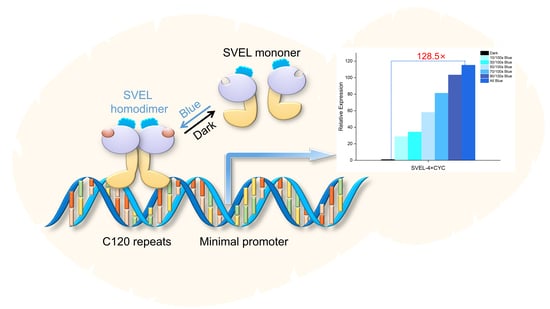
A Single-Component Blue Light-Induced System Based on EL222 in Yarrowia lipolytica
Abstract
:1. Introduction
2. Results
2.1. Design, Optimization and Verification of Blue Light Induction System Based on EL222
2.2. Multi-Copy Construction and Verification of Response Fragments
2.3. Dose-Dependent Activation
2.4. Assess the Performance at the Single-Cell Level
2.5. Light Switchable Spatial Activation
2.6. Temporal Behavior of pYLBI System
2.7. Blue Light-Induced Antibiotic Resistance
3. Discussion
4. Materials and Methods
4.1. Assembly of DNA Bioblocks
4.2. Strains and Growth Conditions
4.3. Microplate Reader Measurements
4.4. Flow Cytometry
4.5. Confocal Fluorescence Imaging
4.6. Imaging of Agar Plates
4.7. Quantitative PCR (qPCR) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Choi, K.R.; Jang, W.D.; Yang, D.; Cho, J.S.; Park, D.; Lee, S.Y. Systems metabolic engineering strategies: Integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 2019, 37, 817–837. [Google Scholar] [CrossRef]
- Woolston, B.M.; Edgar, S.; Stephanopoulos, G. Metabolic Engineering: Past and Future. Annu. Rev. Chem. Biomol. Eng. 2013, 4, 259–288. [Google Scholar] [CrossRef]
- Muhammad, A.; Feng, X.; Rasool, A.; Sun, W.; Li, C. Production of plant natural products through engineered Yarrowia lipolytica. Biotechnol. Adv. 2020, 43, 107555. [Google Scholar] [CrossRef]
- E Toettcher, J.; A Voigt, C.; Weiner, O.; A Lim, W. The promise of optogenetics in cell biology: Interrogating molecular circuits in space and time. Nat. Methods 2010, 8, 35–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deisseroth, K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 2015, 18, 1213–1225. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Yang, J.; Cho, E.; Lu, Y. Bringing Light into Cell-Free Expression. ACS Synth. Biol. 2020, 9, 2144–2153. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tonegawa, S. Optogenetics 3.0. Cell 2010, 141, 22–24. [Google Scholar] [CrossRef] [Green Version]
- Huala, E.; Oeller, P.W.; Liscum, E.; Han, I.-S.; Larsen, E.; Briggs, W.R. Arabidopsis NPH1: A Protein Kinase with a Putative Redox-Sensing Domain. Science 1997, 278, 2120–2123. [Google Scholar] [CrossRef]
- Zoltowski, B.D.; Schwerdtfeger, C.; Widom, J.; Loros, J.J.; Bilwes, A.M.; Dunlap, J.C.; Crane, B.R. Conformational Switching in the Fungal Light Sensor Vivid. Science 2007, 316, 1054–1057. [Google Scholar] [CrossRef] [Green Version]
- Halavaty, A.S.; Moffat, K. N- and C-Terminal Flanking Regions Modulate Light-Induced Signal Transduction in the LOV2 Domain of the Blue Light Sensor Phototropin 1 from Avena sativa. Biochemistry 2007, 46, 14001–14009. [Google Scholar] [CrossRef]
- Nash, A.I.; McNulty, R.; Shillito, M.E.; Swartz, T.E.; Bogomolni, R.A.; Luecke, H.; Gardner, K.H. Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc. Natl. Acad. Sci. USA 2011, 108, 9449–9454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pudasaini, A.; El-Arab, K.K.; Zoltowski, B.D. LOV-based optogenetic devices: Light-driven modules to impart photoregulated control of cellular signaling. Front. Mol. Biosci. 2015, 2, 18. [Google Scholar] [CrossRef] [Green Version]
- Beel, B.; Prager, K.; Spexard, M.; Sasso, S.; Weiss, D.; Müller, N.; Heinnickel, M.; Dewez, D.; Ikoma, D.; Grossman, A.R.; et al. A Flavin Binding Cryptochrome Photoreceptor Responds to Both Blue and Red Light in Chlamydomonas reinhardtii. Plant Cell 2012, 24, 2992–3008. [Google Scholar] [CrossRef] [Green Version]
- Lutz, R.; Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997, 25, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.R.; Pang, W.L.; Ostroff, N.A.; Baumgartner, B.L.; Nayak, S.; Tsimring, L.S.; Hasty, J. Metabolic gene regulation in a dynamically changing environment. Nature 2008, 454, 1119–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Nair, A.; Hermiston, T.W. A comparative study examining the cytotoxicity of inducible gene expression system ligands in different cell types. Toxicol. Vitr. 2008, 22, 261–266. [Google Scholar] [CrossRef]
- Shimizu-Sato, S.; Huq, E.; Tepperman, J.M.; Quail, P.H. A light-switchable gene promoter system. Nat. Biotechnol. 2002, 20, 1041–1044. [Google Scholar] [CrossRef]
- Mendelsohn, A.R. An enlightened genetic switch. Nat. Biotechnol. 2002, 20, 985–987. [Google Scholar] [CrossRef]
- Zhang, K.; Cui, B. Optogenetic control of intracellular signaling pathways. Trends Biotechnol. 2014, 33, 92–100. [Google Scholar] [CrossRef] [Green Version]
- Müller, K.; Naumann, S.; Weber, W.; Zurbriggen, M.D. Optogenetics for gene expression in mammalian cells. Biol. Chem. 2015, 396, 145–152. [Google Scholar] [CrossRef]
- Dance, A. Micromanagement with light. Nature 2015, 528, 291–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Du, Z.; Liu, R.; Li, T.; Zhao, Y.; Chen, X.; Yang, Y. A Single-Component Optogenetic System Allows Stringent Switch of Gene Expression in Yeast Cells. ACS Synth. Biol. 2018, 7, 2045–2053. [Google Scholar] [CrossRef]
- Zhao, E.M.; Zhang, Y.; Mehl, J.; Park, H.; Lalwani, M.A.; Toettcher, J.E.; Avalos, J.L. Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature 2018, 555, 683–687. [Google Scholar] [CrossRef]
- Strickland, D.; Moffat, K.; Sosnick, T.R. Light-activated DNA binding in a designed allosteric protein. Proc. Natl. Acad. Sci. USA 2008, 105, 10709–10714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Wwe, W.; Zhou, Y.; Ye, B. Construction of a light-controlled expression system and its application in Yarrowia lipolytica. Synth. Biol. J. 2021, 2, 778–791. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, Y.; Zhang, H. Design and Characterization of an Optogenetic System in Pichia pastoris. ACS Synth. Biol. 2022, 11, 297–307. [Google Scholar] [CrossRef]
- Ma, J.; Gu, Y.; Marsafari, M.; Xu, P. Synthetic biology, systems biology, and metabolic engineering of Yarrowia lipolytica toward a sustainable biorefinery platform. J. Ind. Microbiol. Biotechnol. 2020, 47, 845–862. [Google Scholar] [CrossRef]
- Darvishi, F.; Ariana, M.; Marella, E.R.; Borodina, I. Advances in synthetic biology of oleaginous yeast Yarrowia lipolytica for producing non-native chemicals. Appl. Microbiol. Biotechnol. 2018, 102, 5925–5938. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Lazar, Z.; Rakicka, M.; Guo, Z.; Fouchard, F.; Crutz-Le Coq, A.-M.; Nicaud, J.-M. Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab. Eng. 2016, 38, 115–124. [Google Scholar] [CrossRef]
- Li, H.; Alper, H.S. Enabling xylose utilization in Yarrowia lipolytica for lipid production. Biotechnol. J. 2016, 11, 1230–1240. [Google Scholar] [CrossRef]
- Abdel-Mawgoud, A.M.; Markham, K.A.; Palmer, C.; Liu, N.; Stephanopoulos, G.; Alper, H.S. Metabolic engineering in the host Yarrowia lipolytica. Metab. Eng. 2018, 50, 192–208. [Google Scholar] [CrossRef]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; De Montigny, J.; Marck, C.; Neuvéglise, C.; Talla, E.; et al. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef]
- Casare’gola, S.; Feynerol, C.; Diez, M.; Fournier, P.; Gaillardin, C. Genomic organization of the yeast Yarrowia lipolytica. Chromosoma 1997, 106, 380–390. [Google Scholar] [CrossRef]
- Nicaud, J.-M. Yarrowia lipolytica . Yeast 2012, 29, 409–418. [Google Scholar] [CrossRef]
- Sassi, H.; Delvigne, F.; Kar, T.; Nicaud, J.-M.; Coq, A.-M.C.-L.; Steels, S.; Fickers, P. Deciphering how LIP2 and POX2 promoters can optimally regulate recombinant protein production in the yeast Yarrowia lipolytica. Microb. Cell Fact. 2016, 15, 159. [Google Scholar] [CrossRef]
- Novikova, L.A.; Yovkova, V.; Luzikov, V.N.; Barth, G.; Mauersberger, S. Recombinant Yarrowia lipolytica strains for the heterologous expression of multi-component enzyme systems: Expression of mammalian steroidogenic proteins. J. Biotechnol. 2021, 339, 42–52. [Google Scholar] [CrossRef]
- Wei, W.-P.; Shang, Y.; Zhang, P.; Liu, Y.; You, D.; Yin, B.-C.; Ye, B.-C. Engineering Prokaryotic Transcriptional Activator XylR as a Xylose-Inducible Biosensor for Transcription Activation in Yeast. ACS Synth. Biol. 2020, 9, 1022–1029. [Google Scholar] [CrossRef]
- Larroude, M.; Rossignol, T.; Nicaud, J.-M.; Ledesma-Amaro, R. Synthetic biology tools for engineering Yarrowia lipolytica. Biotechnol. Adv. 2018, 36, 2150–2164. [Google Scholar] [CrossRef]
- Park, Y.-K.; Korpys, P.; Kubiak, M.; Celinska, E.; Soudier, P.; Trébulle, P.; Larroude, M.; Rossignol, T.; Nicaud, J.-M. Engineering the architecture of erythritol-inducible promoters for regulated and enhanced gene expression in Yarrowia lipolytica. FEMS Yeast Res. 2018, 19, foy105. [Google Scholar] [CrossRef]
- Park, Y.-K.; Vandermies, M.; Soudier, P.; Telek, S.; Thomas, S.; Nicaud, J.-M.; Fickers, P. Efficient expression vectors and host strain for the production of recombinant proteins by Yarrowia lipolytica in process conditions. Microb. Cell Fact. 2019, 18, 167. [Google Scholar] [CrossRef]
- Trassaert, M.; Vandermies, M.; Carly, F.; Denies, O.; Thomas, S.; Fickers, P.; Nicaud, J.-M. New inducible promoter for gene expression and synthetic biology in Yarrowia lipolytica. Microb. Cell Fact. 2017, 16, 141. [Google Scholar] [CrossRef]
- Takakado, A.; Nakasone, Y.; Terazima, M. Photoinduced dimerization of a photosensory DNA-binding protein EL222 and its LOV domain. Phys. Chem. Chem. Phys. 2017, 19, 24855–24865. [Google Scholar] [CrossRef]
- Rivera-Cancel, G.; Motta-Mena, L.B.; Gardner, K.H. Identification of Natural and Artificial DNA Substrates for Light-Activated LOV–HTH Transcription Factor EL222. Biochemistry 2012, 51, 10024–10034. [Google Scholar] [CrossRef] [PubMed]
- Takakado, A.; Nakasone, Y.; Terazima, M. Sequential DNA Binding and Dimerization Processes of the Photosensory Protein EL222. Biochemistry 2018, 57, 1603–1610. [Google Scholar] [CrossRef]
- Jayaraman, P.; Devarajan, K.; Chua, T.K.; Zhang, H.; Gunawan, E.; Poh, C.L. Blue light-mediated transcriptional activation and repression of gene expression in bacteria. Nucleic Acids Res. 2016, 44, 6994–7005. [Google Scholar] [CrossRef]
- Zhao, E.M.; Lalwani, M.A.; Chen, J.-M.; Orillac, P.; Toettcher, J.E.; Avalos, J.L. Optogenetic Amplification Circuits for Light-Induced Metabolic Control. ACS Synth. Biol. 2021, 10, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- LaBelle, J.; Ramos-Martinez, A.; Shen, K.; Motta-Mena, L.B.; Gardner, K.H.; Materna, S.C.; Woo, S. TAEL 2.0: An Improved Optogenetic Expression System for Zebrafish. Zebrafish 2021, 18, 20–28. [Google Scholar] [CrossRef]
- Rullan, M.; Benzinger, D.; Schmidt, G.W.; Milias-Argeitis, A.; Khammash, M. An Optogenetic Platform for Real-Time, Single-Cell Interrogation of Stochastic Transcriptional Regulation. Mol. Cell 2018, 70, 745–756.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhao, E.M.; Lalwani, M.A.; Lovelett, R.J.; García-Echauri, S.A.; Hoffman, S.M.; Gonzalez, C.L.; Toettcher, J.E.; Kevrekidis, I.G.; Avalos, J.L. Design and Characterization of Rapid Optogenetic Circuits for Dynamic Control in Yeast Metabolic Engineering. ACS Synth. Biol. 2020, 9, 3254–3266. [Google Scholar] [CrossRef]
- Kim, B.-K.; Kang, H.; Doh, K.-O.; Lee, S.-H.; Park, J.-W.; Lee, S.-J.; Lee, T.-J. Homodimeric SV40 NLS peptide formed by disulfide bond as enhancer for gene delivery. Bioorganic Med. Chem. Lett. 2012, 22, 5415–5418. [Google Scholar] [CrossRef]
- Reade, A.; Motta-Mena, L.B.; Gardner, K.H.; Stainier, D.Y.; Weiner, O.D.; Woo, S. TAEL: A zebrafish-optimized optogenetic gene expression system with fine spatial and temporal control. Development 2016, 144, 345–355. [Google Scholar] [CrossRef] [Green Version]
- Madzak, C.; Tréton, B.; Blanchin-Roland, S. Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. J. Mol. Microbiol. Biotechnol. 2000, 2, 207–216. [Google Scholar] [PubMed]
- Ogrydziak, D.M.; Scharf, S.J. Alkaline Extracellular Protease Produced by Saccharomycopsis lipolytica CX161-1B. Microbiology 1982, 128, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S.; Hoar, E.T.; Guarente, L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1985, 82, 8562–8566. [Google Scholar] [CrossRef] [Green Version]
- Várnai, A.; Tang, C.; Bengtsson, O.; Atterton, A.; Mathiesen, G.; Eijsink, V.G.H. Expression of endoglucanases in Pichia pastoris under control of the GAP promoter. Microb. Cell Fact. 2014, 13, 57. [Google Scholar] [CrossRef] [Green Version]
- Müller, S.; Sandal, T.; Kamp-Hansen, P.; Dalbøge, H. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. cloning of two novel promoters from Yarrowia lipolytica. Yeast 1998, 14, 1267–1283. [Google Scholar] [CrossRef]
- He, W.; Mu, W.; Jiang, B.; Yan, X.; Zhang, T. Food-Grade Expression of d-Psicose 3-Epimerase with Tandem Repeat Genes in Bacillus subtilis. J. Agric. Food Chem. 2016, 64, 5701–5707. [Google Scholar] [CrossRef]
- García-Granados, R.; Lerma-Escalera, J.A.; Morones-Ramírez, J.R. Metabolic Engineering and Synthetic Biology: Synergies, Future, and Challenges. Front. Bioeng. Biotechnol. 2019, 7, 36. [Google Scholar] [CrossRef]
- Wang, H.H.; Isaacs, F.J.; Carr, P.A.; Sun, Z.Z.; Xu, G.; Forest, C.R.; Church, G. Programming cells by multiplex genome engineering and accelerated evolution. Nature 2009, 460, 894–898. [Google Scholar] [CrossRef]
- Si, T.; Chao, R.; Min, Y.; Wu, Y.; Ren, W.; Zhao, H. Automated multiplex genome-scale engineering in yeast. Nat. Commun. 2017, 8, 15187. [Google Scholar] [CrossRef] [PubMed]
- Geijer, C.; Ledesma-Amaro, R.; Tomás-Pejó, E. Unraveling the potential of non-conventional yeasts in biotechnology. FEMS Yeast Res. 2022, 22, foab071. [Google Scholar] [CrossRef] [PubMed]
- Burini, J.A.; Eizaguirre, J.I.; Loviso, C.; Libkind, D. Levaduras no convencionales como herramientas de innovación y diferenciación en la producción de cerveza. Rev. Argent. Microbiol. 2021, 53, 359–377. [Google Scholar] [CrossRef]
- Binati, R.L.; Salvetti, E.; Bzducha-Wróbel, A.; Bašinskienė, L.; Čižeikienė, D.; Bolzonella, D.; E Felis, G. Non-conventional yeasts for food and additives production in a circular economy perspective. FEMS Yeast Res. 2021, 21, foab052. [Google Scholar] [CrossRef] [PubMed]
- Wendland, J. Special Issue: Non-Conventional Yeasts: Genomics and Biotechnology. Microorganisms 2019, 8, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jost, M.; Fernández-Zapata, J.; Polanco, M.C.; Ortiz-Guerrero, J.M.; Chen, P.Y.-T.; Kang, G.; Padmanabhan, S.; Elías-Arnanz, M.; Drennan, C.L. Structural basis for gene regulation by a B12-dependent photoreceptor. Nature 2015, 526, 536–541. [Google Scholar] [CrossRef] [Green Version]
- Nicaud, J.-M.; Madzak, C.; van den Broek, P.; Gysler, C.; Duboc, P.; Niederberger, P.; Gaillardin, C. Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Res. 2002, 2, 371–379. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Yan, Y.; Zhang, H. A Single-Component Blue Light-Induced System Based on EL222 in Yarrowia lipolytica. Int. J. Mol. Sci. 2022, 23, 6344. https://doi.org/10.3390/ijms23116344
Wang Z, Yan Y, Zhang H. A Single-Component Blue Light-Induced System Based on EL222 in Yarrowia lipolytica. International Journal of Molecular Sciences. 2022; 23(11):6344. https://doi.org/10.3390/ijms23116344
Chicago/Turabian StyleWang, Zhiqian, Yunjun Yan, and Houjin Zhang. 2022. "A Single-Component Blue Light-Induced System Based on EL222 in Yarrowia lipolytica" International Journal of Molecular Sciences 23, no. 11: 6344. https://doi.org/10.3390/ijms23116344
APA StyleWang, Z., Yan, Y., & Zhang, H. (2022). A Single-Component Blue Light-Induced System Based on EL222 in Yarrowia lipolytica. International Journal of Molecular Sciences, 23(11), 6344. https://doi.org/10.3390/ijms23116344