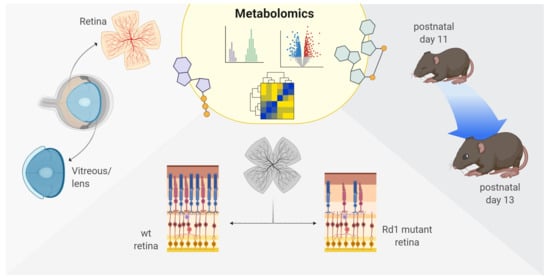
Metabolic Analysis of Vitreous/Lens and Retina in Wild Type and Retinal Degeneration Mice
Abstract
:1. Introduction
2. Results
2.1. More Than Meets The Eye: Metabolic Dysfunctions Underlie Retina Degeneration
2.2. A Comprehensive Metabolomic Analysis Unravels Differences Across Tissues, Age, Health, and Disease
2.3. Eye Opening Results in Minimal Effects on the Wild Type Retina and Vitreous/Lens
2.4. The Rd1 Mutation Induces Changes in the Composition of Vitreous/Lens at Eye Opening
2.5. Differences in Selected Metabolites Underlie The Impact of Degeneration on Vitreous/Lens and Retina Across Time-Points
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Tissue Isolation
4.3. Protein Extraction and MS Measurements
4.4. MS Data Processing
4.5. Metabolite Extraction and Spectroscopy
4.6. Analysis of Metabolic Data
4.7. Immunohistochemistry
4.8. Image Acquisition and Visualization
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IRDs | Inherited retinal disorders |
| RPE | Retinal pigmented epithelium |
| Rd1 | Pde6bRd1 mutation |
| cGMP | Cyclic guanosine monophosphate |
| PCA | Principal component analysis |
| MSEA | Metabolite set enrichment analysis |
| AMP | Cyclic adenosine monophosphate |
| CMP | Cyclic cytidine monophosphate |
| PPP | Pentose phosphate pathway |
| GSH | Reduced Glutathione |
| GSSG | Oxidized Glutathione |
| GSEA | Gene set enrichment analysis |
References
- Henderson, R.H. Inherited retinal dystrophies. Paediatr. Child. Health 2020, 30, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Narayan, D.S.; Chidlow, G.; Wood, J.P.P.M.; Casson, R.J. Glucose metabolism in mammalian photoreceptor inner and outer segments. Clin. Exp. Ophthalmol. 2017, 45, 730–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sancho-Pelluz, J.; Arango-Gonzalez, B.; Kustermann, S.; Romero, F.J.; Van Veen, T.; Zrenner, E.; Ekström, P.; Paquet-Durand, F. Photoreceptor cell death mechanisms in inherited retinal degeneration. Mol. Neurobiol. 2008, 38, 253–269. [Google Scholar] [CrossRef]
- Cheng, S.Y.; Cipi, J.; Ma, S.; Hafler, B.P.; Kanadia, R.N.; Brush, R.S.; Agbaga, M.P.; Punzo, C. Altered photoreceptor metabolism in mouse causes late stage age-related macular degeneration-like pathologies. Proc. Natl. Acad. Sci. USA 2020, 117, 13094–13104. [Google Scholar] [CrossRef] [PubMed]
- Rajappa, M.; Goyal, A.; Kaur, J. Inherited metabolic disorders involving the eye: A clinico-biochemical perspective. Eye 2010, 24, 507–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.; An, J.; Linton, J.D.; Wang, Y.; Hurley, J.B. How Excessive cGMP Impacts Metabolic Proteins in Retinas at the Onset of Degeneration. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2018; Volume 1074, pp. 289–295. [Google Scholar]
- Petit, L.; Ma, S.; Cipi, J.; Cheng, S.Y.; Zieger, M.; Hay, N.; Punzo, C. Aerobic Glycolysis Is Essential for Normal Rod Function and Controls Secondary Cone Death in Retinitis Pigmentosa. Cell Rep. 2018, 23, 2629–2642. [Google Scholar] [CrossRef] [Green Version]
- Chertov, A.O.; Holzhausen, L.; Kuok, I.T.; Couron, D.; Parker, E.; Linton, J.D.; Sadilek, M.; Sweet, I.R.; Hurley, J.B. Roles of Glucose in Photoreceptor Survival. J. Biol. Chem. 2011, 286, 34700–34711. [Google Scholar] [CrossRef] [Green Version]
- Chinchore, Y.; Begaj, T.; Wu, D.; Drokhlyansky, E.; Cepko, C.L. Glycolytic reliance promotes anabolism in photoreceptors. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Kanow, M.A.; Giarmarco, M.M.; Jankowski, C.S.R.; Tsantilas, K.; Engel, A.L.; Du, J.; Linton, J.D.; Farnsworth, C.C.; Sloat, S.R.; Rountree, A.; et al. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- FU, Z.; Kern, T.S.; Hellström, A.; Smith, L. Fatty acid oxidation and photoreceptor metabolic needs. J. Lipid Res. 2020, jlr.TR120000618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Léveillard, T.; Sahel, J.-A.A. Metabolic and redox signaling in the retina. Cell. Mol. Life Sci. 2017, 74, 3649–3665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruschi, M.; Bartolucci, M.; Petretto, A.; Calzia, D.; Caicci, F.; Manni, L.; Traverso, C.E.; Candiano, G.; Panfoli, I. Differential expression of the five redox complexes in the retinal mitochondria or rod outer segment disks is consistent with their different functionality. FASEB BioAdv. 2020, 2, 315–324. [Google Scholar] [CrossRef]
- Calzia, D.; Garbarino, G.; Caicci, F.; Manni, L.; Candiani, S.; Ravera, S.; Morelli, A.; Traverso, C.E.; Panfoli, I. Functional expression of electron transport chain complexes in mouse rod outer segments. Biochimie 2014, 102, 78–82. [Google Scholar] [CrossRef]
- Panfoli, I.; Musante, L.; Bachi, A.; Ravera, S.; Calzia, D.; Cattaneo, A.; Bruschi, M.; Bianchini, P.; Diaspro, A.; Morelli, A.; et al. Proteomic analysis of the retinal rod outer segment disks. J. Proteome Res. 2008, 7, 2654–2669. [Google Scholar] [CrossRef]
- Panfoli, I.; Calzia, D.; Bianchini, P.; Ravera, S.; Diaspro, A.; Candiano, G.; Bachi, A.; Monticone, M.; Aluigi, M.G.; Barabino, S.; et al. Evidence for aerobic metabolism in retinal rod outer segment disks. Int. J. Biochem. Cell Biol. 2009, 41, 2555–2565. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Rountree, A.; Cleghorn, W.M.; Contreras, L.; Lindsay, K.J.; Sadilek, M.; Gu, H.; Djukovic, D.; Raftery, D.; Satrústegui, J.; et al. Phototransduction influences metabolic flux and nucleotide metabolism in mouse retina. J. Biol. Chem. 2016, 291, 4698–4710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linton, J.D.; Holzhausen, L.C.; Babai, N.; Song, H.; Miyagishima, K.J.; Stearns, G.W.; Lindsay, K.; Wei, J.; Chertov, A.O.; Peters, T.A.; et al. Flow of energy in the outer retina in darkness and in light. Proc. Natl. Acad. Sci. USA 2010, 107, 8599–8604. [Google Scholar] [CrossRef] [Green Version]
- Okawa, H.; Sampath, A.P.; Laughlin, S.B.; Fain, G.L. ATP Consumption by Mammalian Rod Photoreceptors in Darkness and in Light. Curr. Biol. 2008, 18, 1917–1921. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, V.B.; Skeie, J.M. Translational vitreous proteomics. PROTEOMICS Clin. Appl. 2014, 8, 204–208. [Google Scholar] [CrossRef] [Green Version]
- Dahm, R.; van Marle, J.; Quinlan, R.A.; Prescott, A.R.; Vrensen, G.F.J.M. Homeostasis in the vertebrate lens: Mechanisms of solute exchange. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 1265–1277. [Google Scholar] [CrossRef] [Green Version]
- Yanshole, V.V.; Snytnikova, O.A.; Kiryutin, A.S.; Yanshole, L.V.; Sagdeev, R.Z.; Tsentalovich, Y.P. Metabolomics of the rat lens: A combined LC-MS and NMR study. Exp. Eye Res. 2014, 125, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Grenell, A.; Zhong, F.; Yam, M.; Hauer, A.; Gregor, E.; Zhu, S.; Lohner, D.; Zhu, J.; Du, J. Metabolic signature of the aging eye in mice. Neurobiol. Aging 2018, 71, 223–233. [Google Scholar] [CrossRef]
- Tan, S.Z.; Mullard, G.; Hollywood, K.A.; Dunn, W.B.; Bishop, P.N. Characterisation of the metabolome of ocular tissues and post-mortem changes in the rat retina. Exp. Eye Res. 2016, 149, 8–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Zhang, T.; Liu, W.; Wang, Y.; Xu, R.; Zeng, S.; Zhang, R.; Zhu, S.; Gillies, M.C.; Zhu, L.; et al. Metabolic Features of Mouse and Human Retinas: Rods versus Cones, Macula versus Periphery, Retina versus RPE. iScience 2020, 23, 101672. [Google Scholar] [CrossRef]
- Paris, L.P.; Johnson, C.H.; Aguilar, E.; Usui, Y.; Cho, K.; Hoang, L.T.; Feitelberg, D.; Benton, H.P.; Westenskow, P.D.; Kurihara, T.; et al. Global metabolomics reveals metabolic dysregulation in ischemic retinopathy. Metabolomics 2016, 12, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schori, C.; Trachsel, C.; Grossmann, J.; Barben, M.; Klee, K.; Storti, F.; Samardzija, M.; Grimm, C. A chronic hypoxic response in photoreceptors alters the vitreous proteome in mice. Exp. Eye Res. 2019, 185, 107690. [Google Scholar] [CrossRef]
- Weiss, E.R.; Osawa, S.; Xiong, Y.; Dhungana, S.; Carlson, J.; McRitchie, S.; Fennell, T.R. Broad spectrum metabolomics for detection of abnormal metabolic pathways in a mouse model for retinitis pigmentosa. Exp. Eye Res. 2019, 184, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Arango-Gonzalez, B.; Trifunović, D.; Sahaboglu, A.; Kranz, K.; Michalakis, S.; Farinelli, P.; Koch, S.; Koch, F.; Cottet, S.; Janssen-Bienhold, U.; et al. Identification of a Common Non-Apoptotic Cell Death Mechanism in Hereditary Retinal Degeneration. PLoS ONE 2014, 9, e112142. [Google Scholar] [CrossRef]
- Power, M.J.; Rogerson, L.E.; Schubert, T.; Berens, P.; Euler, T.; Paquet-Durand, F. Systematic spatiotemporal mapping reveals divergent cell death pathways in three mouse models of hereditary retinal degeneration. J. Comp. Neurol. 2020, 528, 1113–1139. [Google Scholar] [CrossRef] [Green Version]
- Power, M.; Das, S.; Schütze, K.; Marigo, V.; Ekström, P.; Paquet-Durand, F. Cellular mechanisms of hereditary photoreceptor degeneration—Focus on cGMP. Prog. Retin. Eye Res. 2020, 74, 100772. [Google Scholar] [CrossRef] [PubMed]
- Paquet-Durand, F.; Beck, S.; Michalakis, S.; Goldmann, T.; Huber, G.; Mühlfriedel, R.; Trifunović, D.; Fischer, M.D.; Fahl, E.; Duetsch, G.; et al. A key role for cyclic nucleotide gated (CNG) channels in cGMP-related retinitis pigmentosa. Hum. Mol. Genet. 2011, 20, 941–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wert, K.J.; Velez, G.; Kanchustambham, V.L.; Shankar, V.; Evans, L.P.; Sengillo, J.D.; Zare, R.N.; Bassuk, A.G.; Tsang, S.H.; Mahajan, V.B. Metabolite therapy guided by liquid biopsy proteomics delays retinal neurodegeneration. EBioMedicine 2020, 52, 102636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurley, J.B.; Chao, J.R. Retinal disease: How to use proteomics to speed up diagnosis and metabolomics to slow down degeneration. EBioMedicine 2020, 53, 102687. [Google Scholar] [CrossRef] [PubMed]
- Duda, M.; Kawula, K.; Pawlak, A.; Sarna, T.; Wisniewska-Becker, A. EPR Studies on the Properties of Model Photoreceptor Membranes Made of Natural and Synthetic Lipids. Cell Biochem. Biophys. 2017, 75, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Reveles, J.; Dhingra, A.; Alexander, D.; Bragin, A.; Philp, N.J.; Boesze-Battaglia, K. Phagocytosis-dependent ketogenesis in retinal pigment epithelium. J. Biol. Chem. 2017, 292, 8038–8047. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Zabel, M.K.; Wang, X.; Ma, W.; Shah, P.; Fariss, R.N.; Qian, H.; Parkhurst, C.N.; Gan, W.-B.; Wong, W.T. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol. Med. 2015, 7, 1179–1197. [Google Scholar] [CrossRef]
- Hamano, F.; Kuribayashi, H.; Iwagawa, T.; Tsuhako, A.; Nagata, K.; Sagara, H.; Shimizu, T.; Shindou, H.; Watanabe, S. Mapping membrane lipids in the developing and adult mouse retina under physiological and pathological conditions using mass spectrometry. J. Biol. Chem. 2021, 100303. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Kini, A.; Wang, Y.; Liu, T.; Chen, Y.; Vukmanic, E.; Emery, D.; Liu, Y.; Lu, X.; Jin, L.; et al. Metabolic Deregulation of the Blood-Outer Retinal Barrier in Retinitis Pigmentosa. Cell Rep. 2019, 28, 1323–1334.e4. [Google Scholar] [CrossRef] [Green Version]
- Vitvitsky, V.; Garg, S.K.; Banerjee, R. Taurine Biosynthesis by Neurons and Astrocytes. J. Biol. Chem. 2011, 286, 32002–32010. [Google Scholar] [CrossRef] [Green Version]
- Skeie, J.M.; Mahajan, V.B. Proteomic Interactions in the Mouse Vitreous-Retina Complex. PLoS ONE 2013, 8, e82140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adijanto, J.; Du, J.; Moffat, C.; Seifert, E.L.; Hurley, J.B.; Philp, N.J. The retinal pigment epithelium utilizes fatty acids for ketogenesis implications for metabolic coupling with the outer retina. J. Biol. Chem. 2014, 289, 20570–20582. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Lee, S.J.; Scott, P.A.; Lu, X.; Emery, D.; Liu, Y.; Ezashi, T.; Roberts, M.R.; Ross, J.W.; Kaplan, H.J.; et al. Two-Step Reactivation of Dormant Cones in Retinitis Pigmentosa. Cell Rep. 2016, 15, 372–385. [Google Scholar] [CrossRef] [Green Version]
- Joyal, J.S.; Gantner, M.L.; Smith, L.E.H. Retinal energy demands control vascular supply of the retina in development and disease: The role of neuronal lipid and glucose metabolism. Prog. Retin. Eye Res. 2018, 64, 131–156. [Google Scholar] [CrossRef]
- Izuta, Y.; Imada, T.; Hisamura, R.; Oonishi, E.; Nakamura, S.; Inagaki, E.; Ito, M.; Soga, T.; Tsubota, K. Ketone body 3-hydroxybutyrate mimics calorie restriction via the Nrf2 activator, fumarate, in the retina. Aging Cell 2018, 17, e12699. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, F.; Galluzzi, L.; Bravo-San Pedro, J.M.; Madeo, F.; Kroemer, G. Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 2015, 21, 805–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grenell, A.; Wang, Y.; Yam, M.; Swarup, A.; Dilan, T.L.; Hauer, A.; Linton, J.D.; Philp, N.J.; Gregor, E.; Zhu, S.; et al. Loss of MPC1 reprograms retinal metabolism to impair visual function. Proc. Natl. Acad. Sci. USA 2019, 116, 3530–3535. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Morales, P.; Pedraza-Chaverri, J.; Tapia, E. Ketone bodies, stress response, and redox homeostasis. Redox Biol. 2020, 29, 101395. [Google Scholar] [CrossRef]
- Shimazu, T.; Hirschey, M.D.; Newman, J.; He, W.; Shirakawa, K.; Le Moan, N.; Grueter, C.A.; Lim, H.; Saunders, L.R.; Stevens, R.D.; et al. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 2013, 339, 211–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newton, F.; Megaw, R. Mechanisms of Photoreceptor Death in Retinitis Pigmentosa. Genes 2020, 11, 1120. [Google Scholar] [CrossRef] [PubMed]
- Domènech, E.B.; Marfany, G. The relevance of oxidative stress in the pathogenesis and therapy of retinal dystrophies. Antioxidants 2020, 9, 347. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Veenstra, A.; Palczewski, K.; Kern, T.S. Photoreceptor cells are major contributors to diabetes-induced oxidative stress and local inflammation in the retina. Proc. Natl. Acad. Sci. USA 2013, 110, 16586–16591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, H.; Ding, M.; Chen, X.X.; Lu, Q. Microglial NADPH oxidase activation mediates rod cell death in the retinal degeneration in rd mice. Neuroscience 2014, 275, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Komeima, K.; Rogers, B.S.; Lu, L.; Campochiaro, P.A. Antioxidants reduce cone cell death in a model of retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 2006, 103, 11300–11305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campochiaro, P.A.; Mir, T.A. The mechanism of cone cell death in Retinitis Pigmentosa. Prog. Retin. Eye Res. 2018, 62, 24–37. [Google Scholar] [CrossRef]
- Trachsel-Moncho, L.; Benlloch-Navarro, S.; Fernández-Carbonell, Á.; Ramírez-Lamelas, D.T.; Olivar, T.; Silvestre, D.; Poch, E.; Miranda, M. Oxidative stress and autophagy-related changes during retinal degeneration and development. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Punzo, C.; Xiong, W.; Cepko, C.L. Loss of daylight vision in retinal degeneration: Are oxidative stress and metabolic dysregulation to blame? J. Biol. Chem. 2012, 287, 1642–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, Y.; Nakabeppu, Y.; Sonoda, K.-H. Oxidative Stress and Microglial Response in Retinitis Pigmentosa. Int. J. Mol. Sci. 2020, 21, 7170. [Google Scholar] [CrossRef] [PubMed]
- Vlachantoni, D.; Bramall, A.N.; Murphy, M.P.; Taylor, R.W.; Shu, X.; Tulloch, B.; van Veen, T.; Turnbull, D.M.; McInnes, R.R.; Wright, A.F. Evidence of severe mitochondrial oxidative stress and a protective effect of low oxygen in mouse models of inherited photoreceptor degeneration. Hum. Mol. Genet. 2011, 20, 322–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanges, D.; Comitato, A.; Tammaro, R.; Marigo, V. Apoptosis in retinal degeneration involves cross-talk between apoptosis-inducing factor (AIF) and caspase-12 and is blocked by calpain inhibitors. Proc. Natl. Acad. Sci. USA 2006, 103, 17366–17371. [Google Scholar] [CrossRef] [Green Version]
- Tokuda, K.; Baron, B.; Yamashiro, C.; Kuramitsu, Y.; Kitagawa, T.; Kobayashi, M.; Sonoda, K.; Kimura, K. Up-regulation of the pentose phosphate pathway and HIF-1α expression during neural progenitor cell induction following glutamate treatment in rat ex vivo retina. Cell Biol. Int. 2020, 44, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Kuehne, A.; Emmert, H.; Soehle, J.; Winnefeld, M.; Fischer, F.; Wenck, H.; Gallinat, S.; Terstegen, L.; Lucius, R.; Hildebrand, J.; et al. Acute Activation of Oxidative Pentose Phosphate Pathway as First-Line Response to Oxidative Stress in Human Skin Cells. Mol. Cell 2015, 59, 359–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Vallejo, V.; Benlloch-Navarro, S.; Trachsel-Moncho, L.; López-Pedrajas, R.; Almansa, I.; Romero, F.J.; Miranda, M. Alterations in glutamate cysteine ligase content in the retina of two retinitis pigmentosa animal models. Free Radic. Biol. Med. 2016, 96, 245–254. [Google Scholar] [CrossRef]
- Demougeot, C.; Garnier, P.; Mossiat, C.; Bertrand, N.; Giroud, M.; Beley, A.; Marie, C. N-Acetylaspartate, a marker of both cellular dysfunction and neuronal loss: Its relevance to studies of acute brain injury. J. Neurochem. 2001, 77, 408–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjartmar, C.; Battistuta, J.; Terada, N.; Dupree, E.; Trapp, B.D. N-acetylaspartate is an axon-specific marker of mature white matter in vivo: A biochemical and immunohistochemical study on the rat optic nerve. Ann. Neurol. 2002, 51, 51–58. [Google Scholar] [CrossRef]
- Moffett, J.R.; Arun, P.; Ariyannur, P.S.; Namboodiri, A.M.A. N-Acetylaspartate reductions in brain injury: Impact on post-injury neuroenergetics, lipid synthesis, and protein acetylation. Front. Neuroenergetics 2013, 5, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Karnovsky, A.; Weymouth, T.; Hull, T.; Glenn Tarcea, V.; Scardoni, G.; Laudanna, C.; Sartor, M.A.; Stringer, K.A.; Jagadish, H.V.; Burant, C.; et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 2012, 28, 373–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kammers, K.; Cole, R.N.; Tiengwe, C.; Ruczinski, I. Detecting significant changes in protein abundance. EuPA Open Proteomics 2015, 7, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, S.; Aftab, W.; Walenta, L.; Strauss, L.; Poutanen, M.; Mayerhofer, A.; Imhof, A. MALDI-IMS combined with shotgun proteomics identify and localize new factors in male infertility. Life Sci. Alliance 2021, 4, e202000672. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murenu, E.; Kostidis, S.; Lahiri, S.; Geserich, A.S.; Imhof, A.; Giera, M.; Michalakis, S. Metabolic Analysis of Vitreous/Lens and Retina in Wild Type and Retinal Degeneration Mice. Int. J. Mol. Sci. 2021, 22, 2345. https://doi.org/10.3390/ijms22052345
Murenu E, Kostidis S, Lahiri S, Geserich AS, Imhof A, Giera M, Michalakis S. Metabolic Analysis of Vitreous/Lens and Retina in Wild Type and Retinal Degeneration Mice. International Journal of Molecular Sciences. 2021; 22(5):2345. https://doi.org/10.3390/ijms22052345
Chicago/Turabian StyleMurenu, Elisa, Sarantos Kostidis, Shibojyoti Lahiri, Anna S. Geserich, Axel Imhof, Martin Giera, and Stylianos Michalakis. 2021. "Metabolic Analysis of Vitreous/Lens and Retina in Wild Type and Retinal Degeneration Mice" International Journal of Molecular Sciences 22, no. 5: 2345. https://doi.org/10.3390/ijms22052345
APA StyleMurenu, E., Kostidis, S., Lahiri, S., Geserich, A. S., Imhof, A., Giera, M., & Michalakis, S. (2021). Metabolic Analysis of Vitreous/Lens and Retina in Wild Type and Retinal Degeneration Mice. International Journal of Molecular Sciences, 22(5), 2345. https://doi.org/10.3390/ijms22052345