-Rich Diet Affects Murine Liver Proteome and S-Palmitoylome" International Journal of Molecular Sciences 22, no. 23: 13094. https://doi.org/10.3390/ijms222313094
APA StyleZiemlińska, E., Sobocińska, J., Świątkowska, A., Hromada-Judycka, A., Traczyk, G., Malinowska, A., Świderska, B., Mietelska-Porowska, A., Ciesielska, A., & Kwiatkowska, K. (2021). Palm Oil-Rich Diet Affects Murine Liver Proteome and S-Palmitoylome. International Journal of Molecular Sciences, 22(23), 13094. https://doi.org/10.3390/ijms222313094