Design of PG-Surfactants Bearing Polyacrylamide Polymer Chain to Solubilize Membrane Proteins in a Surfactant-Free Buffer
Abstract
:1. Introduction
2. Results and Discussion
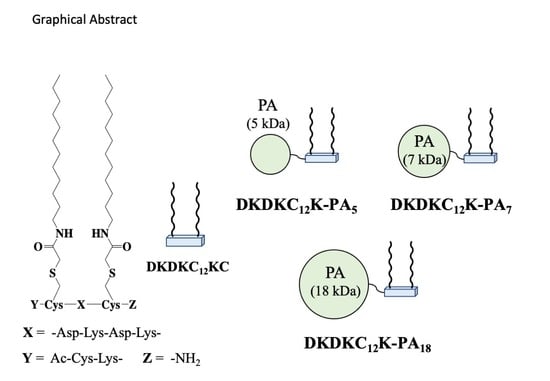
2.1. Design of Solubilization Surfactants Bearing Linear Polyacrylamide (PA) Polymer Chain
2.2. Solubilization of Photosystem I from Thermosynecoccus (T.) Vulcanus by DKDKC12K-PAn (n = 5, 7, and 18)
3. Materials and Methods
3.1. Materials
3.2. Synthesis of PG-Surfactant, DKDKC12KC
3.3. Synthesis of Polyacrylamide Having Dithiopyridyl Terminal Group (PAn-DTP, n = 5, 7, 18) by RAFT Polymerization
3.4. Synthesis of the PA(5, 7, 18 kDa)-Appended DKDKC12KC, DKDKC12K-PAn (n = 5, 7, 18)
3.5. Dynamic Light Scattering Measurements of PG-Surfactant Assemblies
3.6. Critical Aggregation Concentration (CAC) Determination for PG-Surfactants Using 8-Anilino-Naphtharene-1-Sulfonic Acid (ANS)
3.7. Replacement of Solubilization Surfactant via PEG Precipitation
3.8. Isolation of the Conjugates of PSI and DKDKC12K-PAn (n = 5, 7, 18)
3.9. Evaluation of Photo-Induced Electron-Transfer Rate of PSI Based on Decreases in O2 Concentration
3.10. Fluorescence Spectrum of PSI at 77 K
3.11. TEM Measurements of the Conjugates of PSI and DKDKC12K-PAn (n = 5, 7, 18)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dekkera, J.P.; Boekema, E.J. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 2005, 1706, 12–39. [Google Scholar] [CrossRef] [Green Version]
- Marco, P.; Elman, T.; Yacoby, I. Binding of ferredoxin NADP+ oxidoreductase (FNR) to plant photosystem I. Biochim. Biophys. Acta Bioenergy 2019, 1860, 689–698. [Google Scholar] [CrossRef]
- Utschig, L.M.; Soltau, S.R.; Mulfort, K.L.; Niklas, J.; Poluektov, O.G. Z-Scheme solar water splitting via self-assembly of photosystem I-catalyst hybrids in thylakoid membranes. Chem. Sci. 2018, 9, 8504–8512. [Google Scholar] [CrossRef] [Green Version]
- Ciesielski, P.N.; Scott, A.M.; Faulkner, C.J.; Berron, B.J.; Cliffel, D.E.; Jennings, G.K. Functionalized nanoporous gold leaf electrode films for the immobilization of photosystem I. ACS Nano 2008, 23, 2465–2472. [Google Scholar] [CrossRef]
- Utschig, L.M.; Silver, S.C.; Mulfort, K.L.; Tiede, D.M. Nature-Driven photochemistry for catalytic solar hydrogen production: A photosystem I-Transition metal catalyst hybrid. J. Am. Chem. Soc. 2011, 133, 16334–16337. [Google Scholar] [CrossRef]
- Zhao, F.; Conzuelo, F.; Hartmann, V.; Li, H.; Nowaczyk, M.M.; Plumere, N.; Rögner, M.; Schuhmann, W. Light induced H2 evolution from a biophotocathode based on photosystem 1-Pt nanoparticles complexes integrated in solvated redox polymers films. J. Phys. Chem. B 2015, 119, 13726–13731. [Google Scholar] [CrossRef]
- Ihara, M.; Nishihara, H.; Yoon, K.; Lenz, O.; Friedrich, B.; Nakamoto, H.; Kojima, K.; Honma, D.; Kamachi, T.; Okura, I. Light-Driven hydrogen production by a hybrid complex of a [NiFe]-Hydrogenase and the cyanobacterial photosystem I. Photochem. Photobiol. 2006, 82, 676–682. [Google Scholar] [CrossRef]
- Grimme, R.A.; Lubner, C.E.; Bryant, D.A.; Golbeck, J.H. Photosystem I/Molecular wire/metal nanoparticle bioconjugates for the photocatalytic production of H2. J. Am. Chem. Soc. 2008, 130, 6308–6309. [Google Scholar] [CrossRef]
- Nguyen, K.; Bruce, B.D. Growing green electricity: Progress and strategies for use of photosystem I for sustainable photovoltaic energy conversion. Biochim. Biophys. Acta 2014, 1837, 1553–1566. [Google Scholar] [CrossRef] [Green Version]
- Ciornii, D.; Riedel, M.; Stieger, K.R.; Feifel, S.C.; Hejazi, M.; Lokstein, H.; Zouni, A.; Lisdat, F. Bioelectronic circuit on a 3D electrode architecture: Enzymatic catalysis interconnected with photosystem I. J. Am. Chem. Soc. 2017, 139, 16478–16481. [Google Scholar] [CrossRef]
- Misawa, N.; Fujii, S.; Kamiya, K.; Osaki, T.; Takaku, T.; Takahashi, Y.; Takeuchi, S. Construction of a biohybrid odorant sensor using biological olfactory receptors embedded into bilayer lipid membrane on a chip. ACS Sens. 2019, 4, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Nield, J.; Morris, E.P.; Bibby, T.S.; Barber, J. Structural analysis of the photosystem I supercomplex of cyanobacteria induced by iron deficiency. Biochemistry 2003, 42, 3180–3188. [Google Scholar] [CrossRef] [PubMed]
- Elter, S.; Raschle, T.; Arens, S.; Viegas, A.; Gelev, V.; Etzkorn, M.; Wagner, G. The use of amphipols for NMR structural characterization of 7-TM proteins. J. Membr. Biol. 2014, 247, 957–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, P.S.; Rasmussen, S.G.F.; Rana, R.; Gotfryd, K.; Chandra, R.; Goren, M.A.; Kruse, A.C.; Nurva, S.; Loland, C.J.; Pierre, Y.; et al. Maltose-Neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat. Methods 2010, 7, 1003–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tribet, C.; Audebert, R.; Popot, J.-L. Amphipols: Polymers that keep membrane proteins soluble in aqueous solutions. Proc. Natl. Acad. Sci. USA 1996, 93, 15047–15050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dörr, J.M.; Koorengevel, M.C.; Schäfer, M.; Prokofyev, A.V.; Scheidelaar, S.; Cruijsen, E.A.W.; Dafforn, T.R.; Baldus, M.; Killian, J.A. Detergent-Free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. USA 2014, 111, 18607–18612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barniol-Xicota, M.; Verhelst, S.H.L. Stable and functional rhomboid proteases in lipid nanodiscs by using diisobutylene/maleic acid copolymers. J. Am. Chem. Soc. 2018, 140, 14557–14561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Huang, R.; Ackermann, R.; Im, S.-C.; Waskell, L.; Schwendeman, A.; Ramamoorthy, A. Reconstitution of the Cytb5–CytP450 complex in nanodiscs for structural studies using NMR spectroscopy. Angew. Chem. Int. Ed. 2016, 55, 4497–4499. [Google Scholar] [CrossRef] [PubMed]
- Umezaki, K.; Sakai, S.; Koeda, S.; Yamamoto, Y.; Kondo, M.; Ikeda, A.; Dewa, T.; Taga, K.; Tanaka, T.; Mizuno, T. Formation of planar bilayer membranes on solid supports using peptide gemini surfactants. Chem. Lett. 2012, 41, 1430–1432. [Google Scholar] [CrossRef]
- Koeda, S.; Umezaki, K.; Noji, T.; Ikeda, A.; Kawakami, K.; Kondo, M.; Yamamoto, Y.; Shen, J.R.; Taga, K.; Dewa, T.; et al. Application of peptide gemini surfactants as novel solubilization surfactants for photosystems I and II of cyanobacteria. Langmuir 2013, 29, 11667–11680. [Google Scholar] [CrossRef]
- Koeda, S.; Suzuki, T.; Noji, T.; Kawakami, K.; Itoh, S.; Dewa, T.; Kamiya, N.; Mizuno, T. Rational design of novel high molecular weight solubilization surfactants for membrane proteins from the peptide gemini surfactants (PG-Surfactants). Tetrahedron 2016, 72, 6898–6908. [Google Scholar] [CrossRef]
- Shibata, M.; Koeda, S.; Noji, T.; Kawakami, K.; Ido, Y.; Amano, Y.; Umezawa, N.; Higuchi, T.; Dewa, T.; Itoh, S.; et al. Design of new extraction surfactants for membrane proteins from peptide gemini surfactants. Bioconjug. Chem. 2016, 27, 2469–2479. [Google Scholar] [CrossRef]
- Taniguchi, A.; Koeda, S.; Noji, T.; Kawakami, K.; Sumito, N.; Dewa, T.; Itoh, S.; Kamiya, N.; Mizuno, T. Synthesis and characterization of chemically-reactive solubilization surfactants for membrane proteins and preparation of membrane protein hydrogel microfibers. Colloid Interface Sci. Commun. 2019, 32, 100199. [Google Scholar] [CrossRef]
- Reisinger, V.; Eichacker, L.A. Analysis of membrane protein complexes by blue native PAGE. Proteomics 2006, 6 (Suppl. 2), 6–15. [Google Scholar] [CrossRef]
- Sassolas, A.; Hayat, A.; Marty, J.-L. Enzyme immobilization by entrapment within a gel network. Methods Mol. Biol. 2013, 1051, 229–239. [Google Scholar] [PubMed]
- McCormick, C.L.; Lowe, A.B. Aqueous RAFT polymerization: Recent developments in synthesis of functional water-soluble (co)polymers with controlled structures. Acc. Chem. Res. 2004, 37, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bulmus, V.; Herlambang, D.L.; Barner-Kowollik, C.; Stenzel, M.H.; Davis, T.P. In situ formation of protein–polymer conjugates through reversible addition fragmentation chain transfer polymerization. Angew. Chem. Int. Ed. 2007, 46, 3099–3103. [Google Scholar] [CrossRef]
- Jordan, P.; Fromme, P.; Witt, H.T.; Klukas, O.; Saenger, W.; Krauû, N. Three-Dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 2001, 411, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.R. Denshidentatsu kassei. Low Temp. Sci. 2009, 67, 551–560. [Google Scholar]
- De Vendittis, E.; Palumbo, G.; Parlato, G.; Bocchini, V. A fluorimetric method for the estimation of the critical micelle concentration of surfactants. Anal. Biochem. 1981, 115, 278–286. [Google Scholar] [CrossRef]







| PAn-DTP | Mn (PDI) a |
|---|---|
| PA5-DTP | 4500 (1.19) |
| PA7-DTP | 6900 (1.29) |
| PA18-DTP | 17,800 (1.14) |
| CAC (wt%) | CAC (µM) | d (nm) a | |
|---|---|---|---|
| DKDKC12K | --- | 8.3 b | 6 b |
| DKDKC12K-PA5 | 0.030 | 51 c | 13 |
| DKDKC12K-PA7 | 0.038 | 46 c | 25 |
| DKDKC12K-PA18 | 0.090 | 47 c | 29 |
| Solubilization Surfactant | Electron Transfer Rate in PSI (PSI−1s−1) a |
|---|---|
| β-DDM | 32.2 ± 5.9 |
| DKDKC12K-PA5 | 35.8 ± 1.3 |
| DKDKC12K-PA7 | 41.2 ± 0.6 |
| DKDKC12K-PA18 | 38.0 ± 0.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimamoto, T.; Nakakubo, T.; Noji, T.; Koeda, S.; Kawakami, K.; Kamiya, N.; Mizuno, T. Design of PG-Surfactants Bearing Polyacrylamide Polymer Chain to Solubilize Membrane Proteins in a Surfactant-Free Buffer. Int. J. Mol. Sci. 2021, 22, 1524. https://doi.org/10.3390/ijms22041524
Shimamoto T, Nakakubo T, Noji T, Koeda S, Kawakami K, Kamiya N, Mizuno T. Design of PG-Surfactants Bearing Polyacrylamide Polymer Chain to Solubilize Membrane Proteins in a Surfactant-Free Buffer. International Journal of Molecular Sciences. 2021; 22(4):1524. https://doi.org/10.3390/ijms22041524
Chicago/Turabian StyleShimamoto, Taro, Tatsuki Nakakubo, Tomoyasu Noji, Shuhei Koeda, Keisuke Kawakami, Nobuo Kamiya, and Toshihisa Mizuno. 2021. "Design of PG-Surfactants Bearing Polyacrylamide Polymer Chain to Solubilize Membrane Proteins in a Surfactant-Free Buffer" International Journal of Molecular Sciences 22, no. 4: 1524. https://doi.org/10.3390/ijms22041524
APA StyleShimamoto, T., Nakakubo, T., Noji, T., Koeda, S., Kawakami, K., Kamiya, N., & Mizuno, T. (2021). Design of PG-Surfactants Bearing Polyacrylamide Polymer Chain to Solubilize Membrane Proteins in a Surfactant-Free Buffer. International Journal of Molecular Sciences, 22(4), 1524. https://doi.org/10.3390/ijms22041524