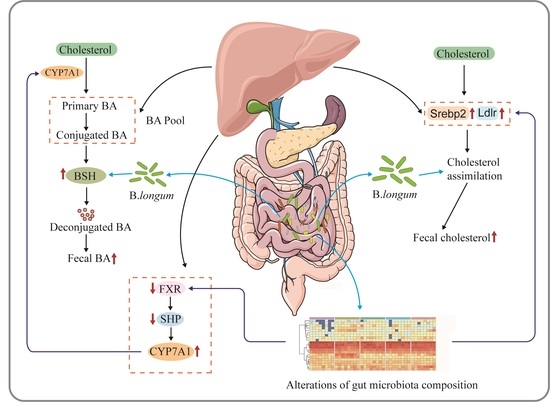
Strain-Specific Effects of Bifidobacterium longum on Hypercholesterolemic Rats and Potential Mechanisms
Abstract
:1. Introduction
2. Results
2.1. Subsection Growth Characteristics of B. longum Strains in Vitro
2.2. Tolerance Ability of B. longum to Simulated Gastroenteric Fluid
2.3. Bile Salt Deconjugation and Cholesterol Assimilation Abilities of the B. longum Strains
2.4. The Effects of B. longum Strains on the Serum Lipids
2.5. Effects of B. longum Strains on Fecal Bile Acid and Cholesterol Levels
2.6. Effects of B. longum Strains on Liver Gene Expression
2.7. Effects of the Four B. longum Strains on Gut Microbiota
2.8. Relationship between the Hypercholesterolemia-Alleviation Effects of B. longum Strains and Their Properties In Vitro
3. Discussion
4. Materials and Methods
4.1. The Culture Conditions and Growth Curve of Bacterial Strains
4.2. The Culture Conditions and Growth Curve of Bacterial Strains
4.3. Quantitative Determination of Bile Salt Deconjugation Ability of B. longum Strains by HPLC
4.4. Cholesterol Assimilation by B. longum Strains
4.5. Animals and Diets
4.6. Analysis of Serum Lipid Levels
4.7. Analysis of Fecal Cholesterol and Bile Acid Contents
4.8. Extraction of Liver RNA and RT-PCR Analysis
4.9. MiSeq Genome Sequencing Analysis of Community Structures
4.10. Correlation Test
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| B. longum | Bifidobacterium longum |
| L. plantarum | Lactobacillus plantarum |
| B. bifidum | Bifidobacterium bifidum |
| L. rhamnosus | Lactobacillus rhamnosus |
| BSH | Bile salt hydrolysis |
| FXR | farnesoid X receptor |
| SHP | small heterodimer partner |
| CYP7A1 | cholesterol 7α-hydroxylase |
| LXR | liver X receptor |
| SREBP2 | sterol regulatory element binding protein 2 |
| LDLR | low density lipoprotein receptor |
References
- Gielen, S.; Landmesser, U. The Year in Cardiology 2013: Cardiovascular disease prevention. Eur. Heart J. 2014, 35, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.S.; Kong, L.Z.; Gao, R.L.; Zhu, M.L.; Wang, W.; Wang, Y.J.; Wu, Z.S.; Chen, W.W.; Liu, M.B. Outline of the Report on Cardiovascular Disease in China, 2010. Biomed. Environ. Sci. 2012, 25, 251–256. [Google Scholar] [PubMed]
- Zhao, D.; Liu, J.; Wang, M.; Zhang, X.; Zhou, M. Epidemiology of cardiovascular disease in China: Current features and implications. Nat. Rev. Cardiol. 2019, 16, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Sayin, S.I.; Wahlstrom, A.; Felin, J.; Jantti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyotylainen, T.; Oresic, M.; Backhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houston, D.K.; Ding, J.; Lee, J.S.; Garcia, M.; Kanaya, A.M.; Tylavsky, F.A.; Newman, A.B.; Visser, M.; Kritchevsky, S.B. Dietary fat and cholesterol and risk of cardiovascular disease in older adults: The Health ABC Study. Nutr. Metab. Cardiovasc. 2011, 21, 430–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, S.C.; Virtamo, J.; Wolk, A. Dietary fats and dietary cholesterol and risk of stroke in women. Atherosclerosis 2012, 221, 282–286. [Google Scholar] [CrossRef]
- Davis, C.E.; Rifkind, B.M.; Brenner, H.; Gordon, D.J. A single cholesterol measurement underestimates the risk of coronary heart disease. An empirical example from the Lipid Research Clinics Mortality Follow-up Study. JAMA 1990, 264, 3044–3046. [Google Scholar] [CrossRef]
- Grundy, S.M. Atherosclerosis imaging and the future of lipid management. Circulation 2004, 110, 3509–3511. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Kim, D.-H.; Seo, K.-H.; Chon, J.-W.; Nah, S.-Y.; Bartley, G.E.; Arvik, T.; Lipson, R.; Yokoyama, W. Modulation of the Intestinal Microbiota Is Associated with Lower Plasma Cholesterol and Weight Gain in Hamsters Fed Chardonnay Grape Seed Flour. J. Agric. Food Chem. 2015, 63, 1460–1467. [Google Scholar] [CrossRef]
- Dunn-Emke, S.; Weidner, G.; Ornish, D. Benefits of a low-fat plant-based diet. Obes. Res. 2001, 9, 731. [Google Scholar] [CrossRef]
- Tong, X.; Xu, J.; Lian, F.; Yu, X.; Zhao, Y.; Xu, L.; Zhang, M.; Zhao, X.; Shen, J.; Wu, S.; et al. Structural Alteration of Gut Microbiota during the Amelioration of Human Type 2 Diabetes with Hyperlipidemia by Metformin and a Traditional Chinese Herbal Formula: A Multicenter, Randomized, Open Label Clinical Trial. mBio 2018, 9, e0239217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.; Zhang, B.; Hu, Y.; Wang, J.; Liu, J.; Qin, R.; Lv, S.; Wang, S. Correlation Analysis of Intestinal Redox State with the Gut Microbiota Reveals the Positive Intervention of Tea Polyphenols on Hyperlipidemia in High Fat Diet Fed Mice. J. Agric. Food Chem. 2019, 67, 7325–7335. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Pan, L.L.; Lv, S.; Yang, Q.; Zhang, H.; Chen, W.; Lv, Z.; Sun, J. Alterations of Gut Microbiota and Blood Lipidome in Gestational Diabetes Mellitus with Hyperlipidemia. Front. Physiol. 2019, 10, 1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costabile, A.; Buttarazzi, I.; Kolida, S.; Quercia, S.; Baldini, J.; Swann, J.R.; Brigidi, P.; Gibson, G.R. An in vivo assessment of the cholesterol-lowering efficacy of Lactobacillus plantarum ECGC 13110402 in normal to mildly hypercholesterolaemic adults. PLoS ONE 2017, 12, e0187964. [Google Scholar] [CrossRef]
- Jones, M.L.; Martoni, C.J.; Parent, M.; Prakash, S. Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Br. J. Nutr. 2012, 107, 1505–1513. [Google Scholar] [CrossRef] [Green Version]
- Al-Sheraji, S.H.; Amin, I.; Azlan, A.; Manap, M.Y.; Hassan, F.A. Effects of Bifidobacterium longum BB536 on lipid profile and histopathological changes in hypercholesterolaemic rats. Benef. Microbes 2015, 6, 661–668. [Google Scholar] [CrossRef]
- Fuentes, M.C.; Lajo, T.; Carrion, J.M.; Cune, J. Cholesterol-lowering efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in hypercholesterolaemic adults. Br. J. Nutr. 2013, 109, 1866–1872. [Google Scholar] [CrossRef] [Green Version]
- Guardamagna, O.; Amaretti, A.; Puddu, P.E.; Raimondi, S.; Abello, F.; Cagliero, P.; Rossi, M. Bifidobacteria supplementation: Effects on plasma lipid profiles in dyslipidemic children. Nutrition 2014, 30, 831–836. [Google Scholar] [CrossRef]
- Hatakka, K.; Mutanen, M.; Holma, R.; Saxelin, M.; Korpela, R. Lactobacillus rhamnosus LC705 Together with Propionibacterium freudenreichii ssp shermanii JS Administered in Capsules Is Ineffective in Lowering Serum Lipids. J. Am. Coll. Nutr. 2008, 27, 441–447. [Google Scholar] [CrossRef]
- Zhao, X.; Higashikawa, F.; Noda, M.; Kawamura, Y.; Matoba, Y.; Kumagai, T.; Sugiyama, M. The Obesity and Fatty Liver Are Reduced by Plant-Derived Pediococcus pentosaceus LP28 in High Fat Diet-Induced Obese Mice. PLoS ONE 2012, 7, 0030696. [Google Scholar] [CrossRef] [Green Version]
- Ruscica, M.; Pavanello, C.; Gandini, S.; Macchi, C.; Botta, M.; Dall’Orto, D.; del Puppo, M.; Bertolotti, M.; Bosisio, R.; Mombelli, G.; et al. Nutraceutical approach for the management of cardiovascular risk—A combination containing the probiotic Bifidobacterium longum BB536 and red yeast rice extract: Results from a randomized, double-blind, placebo-controlled study. Nutr. J. 2019, 18, 13. [Google Scholar] [CrossRef] [PubMed]
- Park, A.J.; Bercik, P.; Huang, X.; Blennerhassett, P.; Sinclair, D.D.; Lu, J.; Deng, Y.; Bergonzelli, G.; McLean, P.; Collins, S.M.; et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut–brain communication. Gastroenterology 2011, 140, S18–S19. [Google Scholar] [CrossRef]
- Wang, B.T.; Ducker, G.S.; Barczak, A.J.; Barbeau, R.; Erle, D.J.; Shokat, K.M. The mammalian target of rapamycin regulates cholesterol biosynthetic gene expression and exhibits a rapamycin-resistant transcriptional profile. Proc. Natl. Acad. Sci. USA 2011, 108, 15201–15206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carstea, E.D.; Morris, J.A.; Coleman, K.G.; Loftus, S.K.; Zhang, D.; Cummings, C.; Gu, J.Z.; Rosenfeld, M.A.; Pavan, W.J.; Krizman, D.B. Niemann-Pick C1 disease gene: Homology to mediators of cholesterol homeostasis. Science 1997, 277, 228–231. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Zhu, Y.; Zhong, S.; Zheng, G. Astilbin lowers the effective caffeine dose for decreasing lipid accumulation via activating AMPK in high-fat diet-induced obese mice. J. Sci. Food Agric. 2020, 573–581. [Google Scholar]
- Schneider, K.M.; Albers, S.; Trautwein, C. Role of bile acids in the gut-liver axis. J. Hepatol. 2018, 68, 1083–1085. [Google Scholar] [CrossRef]
- Michael, D.R.; Davies, T.S.; Moss, J.W.E.; Calvente, D.L.; Ramji, D.P.; Marchesi, J.R.; Pechlivanis, A.; Plummer, S.F.; Hughes, T.R. The anti-cholesterolaemic effect of a consortium of probiotics: An acute study in C57BL/6J mice. Sci. Rep. 2017, 7, 2883. [Google Scholar] [CrossRef] [Green Version]
- Gruber, J.; Kennedy, B.K. Microbiome and Longevity: Gut Microbes Send Signals to Host Mitochondria. Cell 2017, 169, 1168–1169. [Google Scholar] [CrossRef] [Green Version]
- George, K.S.; Munoz, J.; Akhavan, N.S.; Foley, E.M.; Siebert, S.C.; Tenenbaum, G.; Khalil, D.A.; Chai, S.C.; Arjmandi, B.H. Is soy protein effective in reducing cholesterol and improving bone health? Food Funct. 2020, 11, 544–551. [Google Scholar] [CrossRef]
- Chen, X.C.; Huang, L.L.; Chang, T.H.A.; Ong, B.L.; Ong, S.L.; Hu, J.Y. Plant Traits for Phytoremediation in the Tropics. Engineering 2019, 5, 841–848. [Google Scholar] [CrossRef]
- Bove, M.; Cicero, A.F.G.; Borghi, C. Emerging drugs for the treatment of hypercholesterolemia. Expert Opin. Emerg. Drugs 2019, 24, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Araki, E.; Yamashita, S.; Arai, H.; Yokote, K.; Satoh, J.; Inoguchi, T.; Nakamura, J.; Maegawa, H.; Yoshioka, N.; Tanizawa, Y.; et al. Effects of Pemafibrate, a Novel Selective PPARα Modulator, on Lipid and Glucose Metabolism in Patients with Type 2 Diabetes and Hypertriglyceridemia: A Randomized, Double-Blind, Placebo-Controlled, Phase 3 Trial. Diabetes Care 2018, 41, 538–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, D.; Zhu, H.; Gao, F.; Qian, Z.; Mao, W.; Yin, Y.; Tan, J.; Chen, D. Antidiabetic effects of selenium-enriched Bifidobacterium longum DD98 in type 2 diabetes model of mice. Food Funct. 2020, 11, 6528–6541. [Google Scholar] [CrossRef]
- Ballantyne, C.M.; Banach, M.; Mancini, G.B.J.; Lepor, N.E.; Hanselman, J.C.; Zhao, X.; Leiter, L.A. Efficacy and safety of bempedoic acid added to ezetimibe in statin-intolerant patients with hypercholesterolemia: A randomized, placebo-controlled study. Atherosclerosis 2018, 277, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Huang, G.H.; Cai, D.; Li, D.L.; Liang, X.L.; Yu, T.; Shen, P.H.; Su, H.Y.; Liu, J.D.; Gu, H.C.; et al. Qualitative and Semiquantitative Analysis of Fecal Bifidobacterium Species in Centenarians Living in Bama, Guangxi, China. Curr. Microbiol. 2015, 71, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Drago, L.; Toscano, M.; Rodighiero, V.; de Vecchi, E.; Mogna, G. Cultivable and Pyrosequenced Fecal Microflora in Centenarians and Young Subjects. J. Clin. Gastroenterol. 2012, 46, S81–S84. [Google Scholar] [CrossRef]
- Jiang, J.C.; Wu, C.E.; Zhang, C.C.; Zhao, J.X.; Yu, L.L.; Zhang, H.; Arjan, N.; Chen, W.; Zhai, Q.X. Effects of probiotic supplementation on cardiovascular risk factors in hypercholesterolemia: A systematic review and meta-analysis of randomized clinical trial. J. Funct. Foods. 2020, 74, 104177. [Google Scholar] [CrossRef]
- Jones, M.L.; Tomaro-Duchesneau, C.; Martoni, C.J.; Prakash, S. Cholesterol lowering with bile salt hydrolase-active probiotic bacteria, mechanism of action, clinical evidence, and future direction for heart health applications. Expert Opin. Biol. Ther. 2013, 13, 631–642. [Google Scholar] [CrossRef]
- Choi, E.A.; Chang, H.C. Cholesterol-lowering effects of a putative probiotic strain Lactobacillus plantarum EM isolated from kimchi. LWT Food Sci. Technol. 2015, 62, 210–217. [Google Scholar] [CrossRef]
- Song, W.; Song, C.; Shan, Y.; Lu, W.; Zhang, J.; Hu, P.; Wu, X.; Li, L.; Guo, S. The antioxidative effects of three lactobacilli on high-fat diet induced obese mice. RSC Adv. 2016, 6, 65808–65815. [Google Scholar] [CrossRef]
- Wang, G.; Huang, W.; Xia, Y.; Xiong, Z.; Ai, L. Cholesterol-lowering potentials of Lactobacillus strain overexpression of bile salt hydrolase on high cholesterol diet-induced hypercholesterolemic mice. Food Funct. 2019, 10, 1684–1695. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Ono, K.; Horiguchi, M.; Nishi, H.; Nakamura, T.; Nagao, K.; Kinoshita, M.; Kuwabara, Y.; Marusawa, H.; Iwanaga, Y.; et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 17321–17326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.L.; Wu, Q.X.; Qin, X.M. Camellia nitidissima Chi flower extract alleviates obesity and related complications and modulates gut microbiota composition in rats with high-fat-diet-induced obesity. J. Sci. Food Agric. 2020, 100, 4378–4389. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; van Hylckama, J.E.V.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015, 9, 1–15. [Google Scholar] [CrossRef]
- Martinez, I.; Perdicaro, D.J.; Brown, A.W.; Hammons, S.; Carden, T.J.; Carr, T.P.; Eskridge, K.M.; Walter, J. Diet-Induced Alterations of Host Cholesterol Metabolism Are Likely To Affect the Gut Microbiota Composition in Hamsters. Appl. Environ. Microb. 2013, 79, 516–524. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Feng, N.; Zhang, C.; Liu, F.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Lactobacillus reuteri A9 and Lactobacillus mucosae A13 isolated from Chinese superlongevity people modulate lipid metabolism in a hypercholesterolemia rat model. FEMS Microbiol. Lett. 2019, 366, 24. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- Dennis-Wall, J.C.; Culpepper, T.; Nieves, C., Jr.; Rowe, C.C.; Burns, A.M.; Rusch, C.T.; Federico, A.; Ukhanova, M.; Waugh, S.; Mai, V.; et al. Probiotics (Lactobacillus gasseri KS-13, Bifidobacterium bifidum G9-1, and Bifidobacterium longum MM-2) improve rhinoconjunctivitis-specific quality of life in individuals with seasonal allergies: A double-blind, placebo-controlled, randomized trial. Am. J. Clin. Nutr. 2017, 105, 758–767. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.F.; Li, J.Y. A combination of Tween 80 with CaCl2 enhances the hypocholesterolemic activity of bile salt hydrolase-active Lactobacillus casei F0422 in rats fed a cholesterol-rich diet. J. Funct. Foods 2014, 9, 131–140. [Google Scholar] [CrossRef]
- Miremadi, F.; Ayyash, M.; Sherkat, F.; Stojanovska, L. Cholesterol reduction mechanisms and fatty acid composition of cellular membranes of probiotic Lactobacilli and Bifidobacteria. J. Funct. Foods 2014, 9, 295–305. [Google Scholar] [CrossRef]
- Tulumoglu, S.; Kaya, H.I.; Simsek, O. Probiotic characteristics of Lactobacillus fermentum strains isolated from tulum cheese. Anaerobe 2014, 30, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Franz, C.M.; Specht, I.; Haberer, P.; Holzapfel, W.H. Bile salt hydrolase activity of Enterococci isolated from food: Screening and quantitative determination. J. Food Prot. 2001, 64, 725–729. [Google Scholar] [CrossRef] [PubMed]
- Elizabeth Hansbury, T.J.S. Resolution of desmosterol, cholesterol, and other sterol intermediates by reverse-phase highpressure liquid chromatography. J. Lipid Res. 1978, 19, 742–746. [Google Scholar] [CrossRef]
- Kuo, S.-M.; Merhige, P.M.; Hagey, L.R. The Effect of Dietary Prebiotics and Probiotics on Body Weight, Large Intestine Indices, and Fecal Bile Acid Profile in Wild Type and IL10-/- Mice. PLoS ONE 2013, 8, 0060270. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [Green Version]








| Strain | Initial Survival in PBS | Survival after 2 h at pH 2.5 in SGJ | Survival after 3 h at pH 8.0 in 0.3% Oxigall | ||
|---|---|---|---|---|---|
| Mean Counts (Log CFU/mL) | Mean Counts (Log CFU/mL) | Survival Rate | Mean Counts (Log CFU/mL) | Survival Rate | |
| B. longum CCFM 1077 | 9.76 ± 0.06 a | 7.97 ± 0.18 b | 81.66% | 6.36 ± 0.12c | 65.16% |
| B. longum I3 | 9.23 ± 0.24 a | 7.56 ± 0.14 b | 81.91% | 6.08 ± 0.08 c | 65.87% |
| B. longum J3 | 9.32 ± 0.18 a | 7.86 ± 0.26 b | 84.33% | 5.95 ± 0.12 c | 63.84% |
| B. longum B3 | 9.58 ± 0.08 a | 6.98 ± 0.23 b | 72.86% | 6.24 ± 0.18 c | 65.14% |
| Strain | Bile Salt Hydrolysis Ability (%) | Cholesterol Assimilation Ability (%) |
|---|---|---|
| B. longum CCFM 1077 | 98.66 ± 0.65 a | 97.68 ± 1.03 a |
| B. longum I3 | 97.36 ± 0.36 a | 0.96 ± 0.16 b |
| B. longum J3 | 1.01 ± 0.02 b | 99.36 ± 0.32 a |
| B. longum B3 | 1.12 ± 0.06 b | 0.82 ± 0.15 b |
| Ingredient | Cholesterol-Free Diet (g/kg) | Cholesterol-Enriched Diet (g/kg) |
|---|---|---|
| Cornstarch | 465.692 | 459.442 |
| Dextrinized cornstarch | 155 | 155 |
| Casein | 140 | 140 |
| Sucrose | 100 | 100 |
| Soybean oil | 40 | 40 |
| Cellulose | 50 | 50 |
| Choline biartrate | 2.5 | 2.5 |
| L-Systine | 1.8 | 1.8 |
| t-Butylhydroquinone | 0.008 | 0.008 |
| Mineral | 35 | 35 |
| Vitamin | 10 | 10 |
| Cholesterol | - | 5 |
| Sodium cholate | - | 1.25 |
| Gene | Forward Primers (5′-3′) | Reverse Primers (5′-3′) |
|---|---|---|
| FXR | CCAACCTGGGCTTCTACCC | CACACAGCTCATCCCCTTT |
| SHP | TCTGCAGGTCGTCCGACTATTC | AGGCAGTGGCTGTGAGATGC |
| CYP7A1 | ATTCCATACCTGGGCTGTGC | ATGTTTTCAGTGGTATTTCC |
| LXR | CTCTTCTTGCCGCTTCAGTT | AGGAGTGTCGACTTCGCAAA |
| Srebp 2 | AGCAGCAGGTGCAGACGGTA | CATCTGTCTTCAGCGTGGTC |
| LDLR | AGCAGTGAGTGTATCCATCG | AATGCAGGAGCCATCTGCAC |
| β-actin | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Wu, C.; Zhang, C.; Zhang, Q.; Yu, L.; Zhao, J.; Zhang, H.; Narbad, A.; Chen, W.; Zhai, Q. Strain-Specific Effects of Bifidobacterium longum on Hypercholesterolemic Rats and Potential Mechanisms. Int. J. Mol. Sci. 2021, 22, 1305. https://doi.org/10.3390/ijms22031305
Jiang J, Wu C, Zhang C, Zhang Q, Yu L, Zhao J, Zhang H, Narbad A, Chen W, Zhai Q. Strain-Specific Effects of Bifidobacterium longum on Hypercholesterolemic Rats and Potential Mechanisms. International Journal of Molecular Sciences. 2021; 22(3):1305. https://doi.org/10.3390/ijms22031305
Chicago/Turabian StyleJiang, Jinchi, Caie Wu, Chengcheng Zhang, Qingsong Zhang, Leilei Yu, Jianxin Zhao, Hao Zhang, Arjan Narbad, Wei Chen, and Qixiao Zhai. 2021. "Strain-Specific Effects of Bifidobacterium longum on Hypercholesterolemic Rats and Potential Mechanisms" International Journal of Molecular Sciences 22, no. 3: 1305. https://doi.org/10.3390/ijms22031305
APA StyleJiang, J., Wu, C., Zhang, C., Zhang, Q., Yu, L., Zhao, J., Zhang, H., Narbad, A., Chen, W., & Zhai, Q. (2021). Strain-Specific Effects of Bifidobacterium longum on Hypercholesterolemic Rats and Potential Mechanisms. International Journal of Molecular Sciences, 22(3), 1305. https://doi.org/10.3390/ijms22031305