The Impact of the Anticoagulant Type in Blood Collection Tubes on Circulating Extracellular Plasma MicroRNA Profiles Revealed by Small RNA Sequencing
Abstract
:1. Introduction
2. Results
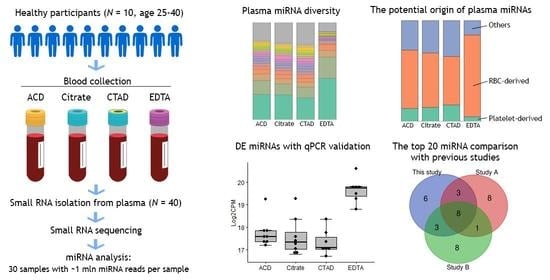
2.1. Study Sample Characteristics
- ACD: 10 samples obtained using acid citrate dextrose (ACD-B) blood tubes;
- Citrate: 10 samples obtained using sodium citrate 3.2% blood tubes;
- CTAD: 10 samples obtained using citrate-theophylline-adenosine-dipyridamole (CTAD) blood tubes;
- EDTA: 10 samples obtained using dipotassium-ethylenediaminetetraacetic acid (K2 EDTA) blood tubes.
2.2. miRNA Sequencing Statistics
2.3. The Impact of Anticoagulation on Circulating miRNA Profiles
2.3.1. The Diversity of miRNAs in Plasma
2.3.2. miRNA Profiles by PCA
2.3.3. Differential Expression of miRNAs
2.3.4. The Impact of Hemolysis and the Contribution of RBC-Derived and Platelet-Derived miRNAs
2.4. Validation of miRNA Sequencing Data by Quantitative PCR (qPCR)
3. Discussion
4. Materials and Methods
4.1. Study Sample
4.2. Plasma Preparation
4.3. Hemolysis Assessment of Plasma Samples
4.4. Plasma miRNA Isolation
4.5. miRNA Sequencing
4.6. Sequencing Data Analysis
4.7. Validation of miRNA Expression by qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Individual ID | Sample ID | Anticoagulant Type ID (Sample Group) | Hemolysis Score (HS) | miRNA Sample Concentration, ng/μL | miRNA Library Successfully Prepared |
|---|---|---|---|---|---|
| miR_PA_01 | 01A | ACD | 0.041 | 1.95 | yes |
| miR_PA_01 | 01B | Citrate | 0.043 | 1.77 | no |
| miR_PA_01 | 01C | CTAD | 0.037 | 1.76 | yes |
| miR_PA_01 | 01D | EDTA | 0.110 | 1.77 | yes |
| miR_PA_02 | 02A | ACD | 0.017 | 1.21 | no |
| miR_PA_02 | 02B | Citrate | 0.047 | 1.29 | yes |
| miR_PA_02 | 02C | CTAD | 0.023 | 0.92 | yes |
| miR_PA_02 | 02D | EDTA | 0.112 | 1.55 | yes |
| miR_PA_03 | 03A | ACD | 0.041 | 1.25 | yes |
| miR_PA_03 | 03B | Citrate | 0.047 | 1.43 | yes |
| miR_PA_03 | 03C | CTAD | 0.054 | 1.12 | yes |
| miR_PA_03 | 03D | EDTA | 0.112 | 1.62 | yes |
| miR_PA_04 | 04A | ACD | 0.031 | 0.94 | yes |
| miR_PA_04 | 04B | Citrate | 0.033 | 1.33 | yes |
| miR_PA_04 | 04C | CTAD | 0.039 | 1.23 | yes |
| miR_PA_04 | 04D | EDTA | 0.098 | 0.97 | yes |
| miR_PA_05 | 05A | ACD | 0.049 | 1.11 | yes |
| miR_PA_05 | 05B | Citrate | 0.051 | 1.47 | yes |
| miR_PA_05 | 05C | CTAD | 0.057 | 1.10 | yes |
| miR_PA_05 | 05D | EDTA | 0.093 | 1.49 | yes |
| miR_PA_06 | 06A | ACD | 0.034 | 1.34 | yes |
| miR_PA_06 | 06B | Citrate | 0.033 | 1.51 | yes |
| miR_PA_06 | 06C | CTAD | 0.040 | 1.49 | yes |
| miR_PA_06 | 06D | EDTA | 0.085 | 1.58 | no |
| miR_PA_07 | 07A | ACD | 0.040 | 1.23 | yes |
| miR_PA_07 | 07B | Citrate | 0.039 | 1.35 | yes |
| miR_PA_07 | 07C | CTAD | 0.045 | 1.23 | yes |
| miR_PA_07 | 07D | EDTA | 0.088 | 1.26 | yes |
| miR_PA_08 | 08A | ACD | 0.034 | 0.99 | yes |
| miR_PA_08 | 08B | Citrate | 0.034 | 1.50 | yes |
| miR_PA_08 | 08C | CTAD | 0.038 | 1.58 | yes |
| miR_PA_08 | 08D | EDTA | 0.082 | 1.65 | yes |
| miR_PA_09 | 09A | ACD | 0.017 | 1.28 | yes |
| miR_PA_09 | 09B | Citrate | 0.019 | 1.11 | yes |
| miR_PA_09 | 09C | CTAD | 0.023 | 0.98 | no |
| miR_PA_09 | 09D | EDTA | 0.048 | 1.64 | yes |
| miR_PA_10 | 10A | ACD | 0.044 | 1.37 | yes |
| miR_PA_10 | 10B | Citrate | 0.049 | 1.39 | yes |
| miR_PA_10 | 10C | CTAD | 0.056 | 1.06 | yes |
| miR_PA_10 | 10D | EDTA | 0.088 | 0.62 | yes |
| Sample ID | Total Reads | Mapping of Reads | miRNA Reads with Mismatches | miRNA Reads Percent, % | Number of Detected miRNAs | ||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | Hairpin | Small RNA | mRNA | Unaligned | |||||
| s01A | 1,856,550 | 1,329,347 | 4737 | 364,976 | 45,293 | 112,197 | 138,099 | 71.6 | 330 |
| s01C | 3,346,595 | 2,206,294 | 10,079 | 701,448 | 107,118 | 321,656 | 257,081 | 65.9 | 374 |
| s01D | 1,156,024 | 1,099,843 | 1246 | 28,194 | 6144 | 20,597 | 67,757 | 95.1 | 355 |
| s02B | 2,550,611 | 1,760,884 | 8874 | 154,260 | 138,024 | 488,569 | 215,528 | 69.0 | 295 |
| s02C | 2,343,118 | 1,228,110 | 8473 | 76,887 | 222,822 | 806,826 | 150,931 | 52.4 | 235 |
| s02D | 2,346,558 | 2,151,181 | 3979 | 27,003 | 21,006 | 143,389 | 231,080 | 91.7 | 278 |
| s03A | 3,899,934 | 2,022,417 | 11,824 | 278,091 | 580,776 | 1,006,826 | 244,680 | 51.9 | 374 |
| s03B | 4,211,248 | 1,881,747 | 13,266 | 295,537 | 753,824 | 1,266,874 | 234,257 | 44.7 | 343 |
| s03C | 2,128,083 | 54,065 | 5147 | 29,729 | 215,321 | 1,823,821 | 8888 | 2.5 | 67 |
| s03D | 1,686,441 | 1,593,459 | 1770 | 15,389 | 22,828 | 52,995 | 169,256 | 94.5 | 335 |
| s04A | 2,001,761 | 1,270,473 | 7911 | 68,452 | 166,763 | 488,162 | 156,889 | 63.5 | 258 |
| s04B | 1,355,907 | 1,217,903 | 3261 | 44,288 | 29,701 | 60,754 | 112,103 | 89.8 | 315 |
| s04C | 1,403,441 | 1,068,112 | 6100 | 54,307 | 87,363 | 187,559 | 118,182 | 76.1 | 280 |
| s04D | 3,443,950 | 957,956 | 11,230 | 57,283 | 269,028 | 2,148,453 | 96,620 | 27.8 | 117 |
| s05A | 3,567,018 | 827,340 | 15,716 | 115,385 | 387,345 | 2,221,232 | 120,655 | 23.2 | 146 |
| s05B | 2,374,559 | 1,706,243 | 9137 | 239,953 | 127,695 | 291,531 | 203,185 | 71.9 | 339 |
| s05C | 2,544,816 | 284,216 | 10,921 | 49,408 | 243,488 | 1,956,783 | 45,297 | 11.2 | 108 |
| s05D | 2,699,302 | 2,504,171 | 4527 | 50,205 | 29,857 | 110,542 | 253,338 | 92.8 | 340 |
| s06A | 1,406,142 | 1,067,283 | 4612 | 41,878 | 60,932 | 231,437 | 143,799 | 75.9 | 298 |
| s06B | 1,571,679 | 921,115 | 5632 | 52,268 | 168,233 | 424,431 | 111,680 | 58.6 | 315 |
| s06C | 2,009,387 | 1,551,149 | 7198 | 75,373 | 112,643 | 263,024 | 186,804 | 77.2 | 328 |
| s07A | 2,575,875 | 2,009,549 | 6715 | 59,721 | 99,934 | 399,956 | 234,586 | 78.0 | 284 |
| s07B | 3,277,986 | 2,350,086 | 8696 | 243,336 | 163,663 | 512,205 | 278,709 | 71.7 | 310 |
| s07C | 2,311,486 | 1,727,119 | 8747 | 62,836 | 117,437 | 395,347 | 197,686 | 74.7 | 318 |
| s07D | 2,943,326 | 2,502,394 | 5966 | 47,772 | 80,190 | 307,004 | 322,713 | 85.0 | 268 |
| s08A | 3,582,150 | 1,949,348 | 8919 | 305,199 | 223,992 | 1,094,692 | 235,363 | 54.4 | 244 |
| s08B | 1,380,417 | 983,151 | 5459 | 54,559 | 105,758 | 231,490 | 108,837 | 71.2 | 285 |
| s08C | 1,880,532 | 994,126 | 8633 | 140,958 | 225,430 | 511,385 | 119,564 | 52.9 | 274 |
| s08D | 1,383,647 | 1,239,228 | 3739 | 34,771 | 31,750 | 74,159 | 104,014 | 89.6 | 303 |
| s09A | 1,505,063 | 1,088,615 | 3362 | 188,554 | 51,334 | 173,198 | 142,186 | 72.3 | 325 |
| s09B | 1,207,691 | 115,606 | 4167 | 17,193 | 164,868 | 905,857 | 16,457 | 9.6 | 87 |
| s09D | 1,715,204 | 1,596,214 | 3337 | 16,329 | 19,895 | 79,429 | 152,743 | 93.1 | 294 |
| s10A | 1,796,035 | 1,605,546 | 4757 | 28,425 | 30,322 | 126,985 | 133,648 | 89.4 | 335 |
| s10B | 1,308,401 | 1,020,862 | 4126 | 38,673 | 46,426 | 198,314 | 91,551 | 78.0 | 300 |
| s10C | 1,265,292 | 1,029,519 | 3707 | 32,936 | 36,423 | 162,707 | 95,339 | 81.4 | 311 |
| s10D | 2,040,931 | 184,828 | 2815 | 25,933 | 222,516 | 1,604,839 | 24,973 | 9.1 | 101 |
| Mean | 2,224,366 | 1,363,875 | 6633 | 114,375 | 150,448 | 589,034 | 153,458 | 64.4 | 274 |
| SD | 843,647 | 640,148 | 3368 | 139,038 | 156,253 | 637,989 | 76,858 | 26.6 | 84 |
| miRNA Name | Stability Value |
|---|---|
| miR-16-5p | 0.822 |
| miR-126-3p | 0.517 |
| miR-145-5p | 0.674 |
| miR-146a-5p | 0.661 |
| miR-150-5p | 0.707 |
| miR-21-5p | 0.316 |
| miR-223-3p | 0.636 |
| miR-92a-3p | 0.668 |
| miR-23a-3p | 0.534 |
| miR-451a | 0.932 |
| miR-30e-5p | 0.308 |
| miR-17-5p | 0.692 |
References
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical Utility of Circulating Non-Coding RNAs—An Update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular MiRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Burwinkel, B. Extracellular MiRNAs: The Mystery of Their Origin and Function. Trends Biochem. Sci. 2012, 37, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 Complexes Carry a Population of Circulating MicroRNAs Independent of Vesicles in Human Plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. MiRBase: Tools for MicroRNA Genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef]
- Da Silva, F.C.; Iop, R.d.R.; Andrade, A.; Costa, V.P.; Gutierres Filho, P.J.B.; da Silva, R. Effects of Physical Exercise on the Expression of MicroRNAs: A Systematic Review. J. Strength Cond. Res. 2020, 34, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Laterza, O.F.; Lim, L.; Garrett-Engele, P.W.; Vlasakova, K.; Muniappa, N.; Tanaka, W.K.; Johnson, J.M.; Sina, J.F.; Fare, T.L.; Sistare, F.D.; et al. Plasma MicroRNAs as Sensitive and Specific Biomarkers of Tissue Injury. Clin. Chem. 2009, 55, 1977–1983. [Google Scholar] [CrossRef] [PubMed]
- Redell, J.B.; Moore, A.N.; Ward, N.H.; Hergenroeder, G.W.; Dash, P.K. Human Traumatic Brain Injury Alters Plasma MicroRNA Levels. J. Neurotrauma 2010, 27, 2147–2156. [Google Scholar] [CrossRef] [PubMed]
- Toffolo, K.; Osei, J.; Kelly, W.; Poulsen, A.; Donahue, K.; Wang, J.; Hunter, M.; Bard, J.; Wang, J.; Poulsen, D. Circulating MicroRNAs as Biomarkers in Traumatic Brain Injury. Neuropharmacology 2019, 145, 199–208. [Google Scholar] [CrossRef]
- Czajka, P.; Fitas, A.; Jakubik, D.; Eyileten, C.; Gasecka, A.; Wicik, Z.; Siller-Matula, J.M.; Filipiak, K.J.; Postula, M. MicroRNA as Potential Biomarkers of Platelet Function on Antiplatelet Therapy: A Review. Front. Physiol. 2021, 12, 652579. [Google Scholar] [CrossRef]
- Rounge, T.B.; Umu, S.U.; Keller, A.; Meese, E.; Ursin, G.; Tretli, S.; Lyle, R.; Langseth, H. Circulating Small Non-Coding RNAs Associated with Age, Sex, Smoking, Body Mass and Physical Activity. Sci. Rep. 2018, 8, 17650. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Cai, H.; Xiang, Y.-B.; Matthews, C.E.; Ye, F.; Zheng, W.; Cai, Q.; Shu, X.-O. Intra-Individual Variation of MiRNA Expression Levels in Human Plasma Samples. Biomarkers 2018, 23, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Felekkis, K.; Papaneophytou, C. Challenges in Using Circulating Micro-RNAs as Biomarkers for Cardiovascular Diseases. Int. J. Mol. Sci. 2020, 21, 561. [Google Scholar] [CrossRef] [PubMed]
- Glinge, C.; Clauss, S.; Boddum, K.; Jabbari, R.; Jabbari, J.; Risgaard, B.; Tomsits, P.; Hildebrand, B.; Kääb, S.; Wakili, R.; et al. Stability of Circulating Blood-Based MicroRNAs—Pre-Analytic Methodological Considerations. PLoS ONE 2017, 12, e0167969. [Google Scholar] [CrossRef] [PubMed]
- Markou, A.N.; Lianidou, E.S. The Impact of Pre-Analytical Factors on the Reliability of MiRNA Measurements. Curr. Pathobiol. Rep. 2019, 7, 29–33. [Google Scholar] [CrossRef]
- McDonald, J.S.; Milosevic, D.; Reddi, H.V.; Grebe, S.K.; Algeciras-Schimnich, A. Analysis of Circulating MicroRNA: Preanalytical and Analytical Challenges. Clin. Chem. 2011, 57, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Khan, J.; Lieberman, J.A.; Lockwood, C.M. Variability in, Variability out: Best Practice Recommendations to Standardize Pre-Analytical Variables in the Detection of Circulating and Tissue MicroRNAs. Clin. Chem. Lab. Med. 2017, 55, 608–621. [Google Scholar] [CrossRef]
- Becker, N.; Lockwood, C.M. Pre-Analytical Variables in MiRNA Analysis. Clin. Biochem. 2013, 46, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood Cell Origin of Circulating MicroRNAs: A Cautionary Note for Cancer Biomarker Studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duttagupta, R.; Jiang, R.; Gollub, J.; Getts, R.C.; Jones, K.W. Impact of Cellular miRNAs on Circulating miRNA Biomarker Signatures. PLoS ONE 2011, 6, e20769. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.H.; Yi, H.S.; Kim, Y.; Kroh, E.M.; Chien, J.W.; Eaton, K.D.; Goodman, M.T.; Tait, J.F.; Tewari, M.; Pritchard, C.C. Plasma Processing Conditions Substantially Influence Circulating MicroRNA Biomarker Levels. PLoS ONE 2013, 8, e64795. [Google Scholar] [CrossRef]
- Basso, D.; Padoan, A.; Laufer, T.; Aneloni, V.; Moz, S.; Schroers, H.; Pelloso, M.; Saiz, A.; Krapp, M.; Fogar, P.; et al. Relevance of Pre-Analytical Blood Management on the Emerging Cardiovascular Protein Biomarkers TWEAK and HMGB1 and on miRNA Serum and Plasma Profiling. Clin. Biochem. 2017, 50, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Ma, J.; Wang, Y.; Biswal, S. Plasma MiRNA as Biomarkers for Assessment of Total-Body Radiation Exposure Dosimetry. PLoS ONE 2011, 6, e22988. [Google Scholar] [CrossRef] [PubMed]
- Chandler, W.L. Microparticle Counts in Platelet-Rich and Platelet-Free Plasma, Effect of Centrifugation and Sample-Processing Protocols. Blood Coagul. Fibrinolysis 2013, 24, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during Sample Preparation Alters MicroRNA Content of Plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef] [PubMed]
- Pizzamiglio, S.; Zanutto, S.; Ciniselli, C.M.; Belfiore, A.; Bottelli, S.; Gariboldi, M.; Verderio, P. A Methodological Procedure for Evaluating the Impact of Hemolysis on Circulating MicroRNAs. Oncol. Lett. 2017, 13, 315–320. [Google Scholar] [CrossRef]
- Shah, J.S.; Soon, P.S.; Marsh, D.J. Comparison of Methodologies to Detect Low Levels of Hemolysis in Serum for Accurate Assessment of Serum MicroRNAs. PLoS ONE 2016, 11, e0153200. [Google Scholar] [CrossRef]
- Appierto, V.; Callari, M.; Cavadini, E.; Morelli, D.; Daidone, M.G.; Tiberio, P. A Lipemia-Independent NanoDrop®-Based Score to Identify Hemolysis in Plasma and Serum Samples. Bioanalysis 2014, 6, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Blondal, T.; Jensby Nielsen, S.; Baker, A.; Andreasen, D.; Mouritzen, P.; Wrang Teilum, M.; Dahlsveen, I.K. Assessing Sample and MiRNA Profile Quality in Serum and Plasma or Other Biofluids. Methods 2013, 59, S1–S6. [Google Scholar] [CrossRef]
- Krammer, T.L.; Mayr, M.; Hackl, M. MicroRNAs as Promising Biomarkers of Platelet Activity in Antiplatelet Therapy Monitoring. Int. J. Mol. Sci. 2020, 21, 3477. [Google Scholar] [CrossRef]
- Krammer, T.L.; Zeibig, S.; Schrottmaier, W.C.; Pirabe, A.; Goebel, S.; Diendorfer, A.B.; Holthoff, H.-P.; Assinger, A.; Hackl, M. Comprehensive Characterization of Platelet-Enriched MicroRNAs as Biomarkers of Platelet Activation. Cells 2022, 11, 1254. [Google Scholar] [CrossRef] [PubMed]
- Sunderland, N.; Skroblin, P.; Barwari, T.; Huntley, R.P.; Lu, R.; Joshi, A.; Lovering, R.C.; Mayr, M. MicroRNA Biomarkers and Platelet Reactivity: The Clot Thickens. Circ. Res. 2017, 120, 418–435. [Google Scholar] [CrossRef]
- Willeit, P.; Zampetaki, A.; Dudek, K.; Kaudewitz, D.; King, A.; Kirkby, N.S.; Crosby-Nwaobi, R.; Prokopi, M.; Drozdov, I.; Langley, S.R.; et al. Circulating MicroRNAs as Novel Biomarkers for Platelet Activation. Circ. Res. 2013, 112, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Söderström, A.C.; Nybo, M.; Nielsen, C.; Vinholt, P.J. The Effect of Centrifugation Speed and Time on Pre-Analytical Platelet Activation. Clin. Chem. Lab. Med. 2016, 54, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Arraud, N.; Linares, R.; Tan, S.; Gounou, C.; Pasquet, J.-M.; Mornet, S.; Brisson, A.R. Extracellular Vesicles from Blood Plasma: Determination of Their Morphology, Size, Phenotype and Concentration. J. Thromb. Haemost. 2014, 12, 614–627. [Google Scholar] [CrossRef]
- Italiano, J.E.; Mairuhu, A.T.A.; Flaumenhaft, R. Clinical Relevance of Microparticles from Platelets and Megakaryocytes. Curr. Opin. Hematol. 2010, 17, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-L.; Li, S.; Han, J.-Y. Platelet Function Tests: A Review of Progresses in Clinical Application. Biomed. Res. Int. 2014, 2014, 456569. [Google Scholar] [CrossRef]
- Mody, M.; Lazarus, A.H.; Semple, J.W.; Freedman, J. Preanalytical Requirements for Flow Cytometric Evaluation of Platelet Activation: Choice of Anticoagulant. Transfus. Med. 1999, 9, 147–154. [Google Scholar] [CrossRef]
- György, B.; Pálóczi, K.; Kovács, A.; Barabás, E.; Bekő, G.; Várnai, K.; Pállinger, É.; Szabó-Taylor, K.; Szabó, T.G.; Kiss, A.A.; et al. Improved Circulating Microparticle Analysis in Acid-Citrate Dextrose (ACD) Anticoagulant Tube. Thromb. Res. 2014, 133, 285–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mussbacher, M.; Schrottmaier, W.C.; Salzmann, M.; Brostjan, C.; Schmid, J.A.; Starlinger, P.; Assinger, A. Optimized Plasma Preparation Is Essential to Monitor Platelet-Stored Molecules in Humans. PLoS ONE 2017, 12, e0188921. [Google Scholar] [CrossRef]
- Mussbacher, M.; Krammer, T.L.; Heber, S.; Schrottmaier, W.C.; Zeibig, S.; Holthoff, H.-P.; Pereyra, D.; Starlinger, P.; Hackl, M.; Assinger, A. Impact of Anticoagulation and Sample Processing on the Quantification of Human Blood-Derived MicroRNA Signatures. Cells 2020, 9, 1915. [Google Scholar] [CrossRef] [PubMed]
- Tomasello, L.; Distefano, R.; Nigita, G.; Croce, C.M. The MicroRNA Family Gets Wider: The IsomiRs Classification and Role. Front. Cell Dev. Biol. 2021, 9, 668648. [Google Scholar] [CrossRef]
- Doss, J.F.; Corcoran, D.L.; Jima, D.D.; Telen, M.J.; Dave, S.S.; Chi, J.-T. A Comprehensive Joint Analysis of the Long and Short RNA Transcriptomes of Human Erythrocytes. BMC Genom. 2015, 16, 952. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Edelman, J.J.B.; Kao, S.C.-H.; Vallely, M.P.; van Zandwijk, N.; Reid, G. The Impact of Hemolysis on Cell-Free MicroRNA Biomarkers. Front. Genet. 2013, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yu, Y.; Niu, B.; Wang, D. Red Blood Cells as Potential Repositories of MicroRNAs in the Circulatory System. Front. Genet. 2020, 11, 442. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.K.Y.; MacMahon, M.; Woodside, J.V.; Simpson, D.A. A Comparison of RNA Extraction and Sequencing Protocols for Detection of Small RNAs in Plasma. BMC Genom. 2019, 20, 446. [Google Scholar] [CrossRef] [PubMed]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Boffelli, D.; Mote, P.; Martin, D.I.K. 5′-YRNA Fragments Derived by Processing of Transcripts from Specific YRNA Genes and Pseudogenes Are Abundant in Human Serum and Plasma. Physiol. Genom. 2013, 45, 990–998. [Google Scholar] [CrossRef] [PubMed]
- Godoy, P.M.; Bhakta, N.R.; Barczak, A.J.; Cakmak, H.; Fisher, S.; MacKenzie, T.C.; Patel, T.; Price, R.W.; Smith, J.F.; Woodruff, P.G.; et al. Large Differences in Small RNA Composition Between Human Biofluids. Cell Rep. 2018, 25, 1346–1358. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.; Bustamante, M.; Hernandez-Ferrer, C.; Fernandez-Orth, D.; Pantano, L.; Sarria, Y.; Piqué-Borras, M.; Vellve, K.; Agramunt, S.; Carreras, R.; et al. Circulating MiRNAs, IsomiRs and Small RNA Clusters in Human Plasma and Breast Milk. PLoS ONE 2018, 13, e0193527. [Google Scholar] [CrossRef]
- Max, K.E.A.; Bertram, K.; Akat, K.M.; Bogardus, K.A.; Li, J.; Morozov, P.; Ben-Dov, I.Z.; Li, X.; Weiss, Z.R.; Azizian, A.; et al. Human Plasma and Serum Extracellular Small RNA Reference Profiles and Their Clinical Utility. Proc. Natl. Acad. Sci. USA 2018, 115, E5334–E5343. [Google Scholar] [CrossRef]
- Godoy, P.M.; Barczak, A.J.; DeHoff, P.; Srinivasan, S.; Etheridge, A.; Galas, D.; Das, S.; Erle, D.J.; Laurent, L.C. Comparison of Reproducibility, Accuracy, Sensitivity, and Specificity of MiRNA Quantification Platforms. Cell Rep. 2019, 29, 4212–4222.e5. [Google Scholar] [CrossRef] [PubMed]
- Dard-Dascot, C.; Naquin, D.; d’Aubenton-Carafa, Y.; Alix, K.; Thermes, C.; van Dijk, E. Systematic Comparison of Small RNA Library Preparation Protocols for Next-Generation Sequencing. BMC Genom. 2018, 19, 118. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Gordon, W.; Du, S.; Zhang, C.; He, W.; Xi, L.; Mathur, S.; Agostino, M.; Paradis, T.; von Schack, D.; et al. QuickMIRSeq: A Pipeline for Quick and Accurate Quantification of Both Known MiRNAs and IsomiRs by Jointly Processing Multiple Samples from MicroRNA Sequencing. BMC Bioinform. 2017, 18, 180. [Google Scholar] [CrossRef]
- Kim, B.-R.; Shin, J.; Guevarra, R.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Konopiński, M.K. Shannon Diversity Index: A Call to Replace the Original Shannon’s Formula with Unbiased Estimator in the Population Genetics Studies. PeerJ 2020, 8, e9391. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Foye, C.; Yan, I.K.; David, W.; Shukla, N.; Habboush, Y.; Chase, L.; Ryland, K.; Kesari, V.; Patel, T. Comparison of MiRNA Quantitation by Nanostring in Serum and Plasma Samples. PLoS ONE 2017, 12, e0189165. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]















| Individual ID | Sex | Age, years | BMI, kg/m2 |
|---|---|---|---|
| s01 | male | 31.2 | 28.3 |
| s02 | female | 26.8 | 20.8 |
| s03 | male | 28.7 | 20.8 |
| s04 | male | 32.5 | 27.4 |
| s05 | male | 34.7 | 22.3 |
| s06 | female | 34.9 | 22.2 |
| s07 | female | 34.1 | 20.4 |
| s08 | male | 33.2 | 20.3 |
| s09 | female | 29.6 | 21.8 |
| s10 | female | 37.7 | 18.7 |
| Assay Name | Assay ID | Mature miRNA Sequence | Type of miRNA |
|---|---|---|---|
| hsa-miR-223-3p | 477983_mir | UGUCAGUUUGUCAAAUACCCCA | Platelet-derived |
| hsa-miR-126-3p | 477887_mir | UCGUACCGUGAGUAAUAAUGCG | Platelet-derived |
| hsa-miR-21-5p | 477975_mir | UAGCUUAUCAGACUGAUGUUGA | Platelet-derived |
| hsa-miR-150-5p | 477918_mir | UCUCCCAACCCUUGUACCAGUG | Platelet-derived |
| hsa-miR-16-5p | 477860_mir | UAGCAGCACGUAAAUAUUGGCG | RBC-derived |
| hsa-miR-92a-3p | 477827_mir | UAUUGCACUUGUCCCGGCCUGU | RBC-derived |
| hsa-miR-451a | 478107_mir | AAACCGUUACCAUUACUGAGUU | RBC-derived/Hemolysis assessment |
| hsa-miR-23a-3p | 478532_mir | AUCACAUUGCCAGGGAUUUCC | Hemolysis assessment |
| hsa-miR-30e-5p | 479235_mir | UGUAAACAUCCUUGACUGGAAG | Normalization control |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhelankin, A.V.; Iulmetova, L.N.; Sharova, E.I. The Impact of the Anticoagulant Type in Blood Collection Tubes on Circulating Extracellular Plasma MicroRNA Profiles Revealed by Small RNA Sequencing. Int. J. Mol. Sci. 2022, 23, 10340. https://doi.org/10.3390/ijms231810340
Zhelankin AV, Iulmetova LN, Sharova EI. The Impact of the Anticoagulant Type in Blood Collection Tubes on Circulating Extracellular Plasma MicroRNA Profiles Revealed by Small RNA Sequencing. International Journal of Molecular Sciences. 2022; 23(18):10340. https://doi.org/10.3390/ijms231810340
Chicago/Turabian StyleZhelankin, Andrey V., Liliia N. Iulmetova, and Elena I. Sharova. 2022. "The Impact of the Anticoagulant Type in Blood Collection Tubes on Circulating Extracellular Plasma MicroRNA Profiles Revealed by Small RNA Sequencing" International Journal of Molecular Sciences 23, no. 18: 10340. https://doi.org/10.3390/ijms231810340
APA StyleZhelankin, A. V., Iulmetova, L. N., & Sharova, E. I. (2022). The Impact of the Anticoagulant Type in Blood Collection Tubes on Circulating Extracellular Plasma MicroRNA Profiles Revealed by Small RNA Sequencing. International Journal of Molecular Sciences, 23(18), 10340. https://doi.org/10.3390/ijms231810340