Hsp90 Activity Is Necessary for the Maturation of Rabies Virus Polymerase
Abstract
:1. Introduction
2. Results
2.1. RABV Infection Does Not Result in the Increased Expression of Hsp90 and Cdc37
2.2. None of the RABV Proteins Are Destabilized by Hsp90 Inhibition
2.3. RABV L Protein Is Expressed and Remains Soluble without the Presence of P Protein
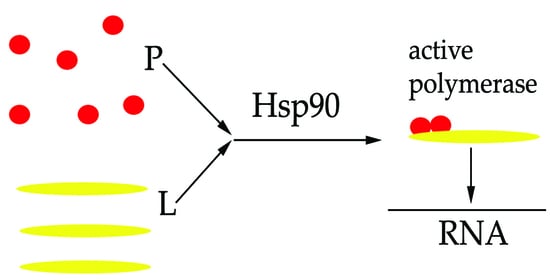
2.4. Hsp90 Is Required for the L and P Proteins Association
2.5. 17-AAG Does Not Inhibit RABV RNA Synthesis Early after Infection
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Virus
4.2. In Vitro Assay
4.3. Virus Titration
4.4. Cell Viability Assay
4.5. Reverse Transcription Quantitative PCR (RT-qPCR)
4.6. Plasmids Construction
4.7. Western Blot Analysis
4.8. Co-Immunoprecipitation (Co-IP)
4.9. Sequence and Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Geller, R.; Taguwa, S.; Frydman, J. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim. Biophys. Acta 2012, 1823, 698–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Jin, F.; Wang, R.; Li, F.; Wu, Y.; Kitazato, K. HSP90: A promising broad-spectrum antiviral drug target. Arch. Virol. 2017, 162, 3269–3282. [Google Scholar] [CrossRef] [PubMed]
- Panaretou, B.; Prodromou, C.; Roe, S.M.; O’Brien, R.; Ladbury, J.E.; Piper, P.W.; Pearl, L.H. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998, 17, 4829–4836. [Google Scholar] [PubMed] [Green Version]
- Obermann, W.M.; Sondermann, H.; Russo, A.A.; Pavletich, N.P.; Hartl, F.U. In Vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J. Cell Biol. 1998, 143, 901–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulte, T.W.; Neckers, L.M. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother. Pharmacol. 1998, 42, 273–279. [Google Scholar] [CrossRef]
- Tatokoro, M.; Koga, F.; Yoshida, S.; Kihara, K. Heat shock protein 90 targeting therapy: State of the art and future perspective. EXCLI J. 2015, 14, 48–58. [Google Scholar]
- Sanchez, J.; Carter, T.R.; Cohen, M.S.; Blagg, B.S.J. Old and new approaches to target the Hsp90 chaperone. Curr. Cancer Drug Targets 2020, 20, 253–270. [Google Scholar] [CrossRef]
- Fooks, A.R.; Banyard, A.C.; Horton, D.L.; Johnson, N.; McElhinney, L.M.; Jackson, A.C. Current status of rabies and prospects for elimination. Lancet 2014, 384, 1389–1399. [Google Scholar] [CrossRef]
- Marosi, A.; Dufkova, L.; Forro, B.; Felde, O.; Erdelyi, K.; Sirmarova, J.; Palus, M.; Honig, V.; Salat, J.; Tikos, R.; et al. Combination therapy of rabies-infected mice with inhibitors of pro-inflammatory host response, antiviral compounds and human rabies immunoglobulin. Vaccine 2019, 37, 4724–4735. [Google Scholar]
- Banyard, A.C.; Mansfield, K.L.; Wu, G.; Selden, D.; Thorne, L.; Birch, C.; Koraka, P.; Osterhaus, A.; Fooks, A.R. Re-evaluating the effect of Favipiravir treatment on rabies virus infection. Vaccine 2019, 37, 4686–4693. [Google Scholar]
- Smreczak, M.; Marzec, A.; Orlowska, A.; Trebas, P.; Reichert, M.; Kycko, A.; Koraka, P.; Osterhaus, A.; Zmudzinski, J.F. The effect of selected molecules influencing the detrimental host immune response on a course of rabies virus infection in a murine model. Vaccine 2019, 37, 4715–4723. [Google Scholar] [CrossRef]
- Liang, B. Structures of the mononegavirales polymerases. J. Virol. 2020, 94, e00175-20. [Google Scholar] [CrossRef]
- Bloyet, L.M.; Welsch, J.; Enchery, F.; Mathieu, C.; de Breyne, S.; Horvat, B.; Grigorov, B.; Gerlier, D. HSP90 Chaperoning in addition to phosphoprotein required for folding but not for supporting enzymatic activities of measles and nipah virus L polymerases. J. Virol. 2016, 90, 6642–6656. [Google Scholar] [CrossRef] [Green Version]
- Katoh, H.; Kubota, T.; Nakatsu, Y.; Tahara, M.; Kidokoro, M.; Takeda, M. Heat shock protein 90 ensures efficient mumps virus replication by assisting with viral polymerase complex formation. J. Virol. 2017, 91, e02220-16. [Google Scholar] [CrossRef] [Green Version]
- Connor, J.H.; McKenzie, M.O.; Parks, G.D.; Lyles, D.S. Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology 2007, 362, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Liu, F.; Liu, J.; Wang, D.; Yan, Y.; Ji, S.; Zan, J.; Zhou, J. The co-chaperone Cdc37 regulates the rabies virus phosphoprotein stability by targeting to Hsp90AA1 machinery. Sci. Rep. 2016, 6, 27123. [Google Scholar] [CrossRef]
- Madhusudana, S.N.; Sundaramoorthy, S.; Ullas, P.T. Utility of human embryonic kidney cell line HEK-293 for rapid isolation of fixed and street rabies viruses: Comparison with Neuro-2a and BHK-21 cell lines. Int. J. Infect. Dis. 2010, 14, e1067-71. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wu, H.; Tao, X.; Li, H.; Rayner, S.; Liang, G.; Tang, Q. Genetic and evolutionary characterization of RABVs from China using the phosphoprotein gene. Virol. J. 2013, 10, 14. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Yan, F.; Tao, J.; Zhu, X.; Liu, J.; Deng, S.; Zhang, X. Cdc37 facilitates cell survival of colorectal carcinoma via activating the CDK4 signaling pathway. Cancer Sci. 2018, 109, 656–665. [Google Scholar] [CrossRef] [Green Version]
- Yufu, Y.; Nishimura, J.; Nawata, H. High constitutive expression of heat shock protein 90 alpha in human acute leukemia cells. Leuk. Res. 1992, 16, 597–605. [Google Scholar] [CrossRef]
- Pick, E.; Kluger, Y.; Giltnane, J.M.; Moeder, C.; Camp, R.L.; Rimm, D.L.; Kluger, H.M. High HSP90 expression is associated with decreased survival in breast cancer. Cancer Res. 2007, 67, 2932–2937. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wang, S.; Ding, D.; Yu, Z.; Sun, W.; Wang, Y. Up-regulation of Cdc37 contributes to schwann cell proliferation and migration after sciatic nerve crush. Neurochem. Res. 2018, 43, 1182–1190. [Google Scholar] [CrossRef]
- Fuoco, N.L.; Dos Ramos Silva, S.; Fernandes, E.R.; Luiz, F.G.; Ribeiro, O.G.; Katz, I.S.S. Infection of neuroblastoma cells by rabies virus is modulated by the virus titer. Antivir. Res. 2018, 149, 89–94. [Google Scholar] [CrossRef]
- Kallel, H.; Rourou, S.; Majoul, S.; Loukil, H. A novel process for the production of a veterinary rabies vaccine in BHK-21 cells grown on microcarriers in a 20-l bioreactor. Appl. Microbiol. Biotechnol. 2003, 61, 441–446. [Google Scholar] [CrossRef]
- Morimoto, K.; Hooper, D.C.; Carbaugh, H.; Fu, Z.F.; Koprowski, H.; Dietzschold, B. Rabies virus quasispecies: Implications for pathogenesis. Proc. Natl. Acad. Sci. USA 1998, 95, 3152–3156. [Google Scholar] [CrossRef] [Green Version]
- Koprowski, H.; Black, J.; Nelsen, D.J. Studies on chick-embryo-adapted-rabies virus. VI. Further changes in pathogenic properties following prolonged cultivation in the developing chick embryo. J. Immunol. 1954, 72, 94–106. [Google Scholar]
- Faul, E.J.; Wanjalla, C.N.; Suthar, M.S.; Gale, M.; Wirblich, C.; Schnell, M.J. Rabies virus infection induces type I interferon production in an IPS-1 dependent manner while dendritic cell activation relies on IFNAR signaling. PLoS Pathog. 2010, 6, e1001016. [Google Scholar] [CrossRef]
- Yang, K.; Shi, H.; Qi, R.; Sun, S.; Tang, Y.; Zhang, B.; Wang, C. Hsp90 regulates activation of interferon regulatory factor 3 and TBK-1 stabilization in Sendai virus-infected cells. Mol. Biol. Cell 2006, 17, 1461–1471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.; Zhu, S.; Hu, L.; Ye, P.; Wang, Y.; Tian, Q.; Mei, M.; Chen, H.; Guo, X. Wild-type rabies virus induces autophagy in human and mouse neuroblastoma cell lines. Autophagy 2016, 12, 1704–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Chen, Z.; Yu, F.; Chen, Q.; Tian, Y.; Ma, S.; Wang, T.; Liu, X. Hsp90 regulates autophagy and plays a role in cancer therapy. Tumour Biol. 2016, 37, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Park, G.; Huuhtanen, J.; Lundgren, S.; Khajuria, R.K.; Hurtado, A.M.; Munoz-Calleja, C.; Cardenoso, L.; Gomez-Garcia de Soria, V.; Chen-Liang, T.H.; et al. Somatic mTOR mutation in clonally expanded T lymphocytes associated with chronic graft versus host disease. Nat. Commun. 2020, 11, 2246. [Google Scholar] [CrossRef]
- Denney, A.S.; Weems, A.D.; McMurray, M.A. Selective functional inhibition of a tumor-derived p53 mutant by cytosolic chaperones identified using split-YFP in budding yeast. G3 2021, 11, jkab230. [Google Scholar] [CrossRef]
- Wang, X.; Venable, J.; LaPointe, P.; Hutt, D.M.; Koulov, A.V.; Coppinger, J.; Gurkan, C.; Kellner, W.; Matteson, J.; Plutner, H.; et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 2006, 127, 803–815. [Google Scholar] [CrossRef] [Green Version]
- Geller, R.; Andino, R.; Frydman, J. Hsp90 inhibitors exhibit resistance-free antiviral activity against respiratory syncytial virus. PLoS ONE 2013, 8, e56762. [Google Scholar] [CrossRef]
- Smallwood, S.; Ryan, K.W.; Moyer, S.A. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology 1994, 202, 154–163. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Banerjee, A.K. Phosphoprotein, P of human parainfluenza virus type 3 prevents self-association of RNA-dependent RNA polymerase, L. Virology 2009, 383, 226–236. [Google Scholar] [CrossRef] [Green Version]
- Canter, D.M.; Jackson, R.L.; Perrault, J. Faithful and efficient in vitro reconstitution of vesicular stomatitis virus transcription using plasmid-encoded L and P proteins. Virology 1993, 194, 518–529. [Google Scholar] [CrossRef]
- Jenni, S.; Bloyet, L.M.; Diaz-Avalos, R.; Liang, B.; Whelan, S.P.J.; Grigorieff, N.; Harrison, S.C. Structure of the vesicular stomatitis virus L protein in complex with its phosphoprotein cofactor. Cell Rep. 2020, 30, 53–60 e5. [Google Scholar] [CrossRef] [Green Version]
- Meslin, F.X.; Kaplan, M.M.; Koprowski, H.; World Health Organization. Laboratory Techniques in Rabies, 4th ed.; World Health Organization: Geneva, Switzerland, 1996; 467p. [Google Scholar]
- Wakeley, P.R.; Johnson, N.; McElhinney, L.M.; Marston, D.; Sawyer, J.; Fooks, A.R. Development of a real-time, TaqMan reverse transcription-PCR assay for detection and differentiation of lyssavirus genotypes 1, 5, and 6. J. Clin. Microbiol. 2005, 43, 2786–2792. [Google Scholar] [CrossRef] [Green Version]
- Synoradzki, K.; Bieganowski, P. Middle domain of human Hsp90 isoforms differentially binds Aha1 in human cells and alters Hsp90 activity in yeast. Biochim. Biophys. Acta 2015, 1853, 445–452. [Google Scholar] [CrossRef] [Green Version]
- Stillman, E.A.; Rose, J.K.; Whitt, M.A. Replication and amplification of novel vesicular stomatitis virus minigenomes encoding viral structural proteins. J. Virol. 1995, 69, 2946–2953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.Z.; Elledge, S.J. SLIC: A method for sequence- and ligation-independent cloning. Methods Mol. Biol 2012, 852, 51–59. [Google Scholar] [PubMed]
- Hall, T.A. A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]






| N-forward | GCTCTGGTACCATGGATGCCGACAAGATTG |
| N-reverse | CGCGAGCGGCCGCAATGAGTCATTCGAATACGTCTTG |
| P-forward | GCTCTGGTACCATGAGCAAGATCTTTGTTAATCC |
| P-reverse | CGCGAGCGGCCGCAAGCAGGATGTATAGCGATTC |
| G-forward | GCTCTGGTACCATGGTTCCTCAGGTTCTTTTG |
| G-reverse | CGCGAGCGGCCGCCACAGTCTGATCTCACCTCC |
| M-forward | GCTCTGGTACCATGAACGTTCTACGCAAGATAG |
| M-reverse | CGCGAGCGGCCGCAATTCTAGAAGCAGAGAAGAGTC |
| RABV L-forward | CGACTGGTACCATGCTAGATCCGGGAGAGGT |
| RABV L-reverse | CGCGAGCGGCCGCAACAAACAACTGTAATCTAGTAGG |
| MeV L-F | ATAGATCTGGATATCGGTACCATGGACTCGCTATCTGTCAAC |
| MeV L-R | CTAGAAGGCACAGTCGAGGCTTAGTCCTTAATCAGGGCACTG |
| VSV L-F | ATAGATCTGGATATCGGTACCATGGAAGTCCACGATTTTGAGAC |
| VSV L-R | CTAGAAGGCACAGTCGAGGCTTAATCTCTCCAAGAGTTTTCCTCG |
| pWSL5-F | GCCTCGACTGTGCCTTCTAG |
| pWSL5-R | GGTACCGATATCCAGATCTATCGA |
| qPCR primer F | ATGTAACACCTCTACAATG |
| qPCR primer R | GCAGGGTATTTRTACTCATA |
| qPCR probe | ACAAGATTGTATTCAAAGTCAATAATCAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dalidowska, I.; Orlowska, A.; Smreczak, M.; Bieganowski, P. Hsp90 Activity Is Necessary for the Maturation of Rabies Virus Polymerase. Int. J. Mol. Sci. 2022, 23, 6946. https://doi.org/10.3390/ijms23136946
Dalidowska I, Orlowska A, Smreczak M, Bieganowski P. Hsp90 Activity Is Necessary for the Maturation of Rabies Virus Polymerase. International Journal of Molecular Sciences. 2022; 23(13):6946. https://doi.org/10.3390/ijms23136946
Chicago/Turabian StyleDalidowska, Iga, Anna Orlowska, Marcin Smreczak, and Pawel Bieganowski. 2022. "Hsp90 Activity Is Necessary for the Maturation of Rabies Virus Polymerase" International Journal of Molecular Sciences 23, no. 13: 6946. https://doi.org/10.3390/ijms23136946
APA StyleDalidowska, I., Orlowska, A., Smreczak, M., & Bieganowski, P. (2022). Hsp90 Activity Is Necessary for the Maturation of Rabies Virus Polymerase. International Journal of Molecular Sciences, 23(13), 6946. https://doi.org/10.3390/ijms23136946