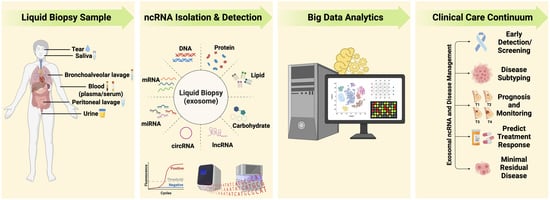
Noncoding RNome as Enabling Biomarkers for Precision Health
Abstract
:1. Introduction
2. ncRNAs Are Disease-Relevant Molecular Analytes
3. Liquid Biopsy as Surrogate for Tissue for Molecular Profiling
4. Extracellular Vesicles/Exosomes: Valuable Biological Information Packages in Biofluids
5. Challenges and Opportunities for Clinical Applications with Exosomal ncRNA
6. Harnessing ncRNAs to Enhance Disease Management
6.1. Early Detection/Screening
6.2. Tumor Subtyping
6.3. Prognosis and Real-Time Monitoring
6.4. Predicting Response to Treatment/Treatment Selection/Precision Oncology
6.5. Minimal Residual Disease
7. A Need for Standardization to Enable Precision Medicine
8. Leveraging Artificial Intelligence/Machine Learning to Drive Precision Health
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The role of non-coding RNAs in oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, G.K.; Khullar, N.; Sidhu, I.S.; Navik, U.S.; Reddy, A.P.; Reddy, P.H.; Bhatti, J.S. Emerging role of non-coding RNA in health and disease. Metab. Brain Dis. 2021, 36, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- Tamgue, O.; Mezajou, C.F.; Ngongang, N.N.; Kameni, C.; Ngum, J.A.; Simo, U.S.F.; Tatang, F.J.; Akami, M.; Ngono, A.N. Non-Coding RNAs in the Etiology and Control of Major and Neglected Human Tropical Diseases. Front. Immunol. 2021, 12, 703936. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Benítez, I.D.; Pinilla, L.; Carratalá, A.; Moncusí-Moix, A.; Gort-Paniello, C.; Molinero, M.; González, J.; Torres, G.; Bernal, M. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl. Res. 2021, 236, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Wang, J.; Wang, Z.; Sun, Y.; Wu, J.; Zhang, Y.; Liu, X.; Zhou, Z.; Zhou, L.; Zhang, C.-Y. A virus-derived microRNA-like small RNA serves as a serum biomarker to prioritize the COVID-19 patients at high risk of developing severe disease. Cell Discov. 2021, 7, 48. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Drula, R.; Mohapatra, S.; Calin, G.A. microRNA in cancer: An overview. In MicroRNA in Human Malignancies; Academic Press: Cambridge, MA, USA, 2022; pp. 21–28. [Google Scholar] [CrossRef]
- Yan, H.; Bu, P. Non-coding RNA in cancer. Essays Biochem. 2021, 65, 625–639. [Google Scholar]
- Bartel, D.P. Metazoan micrornas. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Fehlmann, T.; Kahraman, M.; Ludwig, N.; Backes, C.; Galata, V.; Keller, V.; Geffers, L.; Mercaldo, N.; Hornung, D.; Weis, T. Evaluating the use of circulating microRNA profiles for lung cancer detection in symptomatic patients. JAMA Oncol. 2020, 6, 714–723. [Google Scholar] [CrossRef] [PubMed]
- Ying, L.; Du, L.; Zou, R.; Shi, L.; Zhang, N.; Jin, J.; Xu, C.; Zhang, F.; Zhu, C.; Wu, J. Development of a serum miRNA panel for detection of early stage non-small cell lung cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 25036–25042. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, K.; Hanson, J.; Villeneuve, P.J.; Dimitroulakos, J.; McIntyre, L.; Pilote, L.; Stewart, D.J. Assessment of circulating LncRNAs under physiologic and pathologic conditions in humans reveals potential limitations as biomarkers. Sci. Rep. 2016, 6, 36596. [Google Scholar] [CrossRef]
- Arita, T.; Ichikawa, D.; Konishi, H.; Komatsu, S.; Shiozaki, A.; Shoda, K.; Kawaguchi, T.; Hirajima, S.; Nagata, H.; Kubota, T. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013, 33, 3185–3193. [Google Scholar]
- Tan, Q.; Zuo, J.; Qiu, S.; Yu, Y.; Zhou, H.; Li, N.; Wang, H.; Liang, C.; Yu, M.; Tu, J. Identification of circulating long non-coding RNA GAS5 as a potential biomarker for non-small cell lung cancer diagnosisnon-small cell lung cancer, long non-coding RNA, plasma, GAS5, biomarker. Int. J. Oncol. 2017, 50, 1729–1738. [Google Scholar] [CrossRef]
- Wiedrick, J.T.; Phillips, J.I.; Lusardi, T.A.; McFarland, T.J.; Lind, B.; Sandau, U.S.; Harrington, C.A.; Lapidus, J.A.; Galasko, D.R.; Quinn, J.F. Validation of microRNA biomarkers for Alzheimer’s disease in human cerebrospinal fluid. J. Alzheimer’s Dis. 2019, 67, 875–891. [Google Scholar] [CrossRef]
- Tigchelaar, S.; Gupta, R.; Shannon, C.P.; Streijger, F.; Sinha, S.; Flibotte, S.; Rizzuto, M.A.; Street, J.; Paquette, S.; Ailon, T. MicroRNA biomarkers in cerebrospinal fluid and serum reflect injury severity in human acute traumatic spinal cord injury. J. Neurotrauma 2019, 36, 2358–2371. [Google Scholar] [CrossRef]
- Raoof, R.; Bauer, S.; El Naggar, H.; Connolly, N.M.; Brennan, G.P.; Brindley, E.; Hill, T.; McArdle, H.; Spain, E.; Forster, R.J. Dual-center, dual-platform microRNA profiling identifies potential plasma biomarkers of adult temporal lobe epilepsy. EBioMedicine 2018, 38, 127–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, S.; Haleyurgirisetty, M.; Lee, S.; Hewlett, I.; Devadas, K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine 2019, 43, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liang, H.; Chen, X.; Ke, Y.; Zhou, Z.; Yang, M.; Zen, K.; Yang, R.; Liu, C.; Zhang, C.-Y. An Ebola virus-encoded microRNA-like fragment serves as a biomarker for early diagnosis of Ebola virus disease. Cell Res. 2016, 26, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Hartung, A.; Makarewicz, O.; Egerer, R.; Karrasch, M.; Klink, A.; Sauerbrei, A.; Kentouche, K.; Pletz, M.W. EBV miRNA expression profiles in different infection stages: A prospective cohort study. PLoS ONE 2019, 14, e0212027. [Google Scholar] [CrossRef]
- Li, X.; Fu, Z.; Liang, H.; Wang, Y.; Qi, X.; Ding, M.; Sun, X.; Zhou, Z.; Huang, Y.; Gu, H. H5N1 influenza virus-specific miRNA-like small RNA increases cytokine production and mouse mortality via targeting poly (rC)-binding protein 2. Cell Res. 2018, 28, 157–171. [Google Scholar] [CrossRef]
- Ai, J.; Zhang, R.; Li, Y.; Pu, J.; Lu, Y.; Jiao, J.; Li, K.; Yu, B.; Li, Z.; Wang, R. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem. Biophys. Res. Commun. 2010, 391, 73–77. [Google Scholar] [CrossRef]
- Corsten, M.F.; Dennert, R.; Jochems, S.; Kuznetsova, T.; Devaux, Y.; Hofstra, L.; Wagner, D.R.; Staessen, J.A.; Heymans, S.; Schroen, B. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ. Cardiovasc. Genet. 2010, 3, 499–506. [Google Scholar] [CrossRef]
- Islas, J.F.; Moreno-Cuevas, J.E. A microRNA perspective on cardiovascular development and diseases: An update. Int. J. Mol. Sci. 2018, 19, 2075. [Google Scholar] [CrossRef]
- Mens, M.M.; Heshmatollah, A.; Fani, L.; Ikram, M.A.; Ikram, M.K.; Ghanbari, M. Circulatory MicroRNAs as potential biomarkers for stroke risk: The rotterdam study. Stroke 2021, 52, 945–953. [Google Scholar] [CrossRef]
- Qing, S.; Yuan, S.; Yun, C.; Hui, H.; Mao, P.; Wen, F.; Ding, Y.; Liu, Q. Serum miRNA biomarkers serve as a fingerprint for proliferative diabetic retinopathy. Cell. Physiol. Biochem. 2014, 34, 1733–1740. [Google Scholar] [CrossRef]
- Elemam, N.M.; Hasswan, H.; Aljaibeji, H.; Sulaiman, N. Circulating soluble ACE2 and upstream microRNA expressions in serum of type 2 diabetes mellitus patients. Int. J. Mol. Sci. 2021, 22, 5263. [Google Scholar] [CrossRef] [PubMed]
- Taverner, D.; Llop, D.; Rosales, R.; Ferré, R.; Masana, L.; Vallvé, J.-C.; Paredes, S. Plasma expression of microRNA-425-5p and microRNA-451a as biomarkers of cardiovascular disease in rheumatoid arthritis patients. Sci. Rep. 2021, 11, 15670. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.L.; Zou, R.; Zhou, L.; Lim, J.Y.; Phua, D.C.; Liu, C.; Chong, J.P.; Ng, J.Y.; Liew, O.W.; Chan, S.P. Combining circulating microRNA and NT-proBNP to detect and categorize heart failure subtypes. J. Am. Coll. Cardiol. 2019, 73, 1300–1313. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.S.; Hirsh, V.; Mok, T.; Socinski, M.A.; Gervais, R.; Wu, Y.-L.; Li, L.-Y.; Watkins, C.L.; Sellers, M.V.; Lowe, E.S. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): A randomised phase III trial. Lancet 2008, 372, 1809–1818. [Google Scholar] [CrossRef]
- Quach, N.; Goodman, M.F.; Shibata, D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin. Pathol. 2004, 4, 1. [Google Scholar] [CrossRef]
- Bedard, P.L.; Hansen, A.R.; Ratain, M.J.; Siu, L.L. Tumour heterogeneity in the clinic. Nature 2013, 501, 355–364. [Google Scholar] [CrossRef]
- Ofiara, L.; Navasakulpong, A.; Ezer, N.; Gonzalez, A. The importance of a satisfactory biopsy for the diagnosis of lung cancer in the era of personalized treatment. Curr. Oncol. 2012, 19, 16–23. [Google Scholar] [CrossRef]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef]
- Diaz, L.A., Jr.; Williams, R.T.; Wu, J.; Kinde, I.; Hecht, J.R.; Berlin, J.; Allen, B.; Bozic, I.; Reiter, J.G.; Nowak, M.A. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012, 486, 537–540. [Google Scholar] [CrossRef]
- Gazdar, A. Activating and resistance mutations of EGFR in non-small-cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009, 28, S24–S31. [Google Scholar] [CrossRef]
- Inukai, M.; Toyooka, S.; Ito, S.; Asano, H.; Ichihara, S.; Soh, J.; Suehisa, H.; Ouchida, M.; Aoe, K.; Aoe, M. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non–small cell lung cancer. Cancer Res. 2006, 66, 7854–7858. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, S.; Boggon, T.J.; Dayaram, T.; Jänne, P.A.; Kocher, O.; Meyerson, M.; Johnson, B.E.; Eck, M.J.; Tenen, D.G.; Halmos, B. EGFR mutation and resistance of non–small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005, 352, 786–792. [Google Scholar] [CrossRef]
- Russano, M.; Napolitano, A.; Ribelli, G.; Iuliani, M.; Simonetti, S.; Citarella, F.; Pantano, F.; Dell’Aquila, E.; Anesi, C.; Silvestris, N.; et al. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: The potentiality of blood samples. J. Exp. Clin. Cancer Res. 2020, 39, 95. [Google Scholar] [CrossRef]
- Keup, C.; Suryaprakash, V.; Hauch, S.; Storbeck, M.; Hahn, P.; Sprenger-Haussels, M.; Kolberg, H.C.; Tewes, M.; Hoffmann, O.; Kimmig, R.; et al. Integrative statistical analyses of multiple liquid biopsy analytes in metastatic breast cancer. Genome Med. 2021, 13, 85. [Google Scholar] [CrossRef]
- Freitas, C.; Sousa, C.; Machado, F.; Serino, M.; Santos, V.; Cruz-Martins, N.; Teixeira, A.; Cunha, A.; Pereira, T.; Oliveira, H.P. The role of liquid biopsy in early diagnosis of lung Cancer. Front. Oncol. 2021, 11, 634316. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Cheong, J.K.; Tang, Y.C.; Zhou, L.; Cheng, H.; Too, H.-P. Advances in quantifying circulatory microRNA for early disease detection. Curr. Opin. Biotechnol. 2022, 74, 256–262. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Leite, K.R.; Canavez, J.M.; Reis, S.T.; Tomiyama, A.H.; Piantino, C.B.; Sañudo, A.; Camara-Lopes, L.H.; Srougi, M. miRNA Analysis of Prostate Cancer by Quantitative Real Time PCR: Comparison between Formalin-Fixed Paraffin Embedded and Fresh-Frozen Tissue. Urol. Oncol. Semin. Orig. Investig. 2011, 29, 533–537. [Google Scholar] [CrossRef]
- Nakajima, G.; Hayashi, K.; Xi, Y.; Kudo, K.; Uchida, K.; Takasaki, K.; Yamamoto, M.; Ju, J. Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genom. Proteom. 2006, 3, 317–324. [Google Scholar]
- Lan, H.; Lu, H.; Wang, X.; Jin, H. MicroRNAs as potential biomarkers in cancer: Opportunities and challenges. Biomed. Res. Int. 2015, 2015, 125094. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef]
- Cappelletti, V.; Appierto, V.; Tiberio, P.; Fina, E.; Callari, M.; Daidone, M.G. Circulating biomarkers for prediction of treatment response. J. Natl. Cancer Inst. Monogr. 2015, 2015, 60–63. [Google Scholar] [CrossRef]
- Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding RNAs in extracellular fluids as cancer biomarkers: The new frontier of liquid biopsies. Cancers 2019, 11, 1170. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef]
- Höög, J.L.; Lötvall, J. Diversity of extracellular vesicles in human ejaculates revealed by cryo-electron microscopy. J. Extracell. Vesicles 2015, 4, 28680. [Google Scholar] [CrossRef]
- Chung, K.Y.; Quek, J.M.; Neo, S.H.; Too, H.P. Polymer-based precipitation of extracellular vesicular miRNAs from serum improve gastric cancer miRNA biomarker performance. J. Mol. Diagn. 2020, 22, 610–618. [Google Scholar] [CrossRef]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Yu, Z.; Chen, D.; Wang, Z.; Miao, J.; Li, Q.; Zhang, D.; Song, J.; Cui, D. Progress in microfluidics-based exosome separation and detection technologies for diagnostic applications. Small 2020, 16, e1903916. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Zargartalebi, H.; Salahandish, R.; Aburashed, R.; Wey Yong, K.; Sanati-Nezhad, A. Emerging technologies and commercial products in exosome-based cancer diagnosis and prognosis. Biosens. Bioelectron. 2021, 183, 113176. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, J.B. What are we looking at? Extracellular vesicles, lipoproteins, or both? Circ. Res. 2017, 121, 920–922. [Google Scholar] [CrossRef]
- Sódar, B.W.; Kittel, Á.; Pálóczi, K.; Vukman, K.V.; Osteikoetxea, X.; Szabó-Taylor, K.; Németh, A.; Sperlágh, B.; Baranyai, T.; Giricz, Z.; et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci. Rep. 2016, 6, 24316. [Google Scholar] [CrossRef]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef]
- Wu, D.; Yan, J.; Shen, X.; Sun, Y.; Thulin, M.; Cai, Y.; Wik, L.; Shen, Q.; Oelrich, J.; Qian, X.; et al. Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nat. Commun. 2019, 10, 3854. [Google Scholar] [CrossRef]
- Iliescu, F.S.; Vrtačnik, D.; Neuzil, P.; Iliescu, C. Microfluidic technology for clinical applications of exosomes. Micromachines 2019, 10, 392. [Google Scholar] [CrossRef]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef]
- Rider, M.A.; Hurwitz, S.N.; Meckes, D.G. ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles. Sci. Rep. 2016, 6, 23978. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on strategies and technologies for exosome isolation and purification. Front. Bioeng. Biotechnol. 2022, 9, 811971. [Google Scholar] [CrossRef]
- Kurian, T.K.; Banik, S.; Gopal, D.; Chakrabarti, S.; Mazumder, N. Elucidating methods for isolation and quantification of exosomes: A review. Mol. Biotechnol. 2021, 63, 249–266. [Google Scholar] [CrossRef]
- Lai, J.J.; Chau, Z.L.; Chen, S.Y.; Hill, J.J.; Korpany, K.V.; Liang, N.W.; Lin, L.H.; Lin, Y.H.; Liu, J.K.; Liu, Y.C. Exosome Processing and Characterization Approaches for Research and Technology Development. Adv. Sci. 2022, 9, e2103222. [Google Scholar] [CrossRef]
- Shirejini, S.Z.; Inci, F. The Yin and Yang of exosome isolation methods: Conventional practice, microfluidics, and commercial kits. Biotechnol. Adv. 2021, 54, 107814. [Google Scholar] [CrossRef]
- Xu, K.; Jin, Y.; Li, Y.; Huang, Y.; Zhao, R. Recent Progress of Exosome Isolation and Peptide Recognition-Guided Strategies for Exosome Research. Front. Chem. 2022, 10, 844124. [Google Scholar] [CrossRef]
- Zhu, L.; Sun, H.-T.; Wang, S.; Huang, S.-L.; Zheng, Y.; Wang, C.-Q.; Hu, B.-Y.; Qin, W.; Zou, T.-T.; Fu, Y. Isolation and characterization of exosomes for cancer research. J. Hematol. Oncol. 2020, 13, 152. [Google Scholar] [CrossRef]
- Drula, R.; Ott, L.F.; Berindan-Neagoe, I.; Pantel, K.; Calin, G.A. MicroRNAs from liquid biopsy derived extracellular vesicles: Recent advances in detection and characterization methods. Cancers 2020, 12, 2009. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, Q.; Zhang, J.; Li, C.; Miao, Y.R.; Lei, Q.; Li, Q.; Guo, A.Y. EVmiRNA: A database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019, 47, D89–D93. [Google Scholar] [CrossRef]
- Liu, C.J.; Xie, G.Y.; Miao, Y.R.; Xia, M.; Wang, Y.; Lei, Q.; Zhang, Q.; Guo, A.Y. EVAtlas: A comprehensive database for ncRNA expression in human extracellular vesicles. Nucleic Acids Res. 2022, 50, D111–D117. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhu, Q.; Cheng, L.; Wang, Y.; Li, M.; Yang, Q.; Hu, L.; Lou, D.; Li, J.; Dong, X.; et al. Exosome detection via the ultrafast-isolation system: EXODUS. Nat. Methods 2021, 18, 212–218. [Google Scholar] [CrossRef]
- Zhu, Q.; Cheng, L.; Deng, C.; Huang, L.; Li, J.; Wang, Y.; Li, M.; Yang, Q.; Dong, X.; Su, J. The genetic source tracking of human urinary exosomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2108876118. [Google Scholar] [CrossRef] [PubMed]
- Chronopoulos, A.; Kalluri, R. Emerging role of bacterial extracellular vesicles in cancer. Oncogene 2020, 39, 6951–6960. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; How Huang, K.; Jen Lee, M.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Valencia, K.; Montuenga, L.M. Exosomes in Liquid Biopsy: The Nanometric World in the Pursuit of Precision Oncology. Cancers 2021, 13, 2147. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.M. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [PubMed]
- McAndrews, K.M.; Kalluri, R. Mechanisms associated with biogenesis of exosomes in cancer. Mol Cancer 2019, 18, 52. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, C.; Brex, D.; Caponnetto, A.; Cirnigliaro, M.; Scalia, M.; Magnano, A.; Caltabiano, R.; Barbagallo, D.; Biondi, A.; Cappellani, A. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol. Ther. Nucleic Acids 2018, 12, 229–241. [Google Scholar] [CrossRef]
- Castellanos-Rizaldos, E.; Grimm, D.G.; Tadigotla, V.; Hurley, J.; Healy, J.; Neal, P.L.; Sher, M.; Venkatesan, R.; Karlovich, C.; Raponi, M. Exosome-based detection of EGFR T790M in plasma from non–small cell lung cancer patients. Clin. Cancer Res. 2018, 24, 2944–2950. [Google Scholar] [CrossRef]
- Dong, L.; Lin, W.; Qi, P.; Xu, M.-D.; Wu, X.; Ni, S.; Huang, D.; Weng, W.-W.; Tan, C.; Sheng, W. Circulating long RNAs in serum extracellular vesicles: Their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1158–1166. [Google Scholar] [CrossRef]
- Krug, A.; Enderle, D.; Karlovich, C.; Priewasser, T.; Bentink, S.; Spiel, A.; Brinkmann, K.; Emenegger, J.; Grimm, D.; Castellanos-Rizaldos, E. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. 2018, 29, 700–706. [Google Scholar] [CrossRef]
- Krug, A.K.; Karlovich, C.; Koestler, T.; Brinkmann, K.; Spiel, A.; Emenegger, J.; Noerholm, M.; O’Neill, V.; Sequist, L.V.; Soria, J.-C.; et al. Abstract B136: Plasma EGFR mutation detection using a combined exosomal RNA and circulating tumor DNA approach in patients with acquired resistance to first-generation EGFR-TKIs. Mol. Cancer Ther. 2015, 14, B136. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, Z.; Hu, J.; Yang, D.; Chen, X.; Wang, Q.; Liu, J.; Dou, M.; Peng, W.; Wu, Y. High-throughput single-EV liquid biopsy: Rapid, simultaneous, and multiplexed detection of nucleic acids, proteins, and their combinations. Sci. Adv. 2020, 6, eabc1204. [Google Scholar] [CrossRef] [PubMed]
- Zocco, D.; Bernardi, S.; Novelli, M.; Astrua, C.; Fava, P.; Zarovni, N.; Carpi, F.M.; Bianciardi, L.; Malavenda, O.; Quaglino, P. Isolation of extracellular vesicles improves the detection of mutant DNA from plasma of metastatic melanoma patients. Sci. Rep. 2020, 10, 15745. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.-L.; Chen, Y.-L.; Ge, Z.-Z.; Qu, Y.-Y.; Cao, Y.; Kang, Z.-X. Downregulation of serum exosomal miR-150-5p is associated with poor prognosis in patients with colorectal cancer. Cancer Biomark. 2019, 26, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Boyiadzis, M.; Whiteside, T.L. Plasma-derived exosomes in acute myeloid leukemia for detection of minimal residual disease: Are we ready? Expert Rev. Mol. Diagn. 2016, 16, 623–629. [Google Scholar] [CrossRef]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.; Enderle, D.; Noerholm, M.; Breakefield, X.; Skog, J. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Krol, T.; West, J.; Hayden, J.; He, M. Exosomes-The Good, Bad, Ugly and Current State. Am. Pharm. Rev. 2021. Available online: https://www.americanpharmaceuticalreview.com/Featured-Articles/575432-Exosomes-The-Good-Bad-Ugly-and-Current-State/ (accessed on 10 August 2022).
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Witwer, K.W.; Goberdhan, D.C.; O’Driscoll, L.; Théry, C.; Welsh, J.A.; Blenkiron, C.; Buzás, E.I.; Di Vizio, D.; Erdbrügger, U.; Falcón-Pérez, J.M.; et al. Updating MISEV: Evolving the minimal requirements for studies of extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12182. [Google Scholar] [CrossRef]
- Hilton, S.H.; White, I.M. Advances in the analysis of single extracellular vesicles: A critical review. Sens. Actuators Rep. 2021, 3, 100052. [Google Scholar] [CrossRef]
- Islam, S.; Kjällquist, U.; Moliner, A.; Zajac, P.; Fan, J.B.; Lönnerberg, P.; Linnarsson, S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011, 21, 1160–1167. [Google Scholar] [CrossRef]
- Arab, A.; Karimipoor, M.; Irani, S.; Kiani, A.; Zeinali, S.; Tafsiri, E.; Sheikhy, K. Potential circulating miRNA signature for early detection of NSCLC. Cancer Genet. 2017, 216, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Geng, Q.; Fan, T.; Zhang, B.; Wang, W.; Xu, Y.; Hu, H. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir. Res. 2014, 15, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sromek, M.; Glogowski, M.; Chechlinska, M.; Kulinczak, M.; Szafron, L.; Zakrzewska, K.; Owczarek, J.; Wisniewski, P.; Wlodarczyk, R.; Talarek, L. Changes in plasma miR-9, miR-16, miR-205 and miR-486 levels after non-small cell lung cancer resection. Cell. Oncol. 2017, 40, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Xiang, Y.; Guo, X.; Zhang, Y.; Li, C.; Xie, W.; Wu, N.; Wu, L.; Cai, T.; Ma, X. Circulating long noncoding RNAs act as diagnostic biomarkers in non-small cell lung cancer. Front. Oncol. 2020, 10, 537120. [Google Scholar] [CrossRef]
- Zhang, R.; Xia, Y.; Wang, Z.; Zheng, J.; Chen, Y.; Li, X.; Wang, Y.; Ming, H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017, 490, 406–414. [Google Scholar] [CrossRef]
- Hamam, R.; Ali, A.M.; Alsaleh, K.A.; Kassem, M.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci. Rep. 2016, 6, 25997. [Google Scholar] [CrossRef]
- Shin, V.; Siu, J.; Cheuk, I.; Ng, E.; Kwong, A. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br. J. Cancer 2015, 112, 1751–1759. [Google Scholar] [CrossRef]
- Zou, R.; Loke, S.Y.; Tang, Y.C.; Too, H.P.; Zhou, L.; Lee, A.S.G.; Hartman, M. Development and validation of a circulating microRNA panel for the early detection of breast cancer. Br. J. Cancer 2022, 126, 472–481. [Google Scholar] [CrossRef]
- Dong, L.; Di Liu, D.J.; Xu, H.; Zhang, C.; Qi, D.; Liu, D. LncRNA ARST is a Novel Prognostic and Diagnostic Biomarker for Colorectal Cancer. Cancer Manag. Res. 2022, 14, 19. [Google Scholar] [CrossRef]
- Vychytilova-Faltejskova, P.; Radova, L.; Sachlova, M.; Kosarova, Z.; Slaba, K.; Fabian, P.; Grolich, T.; Prochazka, V.; Kala, Z.; Svoboda, M. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis 2016, 37, 941–950. [Google Scholar] [CrossRef]
- So, J.B.Y.; Kapoor, R.; Zhu, F.; Koh, C.; Zhou, L.; Zou, R.; Tang, Y.C.; Goo, P.C.; Rha, S.Y.; Chung, H.C. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut 2021, 70, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Nweke, E.E.; Brand, M. Downregulation of the let-7 family of microRNAs may promote insulin receptor/insulin-like growth factor signalling pathways in pancreatic ductal adenocarcinoma. Oncol. Lett. 2020, 20, 2613–2620. [Google Scholar] [CrossRef] [PubMed]
- Youssef, Y.M.; White, N.M.; Grigull, J.; Krizova, A.; Samy, C.; Mejia-Guerrero, S.; Evans, A.; Yousef, G.M. Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur. Urol. 2011, 59, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Gilad, S.; Lithwick-Yanai, G.; Barshack, I.; Benjamin, S.; Krivitsky, I.; Edmonston, T.B.; Bibbo, M.; Thurm, C.; Horowitz, L.; Huang, Y. Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J. Mol. Diagn. 2012, 14, 510–517. [Google Scholar] [CrossRef]
- Samsonov, R.; Burdakov, V.; Shtam, T.; Radzhabova, Z.; Vasilyev, D.; Tsyrlina, E.; Titov, S.; Ivanov, M.; Berstein, L.; Filatov, M. Plasma exosomal miR-21 and miR-181a differentiates follicular from papillary thyroid cancer. Tumor Biol. 2016, 37, 12011–12021. [Google Scholar] [CrossRef]
- McAnena, P.; Tanriverdi, K.; Curran, C.; Gilligan, K.; Freedman, J.E.; Brown, J.A.; Kerin, M.J. Circulating microRNAs miR-331 and miR-195 differentiate local luminal a from metastatic breast cancer. BMC Cancer 2019, 19, 436. [Google Scholar] [CrossRef]
- Yerukala Sathipati, S.; Ho, S.-Y. Identifying a miRNA signature for predicting the stage of breast cancer. Sci. Rep. 2018, 8, 16138. [Google Scholar] [CrossRef]
- Jiang, H.; Guo, S.; Zhao, Y.; Wang, Y.; Piao, H.-Y.; Wu, Y.; Zhang, J. Circulating long non-coding RNA PCGEM1 as a novel biomarker for gastric cancer diagnosis. Pathol. Res. Pract. 2019, 215, 152569. [Google Scholar] [CrossRef]
- Zhang, X.; Sai, B.; Wang, F.; Wang, L.; Wang, Y.; Zheng, L.; Li, G.; Tang, J.; Xiang, J. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer 2019, 18, 40. [Google Scholar] [CrossRef]
- Dejima, H.; Iinuma, H.; Kanaoka, R.; Matsutani, N.; Kawamura, M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol. Lett. 2017, 13, 1256–1263. [Google Scholar] [CrossRef]
- Liu, Q.; Yu, Z.; Yuan, S.; Xie, W.; Li, C.; Hu, Z.; Xiang, Y.; Wu, N.; Wu, L.; Bai, L. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 2017, 8, 13048. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Liu, X.; He, J.; Chen, D.; Hunag, Y.; Zhang, Y.K. Overexpression of members of the microRNA-183 family is a risk factor for lung cancer: A case control study. BMC Cancer 2011, 11, 393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Zhang, P.; Wang, F.; Zhang, H.; Yang, J.; Peng, J.; Liu, W.; Qin, H. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut 2012, 61, 1447–1453. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Shuang, D.; Yi, Z.; Sheng, H.; Liu, Y. Up-regulated microRNA-155 expression is associated with poor prognosis in cervical cancer patients. Biomed. Pharmacother. 2016, 83, 64–69. [Google Scholar] [CrossRef]
- Pang, P.; Shi, X.; Huang, W.; Sun, K. miR-497 as a potential serum biomarker for the diagnosis and prognosis of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3765–3769. [Google Scholar]
- Gong, C.; Tan, W.; Chen, K.; You, N.; Zhu, S.; Liang, G.; Xie, X.; Li, Q.; Zeng, Y.; Ouyang, N. Prognostic value of a BCSC-associated microRNA signature in hormone receptor-positive HER2-negative breast cancer. EBioMedicine 2016, 11, 199–209. [Google Scholar] [CrossRef]
- Chen, W.; Cai, F.; Zhang, B.; Barekati, Z.; Zhong, X.Y. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: Potential biomarkers. Tumour Biol. 2013, 34, 455–462. [Google Scholar] [CrossRef]
- Rothe, F.; Ignatiadis, M.; Chaboteaux, C.; Haibe-Kains, B.; Kheddoumi, N.; Majjaj, S.; Badran, B.; Fayyad-Kazan, H.; Desmedt, C.; Harris, A.L. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS ONE 2011, 6, e20980. [Google Scholar] [CrossRef]
- Giovannetti, E.; Funel, N.; Peters, G.J.; Del Chiaro, M.; Erozenci, L.A.; Vasile, E.; Leon, L.G.; Pollina, L.E.; Groen, A.; Falcone, A. MicroRNA-21 in pancreatic cancer: Correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010, 70, 4528–4538. [Google Scholar] [CrossRef]
- Mosakhani, N.; Sarhadi, V.K.; Borze, I.; Karjalainen-Lindsberg, M.L.; Sundström, J.; Ristamäki, R.; Österlund, P.; Knuutila, S. MicroRNA profiling differentiates colorectal cancer according to KRAS status. Genes Chromosomes Cancer 2012, 51, 1–9. [Google Scholar] [CrossRef]
- Cervena, K.; Novosadova, V.; Pardini, B.; Naccarati, A.; Opattova, A.; Horak, J.; Vodenkova, S.; Buchler, T.; Skrobanek, P.; Levy, M. Analysis of MicroRNA Expression Changes During the Course of Therapy in Rectal Cancer Patients. Front. Oncol. 2021, 11, 702258. [Google Scholar] [CrossRef] [PubMed]
- Kheirelseid, E.A.; Miller, N.; Chang, K.H.; Curran, C.; Hennessey, E.; Sheehan, M.; Newell, J.; Lemetre, C.; Balls, G.; Kerin, M.J. miRNA expressions in rectal cancer as predictors of response to neoadjuvant chemoradiation therapy. Int. J. Colorectal. Dis. 2013, 28, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Scarpati, G.D.V.; Falcetta, F.; Carlomagno, C.; Ubezio, P.; Marchini, S.; De Stefano, A.; Singh, V.K.; D’Incalci, M.; De Placido, S.; Pepe, S. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int. J. Radiat. Oncol. Biol. Phys. 2012, 83, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, K.; Bi, M.; Jiao, X.; Zhang, D.; Dong, Q. Circulating microRNA expressions in colorectal cancer as predictors of response to chemotherapy. Anticancer. Drugs 2014, 25, 346–352. [Google Scholar] [CrossRef]
- Weiss, G.; Bemis, L.; Nakajima, E.; Sugita, M.; Birks, D.; Robinson, W.; Varella-Garcia, M.; Bunn Jr, P.; Haney, J.; Helfrich, B. EGFR regulation by microRNA in lung cancer: Correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann. Oncol. 2008, 19, 1053–1059. [Google Scholar] [CrossRef] [PubMed]
- Tumilson, C.A.; Lea, R.W.; Alder, J.E.; Shaw, L. Circulating microRNA biomarkers for glioma and predicting response to therapy. Mol. Neurobiol. 2014, 50, 545–558. [Google Scholar] [CrossRef]
- Lu, L.; Schwartz, P.; Scarampi, L.; Rutherford, T.; Canuto, E.M.; Yu, H.; Katsaros, D. MicroRNA let-7a: A potential marker for selection of paclitaxel in ovarian cancer management. Gynecol. Oncol. 2011, 122, 366–371. [Google Scholar] [CrossRef]
- Rzepiel, A.; Kutszegi, N.; Gézsi, A.; Sági, J.C.; Egyed, B.; Péter, G.; Butz, H.; Nyírő, G.; Müller, J.; Kovács, G.T. Circulating microRNAs as minimal residual disease biomarkers in childhood acute lymphoblastic leukemia. J. Transl. Med. 2019, 17, 372. [Google Scholar] [CrossRef]
- Ohyashiki, J.H.; Ohtsuki, K.; Mizoguchi, I.; Yoshimoto, T.; Katagiri, S.; Umezu, T.; Ohyashiki, K. Downregulated microRNA-148b in circulating PBMCs in chronic myeloid leukemia patients with undetectable minimal residual disease: A possible biomarker to discontinue imatinib safely. Drug Des. Dev. Ther. 2014, 8, 1151. [Google Scholar] [CrossRef]
- Stang, A.; Jöckel, K.H. The Impact of Cancer Screening on All-Cause Mortality. Dtsch. Arztebl. Int. 2018, 115, 481–486. [Google Scholar] [CrossRef]
- Keller, A.; Fehlmann, T.; Backes, C.; Kern, F.; Gislefoss, R.; Langseth, H.; Rounge, T.B.; Ludwig, N.; Meese, E. Competitive learning suggests circulating miRNA profiles for cancers decades prior to diagnosis. RNA Biol. 2020, 17, 1416–1426. [Google Scholar] [CrossRef] [PubMed]
- Fayyad-Kazan, H.; Bitar, N.; Najar, M.; Lewalle, P.; Fayyad-Kazan, M.; Badran, R.; Hamade, E.; Daher, A.; Hussein, N.; ElDirani, R.; et al. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J. Transl. Med. 2013, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, Z.; Li, Y.; Xu, R.; Wang, Y.; Tian, Y.; Chen, W. Long non-coding RNA NEAT1 overexpression is associated with poor prognosis in cancer patients: A systematic review and meta-analysis. Oncotarget 2017, 8, 2672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Fu, L.; Lu, T.; Zhang, G.; Zhang, J.; Zhao, Y.; Jin, H.; Yang, K.; Cai, H. Clinicopathological and Prognostic Significance of Long Non-coding RNA MIAT in Human Cancers: A Review and Meta-Analysis. Front. Genet. 2021, 12, 729768. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Q.; Wang, J.; Feng, W.; Chen, Y.; Liu, J.; Qu, Z.; Li, M. Prognostic and clinicopathological role of long noncoding RNA NORAD in various cancers: A meta-analysis. Biomark. Med. 2021, 15, 427–436. [Google Scholar] [CrossRef]
- Li, W.; Li, N.; Shi, K.; Chen, Q. Systematic review and meta-analysis of the utility of long non-coding RNA GAS5 as a diagnostic and prognostic cancer biomarker. Oncotarget 2017, 8, 66414–66425. [Google Scholar] [CrossRef]
- Tian, G.; Li, G.; Guan, L.; Wang, Z.; Li, N. Prognostic Value of Circular RNA ciRS-7 in Various Cancers: A PRISMA-Compliant Meta-Analysis. Biomed. Res. Int. 2020, 2020, 1487609. [Google Scholar] [CrossRef]
- Luo, D.; Yang, L.; Yu, L.; Chen, Y.; Huang, Z.; Liu, H. Clinicopathological and prognostic significance of long non-coding RNA-ROR in cancer patients: A systematic review and meta-analysis. Medicine 2021, 100, e26535. [Google Scholar] [CrossRef]
- Carden, C.P.; Sarker, D.; Postel-Vinay, S.; Yap, T.A.; Attard, G.; Banerji, U.; Garrett, M.D.; Thomas, G.V.; Workman, P.; Kaye, S.B. Can molecular biomarker-based patient selection in Phase I trials accelerate anticancer drug development? Drug Discov. Today 2010, 15, 88–97. [Google Scholar] [CrossRef]
- Ding, H.; Xu, J.; Lin, Z.; Huang, J.; Wang, F.; Yang, Y.; Cui, Y.; Luo, H.; Gao, Y.; Zhai, X. Minimal residual disease in multiple myeloma: Current status. Biomark. Res. 2021, 9, 75. [Google Scholar] [CrossRef]
- Kerr, K.M. Personalized medicine for lung cancer: New challenges for pathology. Histopathology 2012, 60, 531–546. [Google Scholar] [CrossRef] [PubMed]
- Walter, R.B.; Buckley, S.A.; Pagel, J.M.; Wood, B.L.; Storer, B.E.; Sandmaier, B.M.; Fang, M.; Gyurkocza, B.; Delaney, C.; Radich, J.P.; et al. Significance of minimal residual disease before myeloablative allogeneic hematopoietic cell transplantation for AML in first and second complete remission. Blood 2013, 122, 1813–1821. [Google Scholar] [CrossRef] [PubMed]
- Ivey, A.; Hills, R.K.; Simpson, M.A.; Jovanovic, J.V.; Gilkes, A.; Grech, A.; Patel, Y.; Bhudia, N.; Farah, H.; Mason, J.; et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N. Engl. J. Med. 2016, 374, 422–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, S.; Yokoyama, K.; Shimizu, E.; Yusa, N.; Kondoh, K.; Ogawa, M.; Takei, T.; Kobayashi, A.; Ito, M.; Isobe, M. Prognostic impact of circulating tumor DNA status post–allogeneic hematopoietic stem cell transplantation in AML and MDS. Blood 2019, 133, 2682–2695. [Google Scholar] [CrossRef]
- Rose Brannon, A.; Jayakumaran, G.; Diosdado, M.; Patel, J.; Razumova, A.; Hu, Y.; Meng, F.; Haque, M.; Sadowska, J.; Murphy, B.J. Enhanced specificity of clinical high-sensitivity tumor mutation profiling in cell-free DNA via paired normal sequencing using MSK-ACCESS. Nat. Commun. 2021, 12, 3770. [Google Scholar] [CrossRef] [PubMed]
- Galvão-Lima, L.J.; Morais, A.H.; Valentim, R.A.; Barreto, E.J. miRNAs as biomarkers for early cancer detection and their application in the development of new diagnostic tools. Biomed. Eng. Online 2021, 20, 21. [Google Scholar] [CrossRef]
- Singh, R.; Ramasubramanian, B.; Kanji, S.; Chakraborty, A.R.; Haque, S.J.; Chakravarti, A. Circulating microRNAs in cancer: Hope or hype? Cancer Lett. 2016, 381, 113–121. [Google Scholar] [CrossRef]
- Witwer, K.W. Circulating microRNA biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar] [CrossRef]
- Zhang, M.; Hu, Z.D. Suggestions for designing studies investigating diagnostic accuracy of biomarkers. Ann. Transl. Med. 2019, 7, 788. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Srivastava, S.K.; Khan, M.A.; Prajapati, V.K.; Singh, S.; Carter, J.E.; Singh, A.P. Racial disparities in prostate cancer: A molecular perspective. Front. Biosci. (Landmark. Ed.) 2017, 22, 772. [Google Scholar] [CrossRef]
- Duttagupta, R.; Jiang, R.; Gollub, J.; Getts, R.C.; Jones, K.W. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS ONE 2011, 6, e20769. [Google Scholar] [CrossRef] [PubMed]
- Hasakova, K.; Bezakova, J.; Vician, M.; Reis, R.; Zeman, M.; Herichova, I. Gender-dependent expression of leading and passenger strand of miR-21 and miR-16 in human colorectal cancer and adjacent colonic tissues. Physiol. Res. 2017, 66, S575–S582. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, A.R.; Zheng, H.; Kwok, J.G.; Qu, Y.; Zou, A.E.; Korrapati, A.; Li, P.X.; Califano, J.A.; Hovell, M.F.; Wang-Rodriguez, J. A comprehensive study of smoking-specific microRNA alterations in head and neck squamous cell carcinoma. Oral. Oncol. 2017, 72, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Fehlmann, T.; Ludwig, N.; Backes, C.; Meese, E.; Keller, A. Distribution of microRNA biomarker candidates in solid tissues and body fluids. RNA Biol. 2016, 13, 1084–1088. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef]
- Moher, D.; Hopewell, S.; Schulz, K.F.; Montori, V.; Gøtzsche, P.C.; Devereaux, P.J.; Elbourne, D.; Egger, M.; Altman, D.G. CONSORT 2010 Explanation and Elaboration: Updated guidelines for reporting parallel group randomised trials. BMJ 2010, 340, c869. [Google Scholar] [CrossRef]
- Laterza, O.F.; Lim, L.; Garrett-Engele, P.W.; Vlasakova, K.; Muniappa, N.; Tanaka, W.K.; Johnson, J.M.; Sina, J.F.; Fare, T.L.; Sistare, F.D. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin. Chem. 2009, 55, 1977–1983. [Google Scholar] [CrossRef]
- Dlamini, Z.; Francies, F.Z.; Hull, R.; Marima, R. Artificial intelligence (AI) and big data in cancer and precision oncology. Comput. Struct. Biotechnol. J. 2020, 18, 2300–2311. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Martínez-Camblor, P.; Bär, C.; Duarte, K.; Girerd, N.; Fellström, B.; Schmieder, R.E.; Jardine, A.G.; Massy, Z.A.; Holdaas, H. Improved cardiovascular risk prediction in patients with end-stage renal disease on hemodialysis using machine learning modeling and circulating microribonucleic acids. Theranostics 2020, 10, 8665. [Google Scholar] [CrossRef]
- Shim, M.; Lee, S.H.; Hwang, H.J. Inflated prediction accuracy of neuropsychiatric biomarkers caused by data leakage in feature selection. Sci. Rep. 2021, 11, 7980. [Google Scholar] [CrossRef]
- Poldrack, R.A.; Huckins, G.; Varoquaux, G. Establishment of Best Practices for Evidence for Prediction: A Review. JAMA Psychiatry 2020, 77, 534–540. [Google Scholar] [CrossRef]
- Bone, D.; Goodwin, M.S.; Black, M.P.; Lee, C.C.; Audhkhasi, K.; Narayanan, S. Applying machine learning to facilitate autism diagnostics: Pitfalls and promises. J. Autism Dev. Disord. 2015, 45, 1121–1136. [Google Scholar] [CrossRef] [PubMed]
- Tu, F.; Zhu, J.; Zheng, Q.; Zhou, M. Be careful of when: An empirical study on time-related misuse of issue tracking data. In Proceedings of the 2018 26th ACM Joint Meeting on European Software Engineering Conference and Symposium on the Foundations of Software Engineering, Lake Buena Vista, FL, USA, 4–9 November 2018; pp. 307–318. [Google Scholar]
- Simon, R. Roadmap for developing and validating therapeutically relevant genomic classifiers. J. Clin. Oncol. 2005, 23, 7332–7341. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, S.; Narayanan, A. Leakage and the Reproducibility Crisis in ML-based Science. arXiv 2022, arXiv:2207.07048. [Google Scholar]
- Gado, A.; Ebeid, B. Gastric cancer missed at endoscopy. Alex. J. Med. 2013, 49, 25–27. [Google Scholar] [CrossRef]
- Kautzky, A.; Seiger, R.; Hahn, A.; Fischer, P.; Krampla, W.; Kasper, S.; Kovacs, G.G.; Lanzenberger, R. Prediction of Autopsy Verified Neuropathological Change of Alzheimer’s Disease Using Machine Learning and MRI. Front. Aging Neurosci. 2018, 10, 406. [Google Scholar] [CrossRef]
- Choi, K.S.; Jun, J.K.; Lee, H.Y.; Park, S.; Jung, K.W.; Han, M.A.; Choi, I.J.; Park, E.C. Performance of gastric cancer screening by endoscopy testing through the National Cancer Screening Program of Korea. Cancer Sci. 2011, 102, 1559–1564. [Google Scholar] [CrossRef]


| Sampling Methods and Biomarker | Advantages | Disadvantages |
|---|---|---|
| Tumor biopsy: Detection of cancer cells by histology |
|
|
| Tumor biopsy: Detection of ncRNAs by molecular techniques |
|
|
| Liquid biopsy: Detection of ncRNAs by molecular techniques (including exosome enrichment) |
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheong, J.K.; Rajgor, D.; Lv, Y.; Chung, K.Y.; Tang, Y.C.; Cheng, H. Noncoding RNome as Enabling Biomarkers for Precision Health. Int. J. Mol. Sci. 2022, 23, 10390. https://doi.org/10.3390/ijms231810390
Cheong JK, Rajgor D, Lv Y, Chung KY, Tang YC, Cheng H. Noncoding RNome as Enabling Biomarkers for Precision Health. International Journal of Molecular Sciences. 2022; 23(18):10390. https://doi.org/10.3390/ijms231810390
Chicago/Turabian StyleCheong, Jit Kong, Dimple Rajgor, Yang Lv, Ka Yan Chung, Yew Chung Tang, and He Cheng. 2022. "Noncoding RNome as Enabling Biomarkers for Precision Health" International Journal of Molecular Sciences 23, no. 18: 10390. https://doi.org/10.3390/ijms231810390
APA StyleCheong, J. K., Rajgor, D., Lv, Y., Chung, K. Y., Tang, Y. C., & Cheng, H. (2022). Noncoding RNome as Enabling Biomarkers for Precision Health. International Journal of Molecular Sciences, 23(18), 10390. https://doi.org/10.3390/ijms231810390