Biofilm Spreading by the Adhesin-Dependent Gliding Motility of Flavobacterium johnsoniae. 1. Internal Structure of the Biofilm
Abstract
:1. Introduction
2. Results
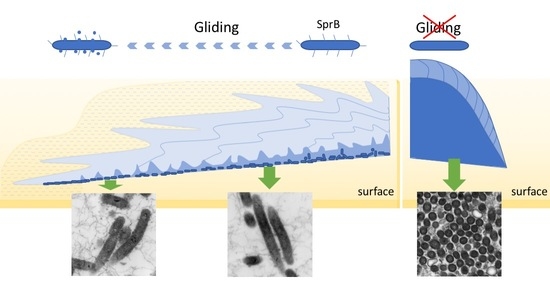
2.1. WT F. johnsoniae Colonies Spread on Nutrient-Poor Agar Surfaces
2.2. Movement of Single Bacterial Cells in the WT Colony Spread on 1% A-PY2
2.3. Static Single Bacterial Cells in the ΔsprB Mutant Colony on 1% A-PY2
2.4. WT F. johnsoniae Colonies Form a Biofilm on Nutrient-Poor Agar Surfaces
2.5. Bacteria at the Advancing Front of WT Colonies Secreted Many Vesicles
2.6. WT Cells form a Queue at the Advancing Vertical Top of the Colony
2.7. Biofilm Maturation Depends on the Cell-Surface Adhesin SprB
2.8. Surface Connecting Structure of F. johnsoniae Cells in Biofilm
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Biofilm Cultivation
4.2. Fixation
4.3. Epon Embedding and Sectioning
4.4. TEM Imaging
4.5. Quantification of Vesicles
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| WT | Wild type |
| ΔsprB | sprB deletion mutant CJ1922 |
| T9SS | Type IX secretion system |
| EPM | Extracellular polymeric matrix |
| TEM | Transmission electron microscopy |
References
- Zhu, Y.; McBride, M.J. Deletion of the Cytophaga hutchinsonii type IX secretion system gene sprP results in defects in gliding motility and cellulose utilization. Appl. Microbiol. Biotechnol. 2014, 98, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Hunnicutt, D.W.; McBride, M.J. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl. Acad. Sci. USA 1997, 94, 12139–12144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBride, M.J. Cytophaga-flavobacterium gliding motility. J. Mol. Microbiol. Biotechnol. 2004, 7, 63–71. [Google Scholar] [CrossRef]
- Shrivastava, A.; Berg, H.C. Towards a model for Flavobacterium gliding. Curr. Opin. Microbiol. 2015, 28, 93–97. [Google Scholar] [CrossRef] [Green Version]
- Nelson, S.S.; Bollampalli, S.; McBride, M.J. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J. Bacteriol. 2008, 190, 2851–2857. [Google Scholar] [CrossRef] [Green Version]
- Nakane, D.; Sato, K.; Wada, H.; McBride, M.J.; Nakayama, K. Helical flow of surface protein required for bacterial gliding motility. Proc. Natl. Acad. Sci. USA 2013, 110, 11145–11150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrivastava, A.; Roland, T.; Berg, H.C. The Screw-Like Movement of a Gliding Bacterium Is Powered by Spiral Motion of Cell-Surface Adhesins. Biophys J. 2016, 111, 1008–1013. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; McBride, M.J. Cell surface Filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J. Bacteriol. 2007, 189, 7503–7506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, K.; Naito, M.; Yukitake, H.; Hirakawa, H.; Shoji, M.; McBride, M.J.; Rhodes, R.G.; Nakayama, K. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 276–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBride, M.J.; Zhu, Y. Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J. Bacteriol. 2013, 195, 270–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, J.J.; Shrivastava, A.; McBride, M.J. Untangling Flavobacterium johnsoniae Gliding Motility and Protein Secretion. J. Bacteriol. 2018, 200, e00362-17. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, S.S.; Johnston, J.J.; Zhu, Y.; Hying, Z.T.; McBride, M.J. The Carboxy-Terminal Region of Flavobacterium johnsoniae SprB Facilitates Its Secretion by the Type IX Secretion System and Propulsion by the Gliding Motility Machinery. J. Bacteriol. 2019, 201, e00218–e00219. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Guan, Z.; Gao, P.; Zhang, W.; Qi, Q.; Lu, X. Cytophaga hutchinsonii gldN, encoding a core component of the type IX secretion system, is essential for ion assimilation, cellulose degradation, and cell motility. Appl. Environ. Microbiol. 2020, 86, e00242-20. [Google Scholar] [CrossRef]
- Gavriilidou, A.; Gutleben, J.; Versluis, D.; Forgiarini, F.; Passel, M.W.J.; Ingham, C.J.; Smidt, H.; Sipkema, D. Comparative genomic analysis of Flavobacteriaceae: Insights into carbohydrate metabolism, gliding motility and secondary metabolite biosynthesis. BMC Genom. 2020, 21, 569. [Google Scholar] [CrossRef]
- Rhodes, R.G.; Samarasam, M.N.; Van Groll, E.J.; McBride, M.J. Mutations in Flavobacterium johnsoniae sprE result in defects in gliding motility and protein secretion. J. Bacteriol. 2011, 193, 5322–5327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, K.; Naya, M.; Hatano, Y.; Kondo, Y.; Sato, M.; Narita, Y.; Nagano, K.; Naito, M.; Nakayama, K.; Sato, C. Colony spreading of the gliding bacterium Flavobacterium johnsoniae in the absence of the motility adhesin SprB. Sci. Rep. 2021, 11, 697. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Bowen, W.H.; Burne, R.A.; Wu, H.; Koo, H. Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments. Trends Microbiol. 2018, 26, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Xue-song, H.; Wen-yuan, S. Oral microbiology: Past, present and future. Int. J. Oral Sci. 2009, 1, 47–58. [Google Scholar]
- Parsek, M.R.; Singh, P.K. Bacterial biofilms: An emerging link to disease pathogenesis. Annu. Rev. Microbiol. 2003, 57, 677–701. [Google Scholar] [CrossRef] [PubMed]
- Brady, R.A.; Leid, J.G.; Calhoun, J.H.; Costerton, J.W.; Shirtliff, M.E. Osteomyelitis and the role of biofilms in chronic infection. FEMS Immunol. Med Microbiol. 2008, 52, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decostere, A.; Haesebrouck, F.; Charlier, G.; Ducatelle, R. The association of Flavobacterium columnare strains of high and low virulence with gill tissue of black mollies (Poecilia sphenops). Vet. Microbiol. 1999, 67, 287–298. [Google Scholar] [CrossRef]
- Levipan, H.A.; Avendaño-Herrera, R. Different Phenotypes of Mature Biofilm in Flavobacterium psychrophilum Share a Potential for Virulence That Differs from Planktonic State. Front. Cell. Infect. Microbiol. 2017, 7, 76. [Google Scholar] [CrossRef]
- Kondo, M.; Kawai, K.; Kurohara, K.; Oshima, S. Adherence of Flavobacterium psychrophilum on the body surface of the ayu Plecoglossus altivelis. Microbes Infect. 2002, 4, 279–283. [Google Scholar] [CrossRef]
- Cai, W.; De La Fuente, L.; Arias, C.R. Biofilm formation by the fish pathogen Flavobacterium columnare: Development and parameters affecting surface attachment. Appl. Environ. Microbiol. 2013, 79, 5633–5642. [Google Scholar] [CrossRef] [Green Version]
- Loch, T.P.; Faisal, M. Emerging flavobacterial infections in fish: A review. J. Adv. Res. 2015, 6, 283–300. [Google Scholar] [CrossRef] [PubMed]
- Hess, E.; Renzi, F.; Karhunen, P.; Do, M.; Lefèvre, A.; Antikainen, J.; Carlier, E.; Hästbacka, J.; Cornelis, G.R. Capnocytophaga canimorsus Capsular Serovar and Disease Severity, Helsinki Hospital District, Finland, 2000–2017. Emerg. Infect. Dis. 2018, 24, 2195–2201. [Google Scholar] [CrossRef] [PubMed]
- Mally, M.; Shin, H.; Paroz, C.; Landmann, R.; Cornelis, G.R. Capnocytophaga canimorsus: A Human Pathogen Feeding at the Surface of Epithelial Cells and Phagocytes. PLoS Pathog. 2008, e1000164. [Google Scholar] [CrossRef]
- Laanto, E.; Penttinen, R.K.; Bamford, J.K.; Sundberg, L.R. Comparing the different morphotypes of a fish pathogen-implications for key virulence factors in Flavobacterium columnare. BMC Microbiol. 2014, 14, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Zhu, Y.; LaFrentz, B.R.; Evenhuis, J.P.; Hunnicutt, D.W.; Conrad, R.A.; Barbier, P.; Gullstrand, C.W.; Roets, J.E.; Powers, J.L.; et al. The Type IX Secretion System Is Required for Virulence of the Fish Pathogen Flavobacterium columnare. Appl. Environ. Microbiol. 2017, 83, e01769-17. [Google Scholar] [CrossRef] [Green Version]
- Barbier, P.; Rochat, T.; Mohammed, H.H.; Wiens, G.D.; Bernardet, J.F.; Halpern, D.; Duchaud, E.; McBride, M.J. The Type IX Secretion System Is Required for Virulence of the Fish Pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 2020, 86, e00799-20. [Google Scholar] [CrossRef]
- Pérez-Pascual, D.; Rochat, T.; Kerouault, B.; Gómez, E.; Neulat-Ripoll, F.; Henry, C.; Quillet, E.; Guijarro, J.A.; Bernardet, J.F.; Duchaud, E. More than gliding: Involvement of GldD and GldG in the virulence of Flavobacterium psychrophilum. Front. Microbiol. 2017, 8, 2168. [Google Scholar] [CrossRef] [Green Version]
- Shrivastava, A.; Patel, V.K.; Tang, Y.; Yost, S.C.; Dewhirst, F.E.; Berg, H.C. Cargo transport shapes the spatial organization of a microbial community. Proc. Natl. Acad. Sci. USA 2018, 115, 8633–8638. [Google Scholar] [CrossRef] [Green Version]
- Rhodes, R.G.; Pucker, H.G.; McBride, M.J. Development and use of a gene deletion strategy for Flavobacterium johnsoniae to identify the redundant gliding motility genes remF, remG, remH, and remI. J. Bacteriol. 2011, 193, 2418–2428. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Kaufman, M.G.; Bagdasarian, M.; Bates, A.K.; Walker, E.D. Development of an efficient expression system for Flavobacterium strains. Gene 2010, 458, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Conover, M.; Lu, H.; Parsek, M.R.; Bayles, K.; Wozniak, D.J. Assembly and Development of the Pseudomonas aeruginosa Biofilm Matrix. PLOS Pathog. 2009, 5, e1000354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, G.; Sharma, S.; Sharma, P.; Chandola, D.; Dang, S.; Gupta, S.; Gabrani, R. Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 2016, 121, 309–319. [Google Scholar] [CrossRef] [Green Version]
- Prigent-Combaret, C.; Prensier, G.; Le Thi, T.T.; Vidal, O.; Lejeune, P.; Dorel, C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: Role of flagella, curli and colanic acid. Environ. Microbiol. 2000, 2, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Roschitzki, B.; Riedel, K.; Eberl, L. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J. Proteome Res. 2012, 11, 4906–4915. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barken, K.B.; Pamp, S.J.; Yang, L.; Gjermansen, M.; Bertrand, J.J.; Klausen, M.; Givskov, M.; Whitchurch, C.B.; Engel, J.N.; Tolker-Nielsen, T. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2008, 10, 2331–2343. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef]
- Wang, S.; Yu, S.; Zhang, Z.; Wei, Q.; Yan, L.; Ai, G.; Liu, H.; Ma, L.Z. Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2014, 80, 6724–6732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omar, A.; Wright, J.B.; Schultz, G.; Burrell, R.; Nadworny, P. Microbial biofilms and chronic wounds. Microorganisms 2017, 5, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimoto, S.; Okuda, K.; Miyakawa, R.; Sato, M.; Arita-Morioka, K.; Chiba, A.; Yamanaka, K.; Ogura, T.; Mizunoe, Y.; Sato, C. Imaging of bacterial multicellular behaviour in biofilms in liquid by atmospheric scanning electron microscopy. Sci. Rep. 2016, 6, 25889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuda, K.I.; Nagahori, R.; Yamada, S.; Sugimoto, S.; Sato, C.; Sato, M.; Iwase, T.; Hashimoto, K.; Mizunoe, Y. The composition and structure of biofilms developed by Propionibacterium acnes Isolated from Cardiac Pacemaker Devices. Front. Microbiol. 2018, 9, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.H.; Gorce, J.B.; Punzmann, H.; Francois, N.; Shats, M.; Xia, H. Surface waves control bacterial attachment and formation of biofilms in thin layers. Sci. Adv. 2020, 6, eaaz9386. [Google Scholar] [CrossRef]
- Bazaka, K.; Jacob, M.V.; Crawford, R.J.; Ivanova, E.P. Efficient surface modification of biomaterial to prevent biofilm formation and the attachment of microorganisms. Appl. Microbiol. Biotechnol. 2012, 95, 299–331. [Google Scholar] [CrossRef]
- Klausen, M.; Heydorn, A.; Ragas, P.; Lambertsen, L.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003, 48, 1511–1524. [Google Scholar] [CrossRef]
- Sato, K.; Naya, M.; Hatano, Y.; Kondo, Y.; Sato, M.; Narita, Y.; Nagano, K.; Naito, M.; Sato, C. Biofilm Spreading by the Adhesin-Dependent Gliding Motility of Flavobacterium johnsoniae. 2. Surface Structure of the Biofilm. submitted.
- Sato, C.; Yamazaki, D.; Sato, M.; Takeshima, H.; Memtily, N.; Hatano, Y.; Tsukuba, T.; Sakai, E. Calcium phosphate mineralization in bone tissues directly observed in aqueous liquid by atmospheric SEM (ASEM) without staining: Microfluidics crystallization chamber and immuno-EM. Sci. Rep. 2019, 9, 7352. [Google Scholar] [CrossRef] [PubMed]
- Naya, M.; Sato, C. Pyrene Excimer-Based Fluorescent Labeling of Neighboring Cysteines by Protein Dynamics: ASEM-Induced Thiol-Ene Click Reaction for High Spatial Resolution CLEM. Int. J. Mol. Sci. 2020, 21, 7550. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.; Puhler, A. A Broad host range mobilization system for In vivo genetic engineering: Transposon mutagenesis in Gram negative bacteria. BioTechnology 1983, 1, 784–791. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, K.; Naya, M.; Hatano, Y.; Kondo, Y.; Sato, M.; Nagano, K.; Chen, S.; Naito, M.; Sato, C. Biofilm Spreading by the Adhesin-Dependent Gliding Motility of Flavobacterium johnsoniae. 1. Internal Structure of the Biofilm. Int. J. Mol. Sci. 2021, 22, 1894. https://doi.org/10.3390/ijms22041894
Sato K, Naya M, Hatano Y, Kondo Y, Sato M, Nagano K, Chen S, Naito M, Sato C. Biofilm Spreading by the Adhesin-Dependent Gliding Motility of Flavobacterium johnsoniae. 1. Internal Structure of the Biofilm. International Journal of Molecular Sciences. 2021; 22(4):1894. https://doi.org/10.3390/ijms22041894
Chicago/Turabian StyleSato, Keiko, Masami Naya, Yuri Hatano, Yoshio Kondo, Mari Sato, Keiji Nagano, Shicheng Chen, Mariko Naito, and Chikara Sato. 2021. "Biofilm Spreading by the Adhesin-Dependent Gliding Motility of Flavobacterium johnsoniae. 1. Internal Structure of the Biofilm" International Journal of Molecular Sciences 22, no. 4: 1894. https://doi.org/10.3390/ijms22041894
APA StyleSato, K., Naya, M., Hatano, Y., Kondo, Y., Sato, M., Nagano, K., Chen, S., Naito, M., & Sato, C. (2021). Biofilm Spreading by the Adhesin-Dependent Gliding Motility of Flavobacterium johnsoniae. 1. Internal Structure of the Biofilm. International Journal of Molecular Sciences, 22(4), 1894. https://doi.org/10.3390/ijms22041894