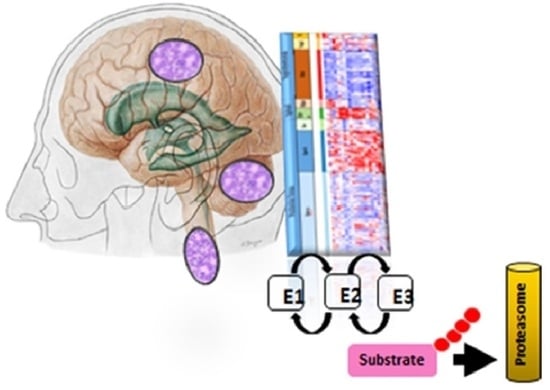
Ubiquitin Proteasome Gene Signatures in Ependymoma Molecular Subtypes
Abstract
:1. Introduction
2. Methods
3. Results
3.1. E1 Ubiquitin Activator Gene Expression in EPN Subtypes
3.2. Ubiquitin Conjugase (E2) Gene Expression in EPN Subtypes
3.3. Ubiquitin E2 Conjugases and Patient Survival Times
3.4. Ubiquitin E3 Ligase Gene Expression in EPN Subtypes
3.5. Differentiation and DNA Repair Factors Are UPS Targets in Selected EPN Subtypes
3.6. Ubiquitin Ligase E3 Adaptor Gene Expression in EPN Subtypes
3.7. Ubiquitin E3 Ligase Adaptors and Viral Markers
3.8. Anaphase Promoting Complex/Cyclosome (APC/c) E3 Ligase Adaptors
4. Discussion
4.1. Setting the UPS Stage: Ubiquitin E1 Activator
4.2. Immune Recognition and Inflammation
4.3. UPS and NOTCH Signaling
4.4. The Anaphase Promoting Complex/Cyclosome (APC/c)
4.5. Neddylation
4.6. Linking UPS with Genomic Stability and DNA Repair in EPN Subtypes
4.7. Developmental Factors in EPN Subtypes
4.8. Summary Table
4.9. Current Targeting Approach for the UPS Pathway
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, T.; Plutynski, A.; Ward, S.; Rubin, J.B. An integrative view on sex differences in brain tumors. Cell Mol. Life Sci. 2015, 72, 3323–3342. [Google Scholar] [CrossRef] [Green Version]
- Taylor, M.D.; Poppleton, H.; Fuller, C.; Su, X.; Liu, Y.; Jensen, P.; Magdaleno, S.; Dalton, J.; Calabrese, C.; Board, J.; et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell 2005, 8, 323–335. [Google Scholar] [PubMed] [Green Version]
- Ouni, I.; Flick, K.; Kaiser, P. Ubiquitin and transcription: The SCF/Met4 pathway, a (protein-) complex issue. Transcription 2011, 2, 135–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pajtler, K.W.; Witt, H.; Sill, M.; Jones, D.T.; Hovestadt, V.; Kratochwil, F.; Wani, K.; Tatevossian, R.; Punchihewa, C.; Johann, P.; et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 2015, 27, 728–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barghout, S.H.; Schimmer, A.D. E1 Enzymes as Therapeutic Targets in Cancer. Pharmacol. Rev. 2021, 73, 1–58. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, Y.; Luo, Q.; Li, L.; Jia, L. Neddylation: A novel modulator of the tumor microenvironment. Mol. Cancer 2019, 18, 77. [Google Scholar] [CrossRef] [Green Version]
- Walden, H.; Podgorski, M.S.; Huang, D.T.; Miller, D.W.; Howard, R.J.; Minor, D.L., Jr.; Holton, J.M.; Schulman, B.A. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell 2003, 12, 1427–1437. [Google Scholar] [CrossRef]
- Baek, K.; Scott, D.C.; Schulman, B.A. NEDD8 and ubiquitin ligation by cullin-RING E3 ligases. Curr. Opin. Struct. Biol. 2021, 67, 101–109. [Google Scholar] [CrossRef]
- Groettrup, M.; Pelzer, C.; Schmidtke, G.; Hofmann, K. Activating the ubiquitin family: UBA6 challenges the field. Trends Biochem. Sci. 2008, 33, 230–237. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Yuan, B.; Zhu, W.; Zhang, R.; Li, L.; Hao, X.; Chen, S.; Hou, F. Ube2D3 and Ube2N are essential for RIG-I-mediated MAVS aggregation in antiviral innate immunity. Nat. Commun. 2017, 8, 15138. [Google Scholar] [CrossRef]
- Cadena, C.; Ahmad, S.; Xavier, A.; Willemsen, J.; Park, S.; Park, J.W.; Oh, S.W.; Fujita, F.; Hou, F.; Binder, M.; et al. Ubiquitin-Dependent and -Independent Roles of E3 Ligase RIPLET in Innate Immunity. Cell 2019, 177, 1187–1200.e16. [Google Scholar] [CrossRef]
- Rice, G.I.; Duany, Y.D.T.; Jenkinson, E.M.; Forte, G.M.; Anderson, B.H.; Ariaudo, G.; Bader-Meunier, B.; Baildam, E.M.; Battini, R.; Beresford, M.W.; et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 2014, 46, 503–509. [Google Scholar] [CrossRef]
- Lin, Y.; Hwang, W.C.; Basavappa, R. Structural and functional analysis of the human mitotic-specific ubiquitin-conjugating enzyme, UbcH10. J. Biol. Chem. 2002, 277, 21913–21921. [Google Scholar] [CrossRef] [Green Version]
- Garnett, M.J.; Mansfeld, J.; Godwin, C.; Matsusaka, T.; Wu, J.; Russell, P.; Pines, J.; Venkitaraman, A.R. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 2009, 11, 1363–1369. [Google Scholar] [CrossRef] [Green Version]
- Watson, E.R.; Brown, N.G.; Peters, J.M.; Stark, H.; Schulman, B.A. Posing the APC/C E3 Ubiquitin Ligase to Orchestrate Cell Division. Trends Cell Biol. 2019, 29, 117–134. [Google Scholar] [CrossRef]
- Martinez-Chacin, R.C.; Bodrug, T.; Bolhuis, D.L.; Kedziora, K.M.; Bonacci, T.; Ordureau, A.; Gibbs, M.E.; Weissmann, F.; Qiao, R.; Grant, G.D.; et al. Ubiquitin chain-elongating enzyme UBE2S activates the RING E3 ligase APC/C for substrate priming. Nat. Struct. Mol. Biol. 2020, 27, 550–560. [Google Scholar] [CrossRef]
- Delcuve, G.P.; Khan, D.H.; Davie, J.R. Roles of histone deacetylases in epigenetic regulation: Emerging paradigms from studies with inhibitors. Clin. Epigenetics 2012, 4, 5. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Tang, J.; Xu, C.; Zhao, H.; Zhou, Y.; Wang, Y.; Yang, M.; Chen, X.; Chen, J. Histone deacetylase 4 inhibits NF-kappaB activation by facilitating IkappaBalpha sumoylation. J. Mol. Cell Biol. 2020, 12, 933–945. [Google Scholar] [CrossRef]
- Zhao, X.; Sternsdorf, T.; Bolger, T.A.; Evans, R.M.; Yao, T.P. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol. Cell Biol. 2005, 25, 8456–8464. [Google Scholar] [CrossRef] [Green Version]
- Packer, J.R.; Hirst, A.M.; Droop, A.P.; Adamson, R.; Simms, M.S.; Mann, V.M.; Frame, F.M.; O’Connell, D.; Maitland, N.J. Notch signalling is a potential resistance mechanism of progenitor cells within patient-derived prostate cultures following ROS-inducing treatments. FEBS Lett. 2020, 594, 209–226. [Google Scholar] [CrossRef]
- Meier-Stiegen, F.; Schwanbeck, R.; Bernoth, K.; Martini, S.; Hieronymus, T.; Ruau, D.; Zenke, M.; Just, J. Activated Notch1 target genes during embryonic cell differentiation depend on the cellular context and include lineage determinants and inhibitors. PLoS ONE 2010, 5, e11481. [Google Scholar] [CrossRef] [Green Version]
- Magalhaes, T.d.A.; Cruzeiro, G.A.V.; de Sousa, G.R.; da Silva, K.R.; Lira, R.C.P.; Scrideli, C.A.; Tone, L.G.; Valera, E.T.; Borges, K.S. Notch pathway in ependymoma RELA-fused subgroup: Upregulation and association with cancer stem cells markers expression. Cancer Gene Ther. 2020, 27, 509–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiefel, H.; Bondong, S.; Hazin, J.; Ridinger, J.; Schirmer, U.; Riedle, S.; Altevogt, P. L1CAM: A major driver for tumor cell invasion and motility. Cell Adh. Migr. 2012, 6, 374–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larrew, T.; Saway, B.F.; Lowe, S.R.; Olar, A. Molecular Classification and Therapeutic Targets in Ependymoma. Cancers 2021, 13, 6218. [Google Scholar] [CrossRef] [PubMed]
- Schafer, M.K.; Schmitz, B.; Diestel, S. L1CAM ubiquitination facilitates its lysosomal degradation. FEBS Lett. 2010, 584, 4475–4480. [Google Scholar] [CrossRef] [Green Version]
- Heliot, C.; Desgrange, A.; Buisson, I.; Prunskaite-Hyyrylainen, R.; Shan, J.; Vainio, S.; Umbhauer, M.; Cereghini, S. HNF1B controls proximal-intermediate nephron segment identity in vertebrates by regulating Notch signalling components and Irx1/2. Development 2013, 140, 873–885. [Google Scholar] [CrossRef] [Green Version]
- Yahoo, N.; Pournasr, B.; Rostamzadeh, J.; Hakhamaneshi, M.S.; Ebadifar, A.; Fathi, F.; Badarvand, H. Forced expression of Hnf1b/Foxa3 promotes hepatic fate of embryonic stem cells. Biochem. Biophys. Res. Commun. 2016, 474, 199–205. [Google Scholar] [CrossRef] [Green Version]
- Lopes-Coelho, F.; Silva, F.; Hipolito, A.; Cardoso, B.A.; Serpa, J. Acetylation drives hepatocyte nuclear factor 1beta stability by blocking proteasome-mediated degradation. J. Cell Biochem. 2019, 120, 9337–9344. [Google Scholar] [CrossRef]
- Zhu, B.; Zheng, Y.; Pham, A.D.; Mandal, S.S.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell 2005, 20, 601–611. [Google Scholar] [CrossRef]
- Barton, V.N.; Donson, A.M.; Kleinschmidt-DeMasters, B.K.; Birks, D.K.; Handler, M.H.; Foreman, N.K. Unique molecular characteristics of pediatric myxopapillary ependymoma. Brain Pathol. 2010, 20, 560–570. [Google Scholar] [CrossRef]
- Balastik, M.; Ferraguti, F.; Pires-da Silva, A.; Lee, T.H.; Alvarez-Bolado, G.; Lu, K.P.; Gruss, P. Deficiency in ubiquitin ligase TRIM2 causes accumulation of neurofilament light chain and neurodegeneration. Proc. Natl. Acad. Sci. USA 2008, 105, 12016–12021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ylikallio, E.; Poyhonen, R.; Zimon, M.; De Vriendt, E.; Hilander, T.; Paetau, A.; Jordanova, A.; Lönnqvist, T.; Tyynismaa, H. Deficiency of the E3 ubiquitin ligase TRIM2 in early-onset axonal neuropathy. Hum. Mol. Genet. 2013, 22, 2975–2983. [Google Scholar] [CrossRef] [Green Version]
- Okumura, F.; Joo-Okumura, A.; Obara, K.; Petersen, A.; Nishikimi, A.; Fukui, Y.; Nakasukasa, K.; Kamura, T. Ubiquitin ligase SPSB4 diminishes cell repulsive responses mediated by EphB2. Mol. Biol. Cell 2017, 28, 3532–3541. [Google Scholar] [CrossRef]
- Glass, M.; Misiak, D.; Bley, N.; Muller, S.; Hagemann, S.; Busch, B.; Rausch, A.; Hüttelmaier, S. IGF2BP1, a Conserved Regulator of RNA Turnover in Cancer. Front. Mol. Biosci. 2021, 8, 632219. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.T.; Yu, H.Q.; Fang, L.; Tan, Y.; Liu, Z.Y.; Wu, D.; Zhang, J.; Xiong, H.J.; Xie, C.M. Elevated FBXO45 promotes liver tumorigenesis through enhancing IGF2BP1 ubiquitination and subsequent PLK1 upregulation. eLife 2021, 10, e70715. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Wang, Z.W.; Zhu, X. FBXO45 is a potential therapeutic target for cancer therapy. Cell Death Discov. 2020, 6, 55. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Feng, X.H.; Schwartz, R.J. SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J. Biol. Chem. 2004, 279, 49091–49098. [Google Scholar] [CrossRef] [Green Version]
- Kolas, N.K.; Chapman, J.R.; Nakada, S.; Ylanko, J.; Chahwan, R.; Sweeney, F.D.; Panier, S.; Mendez, M.; Wildenhain, J.; Thomson, T.M.; et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 2007, 318, 1637–1640. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.R.; Wang, Y.; Shen, Z.J.; Bennett, A.; Hindi, I.; Tyler, J.K.; Sleckman, B.P. The RNF8 and RNF168 Ubiquitin Ligases Regulate Pro- and Anti-Resection Activities at Broken DNA Ends During Non-Homologous End Joining. DNA Repair 2021, 108, 103217. [Google Scholar] [CrossRef]
- Tateishi, K.; Omata, M.; Tanaka, K.; Chiba, T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 2001, 155, 571–579. [Google Scholar] [CrossRef]
- Li, Z.; Cui, Q.; Wang, X.; Li, B.; Zhao, D.; Xia, Q.; Zhao, P. Functions and substrates of NEDDylation during cell cycle in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2017, 90, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.C.; Guo, Y.J.; Wang, B.; Wang, C.; Mamun, M.A.A.; Gao, Y.; Liu, H.M. Targeting neddylation E2s: A novel therapeutic strategy in cancer. J. Hematol. Oncol. 2021, 14, 57. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.Y.; Hwang, B.J.; Ford, J.M.; Hanawalt, P.C.; Chu, G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell 2000, 5, 737–744. [Google Scholar] [CrossRef] [Green Version]
- Inoki, T.; Yamagami, S.; Inoki, Y.; Tsuru, T.; Hamamoto, T.; Kagawa, Y.; Mori, T.; Endo, H. Human DDB2 splicing variants are dominant negative inhibitors of UV-damaged DNA repair. Biochem. Biophys. Res. Commun. 2004, 314, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-Silva, C.; Sabatella, M.; Helfricht, A.; Marteijn, J.A.; Theil, A.F.; Vermeulen, W.; Lans, H. Ubiquitin and TFIIH-stimulated DDB2 dissociation drives DNA damage handover in nucleotide excision repair. Nat. Commun. 2020, 11, 4868. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Stock, C.; Bommelje, C.C.; Weeda, V.B.; Shah, K.; Bains, S.; Buss, E.; Shaha, M.; Rechler, W.; Ramanathan, S.Y.; et al. SCCRO3 (DCUN1D3) antagonizes the neddylation and oncogenic activity of SCCRO (DCUN1D1). J. Biol. Chem. 2014, 289, 34728–34742. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Huang, J.; Shi, T.; Hu, F.; Zhang, L.; Zhou, P.K.; Ma, D.; Ma, T.; Qiu, X. DCUN1D3 activates SCFSKP2 ubiquitin E3 ligase activity and cell cycle progression under UV damage. Oncotarget 2016, 7, 58483–58491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Neilsen, P.M.; Crawford, J.; McKirdy, R.; Lee, J.; Powell, J.A.; Saif, Z.; Martin, J.M.; Lombaerts, M.; Cornelisse, C.J.; et al. FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex. Cancer Res. 2005, 65, 11304–11313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.L.; Jiang, Y.; Wang, Y.H.; Chen, T.; He, H.J.; Liu, T.; Yang, T.; Yang, L.W.; Chen, J.; Song, Z.Q.; et al. FBXO31 promotes cell proliferation, metastasis and invasion in lung cancer. Am. J. Cancer Res. 2015, 5, 1814–1822. [Google Scholar]
- Li, Y.; Jin, K.; Bunker, E.; Zhang, X.; Luo, X.; Liu, X.; Hao, B. Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCF(FBXO31) ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2018, 115, 319–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santra, M.K.; Wajapeyee, N.; Green, M.R. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature 2009, 459, 722–725. [Google Scholar] [CrossRef] [PubMed]
- Cummings, C.M.; Bentley, C.A.; Perdue, S.A.; Baas, P.W.; Singer, J.D. The Cul3/Klhdc5 E3 ligase regulates p60/katanin and is required for normal mitosis in mammalian cells. J. Biol. Chem. 2009, 284, 11663–11675. [Google Scholar] [CrossRef] [Green Version]
- Naucler, C.S.; Geisler, J.; Vetvik, K. The emerging role of human cytomegalovirus infection in human carcinogenesis: A review of current evidence and potential therapeutic implications. Oncotarget 2019, 10, 4333–4347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flatt, J.W.; Greber, U.F. Viral mechanisms for docking and delivering at nuclear pore complexes. Semin. Cell Dev. Biol. 2017, 68, 59–71. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.; Au, S.; Pante, N. How viruses access the nucleus. Biochim. Biophys. Acta 2011, 1813, 1634–1645. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Liu, T. Evaluation of Oncogene NUP37 as a Potential Novel Biomarker in Breast Cancer. Front. Oncol. 2021, 11, 669655. [Google Scholar] [CrossRef]
- He, Y.; Li, J.; Shen, L.; Zhou, H.; Fei, W.; Zhang, G.; Li, Z.; Wang, F.; Wen, Y. Pan-cancer analysis reveals NUP37 as a prognostic biomarker correlated with the immunosuppressive microenvironment in glioma. Aging 2022, 14, 1033–1047. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhu, Y.; Ma, S.; Li, Y.; Liu, K.; Xu, S.; Li, X.; Li, L.; Hu, J.; Liu, Y. Bioinformatics analysis identifies DYNC1I1 as prognosis marker in male patients with liver hepatocellular carcinoma. PLoS ONE 2021, 16, e0258797. [Google Scholar] [CrossRef] [PubMed]
- Hyer, M.L.; Milhollen, M.A.; Ciavarri, J.; Fleming, P.; Traore, T.; Sappal, D.; Huck, J.; Shi, J.; Gavin, J.; Brownell, J.; et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med. 2018, 24, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Barghout, S.H.; Schimmer, A.D. The ubiquitin-activating enzyme, UBA1, as a novel therapeutic target for AML. Oncotarget 2018, 9, 34198–34199. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.W.; Toth, J.I.; da Silva, S.R.; Paiva, S.L.; Lukkarila, J.L.; Hurren, R.; Maclean, N.; Sukhai, M.A.; Bhattacharjee, R.N.; Goard, C.A.; et al. Mutations in UBA3 confer resistance to the NEDD8-activating enzyme inhibitor MLN4924 in human leukemic cells. PLoS ONE 2014, 9, e93530. [Google Scholar] [CrossRef]
- Shah, J.J.; Jakubowiak, A.J.; O’Connor, O.A.; Orlowski, R.Z.; Harvey, R.D.; Smith, M.R.; Lebovic, D.; Diefenbach, C.; Kelly, K.; Hua, Z.; et al. Phase I Study of the Novel Investigational NEDD8-Activating Enzyme Inhibitor Pevonedistat (MLN4924) in Patients with Relapsed/Refractory Multiple Myeloma or Lymphoma. Clin. Cancer Res. 2016, 22, 34–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatia, S.; Pavlick, A.C.; Boasberg, P.; Thompson, J.A.; Mulligan, G.; Pickard, M.D.; Faessel, H.; Dezube, B.J.; Hamid, O. A phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with metastatic melanoma. Investig. New Drugs 2016, 34, 439–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarantopoulos, J.; Shapiro, G.I.; Cohen, R.B.; Clark, J.W.; Kauh, J.S.; Weiss, G.J.; Cleary, J.M.; Mahalingam, D.; Pickard, M.D.; Faessel, H.M.; et al. Phase I Study of the Investigational NEDD8-Activating Enzyme Inhibitor Pevonedistat (TAK-924/MLN4924) in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2016, 22, 847–857. [Google Scholar] [CrossRef] [Green Version]
- Hua, W.; Li, C.; Yang, Z.; Li, L.; Jiang, Y.; Yu, G.; Zhu, W.; Liu, Z.; Duan, S.; Chu, Y.; et al. Suppression of glioblastoma by targeting the overactivated protein neddylation pathway. Neuro Oncol. 2015, 17, 1333–1343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Zhao, X.; Yang, Z.; Yang, R.; Chen, C.; Zhao, K.; Wang, W.; Ma, Y.; Zhang, Q.; Wang, X. Neddylation inhibition upregulates PD-L1 expression and enhances the efficacy of immune checkpoint blockade in glioblastoma. Int. J. Cancer 2019, 145, 763–774. [Google Scholar] [CrossRef]
- Hu, H.; Sun, S.C. Ubiquitin signaling in immune responses. Cell Res. 2016, 26, 457–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kloetzel, P.M. The proteasome and MHC class I antigen processing. Biochim. Biophys. Acta 2004, 1695, 225–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loureiro, J.; Ploegh, H.L. Antigen presentation and the ubiquitin-proteasome system in host-pathogen interactions. Adv. Immunol. 2006, 92, 225–305. [Google Scholar] [PubMed]
- Kim, K.I.; Giannakopoulos, N.V.; Virgin, H.W.; Zhang, D.E. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell Biol. 2004, 24, 9592–9600. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Li, C.; Li, W.; Chen, H.; Fu, Y.; Yi, Z. Increased UBE2L6 regulated by type 1 interferon as potential marker in TB. J. Cell Mol. Med. 2021, 25, 11232–11243. [Google Scholar] [CrossRef] [PubMed]
- Orfali, N.; Shan-Krauer, D.; O’Donovan, T.R.; Mongan, N.P.; Gudas, L.J.; Cahill, M.R.; Tschan, M.P.; McKenna, S.L. Inhibition of UBE2L6 attenuates ISGylation and impedes ATRA-induced differentiation of leukemic cells. Mol. Oncol. 2020, 14, 1297–1309. [Google Scholar] [CrossRef] [Green Version]
- Sijts, A.; Zaiss, D.; Kloetzel, P.M. The role of the ubiquitin-proteasome pathway in MHC class I antigen processing: Implications for vaccine design. Curr. Mol. Med. 2001, 1, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Shamu, C.E.; Flierman, D.; Ploegh, H.L.; Rapoport, T.A.; Chau, V. Polyubiquitination is required for US11-dependent movement of MHC class I heavy chain from endoplasmic reticulum into cytosol. Mol. Biol. Cell 2001, 12, 2546–2555. [Google Scholar] [CrossRef] [PubMed]
- Kikkert, M.; Hassink, G.; Barel, M.; Hirsch, C.; van der Wal, F.J.; Wiertz, E. Ubiquitination is essential for human cytomegalovirus US11-mediated dislocation of MHC class I molecules from the endoplasmic reticulum to the cytosol. Biochem. J. 2001, 358 Pt 2, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Ishido, S.; Goto, E.; Matsuki, Y.; Ohmura-Hoshino, M. E3 ubiquitin ligases for MHC molecules. Curr. Opin. Immunol. 2009, 21, 78–83. [Google Scholar] [CrossRef]
- Jia, X.; Zhao, C.; Zhao, W. Emerging Roles of MHC Class I Region-Encoded E3 Ubiquitin Ligases in Innate Immunity. Front. Immunol. 2021, 12, 687102. [Google Scholar] [CrossRef]
- Hatakeyama, S. TRIM proteins and cancer. Nat. Rev. Cancer 2011, 11, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Boehm, U.; Klamp, T.; Groot, M.; Howard, J.C. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 1997, 15, 749–795. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Mink, S.; Wong, K.A.; Stein, N.; Getman, C.; Dempsey, P.W.; Wu, H.; Shuai, K. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat. Immunol. 2004, 5, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Sarute, N.; Ibrahim, N.; Fagla, B.M.; Lavanya, M.; Cuevas, C.; Stavrou, S.; Otkiran-Clare, G.; Tyynismaa, H.; Henao-Mejia, J.; Ross, S.R. TRIM2, a novel member of the antiviral family, limits New World arenavirus entry. PLoS Biol. 2019, 17, e3000137. [Google Scholar] [CrossRef] [PubMed]
- Pagani, I.; Poli, G.; Vicenzi, E. TRIM22. A Multitasking Antiviral Factor. Cells 2021, 10, 1864. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Wei, L.; Yu, Z.B.; Yao, Z.Y.; Cheng, J.; Wang, Y.T.; Song, X.T.; Li, M. The Roles of TRIMs in Antiviral Innate Immune Signaling. Front. Cell Infect. Microbiol. 2021, 11, 628275. [Google Scholar] [CrossRef]
- Gao, B.; Xu, W.; Wang, Y.; Zhong, L.; Xiong, S. Induction of TRIM22 by IFN-gamma Involves JAK and PC-PLC/PKC, but Not MAPKs and pI3K/Akt/mTOR Pathways. J. Interferon Cytokine Res. 2013, 33, 578–587. [Google Scholar] [CrossRef] [Green Version]
- Gorman, J.A.; Hundhausen, C.; Errett, J.S.; Stone, A.E.; Allenspach, E.J.; Ge, Y.; Arkatkar, T.; Clough, C.; Dai, X.; Khim, S.; et al. The A946T variant of the RNA sensor IFIH1 mediates an interferon program that limits viral infection but increases the risk for autoimmunity. Nat. Immunol. 2017, 18, 744–752. [Google Scholar] [CrossRef]
- Lu, Y.; Stuart, J.H.; Talbot-Cooper, C.; Agrawal-Singh, S.; Huntly, B.; Smid, A.I.; Snowden, J.S.; Dupont, L.; Smith, G.L. Histone deacetylase 4 promotes type I interferon signaling, restricts DNA viruses, and is degraded via vaccinia virus protein C6. Proc. Natl. Acad. Sci. USA 2019, 116, 11997–12006. [Google Scholar] [CrossRef] [Green Version]
- Perla, A.; Fratini, L.; Cardoso, P.S.; Nor, C.; Brunetto, A.T.; Brunetto, A.L.; Brunetto de Farias, C.; Jaeger, M.; Roesler, R. Histone Deacetylase Inhibitors in Pediatric Brain Cancers: Biological Activities and Therapeutic Potential. Front. Cell Dev. Biol. 2020, 8, 546. [Google Scholar] [CrossRef]
- Mielcarek, M.; Zielonka, D.; Carnemolla, A.; Marcinkowski, J.T.; Guidez, F. HDAC4 as a potential therapeutic target in neurodegenerative diseases: A summary of recent achievements. Front. Cell Neurosci. 2015, 9, 42. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Boucher, J.M.; Liaw, L. Histone deacetylase activity selectively regulates notch-mediated smooth muscle differentiation in human vascular cells. J. Am. Heart. Assoc. 2012, 1, e000901. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, P.; Adami, P.V.; Morelli, L. Notch signaling in the pathologic adult brain. Biomol. Concepts 2013, 4, 465–476. [Google Scholar] [CrossRef]
- Lobry, C.; Oh, P.; Mansour, M.R.; Look, A.T.; Aifantis, I. Notch signaling: Switching an oncogene to a tumor suppressor. Blood 2014, 123, 2451–2459. [Google Scholar] [CrossRef]
- Lim, J.S.; Ibaseta, A.; Fischer, M.M.; Cancilla, B.; O’Young, G.; Cristea, S.; Luca, V.C.; Yang, D.; Jahchan, N.S.; Hamard, C.; et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 2017, 545, 360–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bigas, A.; Porcheri, C. Notch and Stem Cells. Adv. Exp. Med. Biol. 2018, 1066, 235–263. [Google Scholar] [PubMed]
- Steinbuck, M.P.; Winandy, S. A Review of Notch Processing With New Insights Into Ligand-Independent Notch Signaling in T-Cells. Front. Immunol. 2018, 9, 1230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuwa, T.J.; Hori, K.; Sasamura, T.; Higgs, J.; Baron, M.; Matsuno, K. The first deltex null mutant indicates tissue-specific deltex-dependent Notch signaling in Drosophila. Mol. Genet. Genom. 2006, 275, 251–263. [Google Scholar] [CrossRef]
- Schnute, B.; Troost, T.; Klein, T. Endocytic Trafficking of the Notch Receptor. Adv. Exp. Med. Biol. 2018, 1066, 99–122. [Google Scholar] [PubMed]
- Antila, C.J.M.; Rraklli, V.; Blomster, H.A.; Dahlstrom, K.M.; Salminen, T.A.; Holmberg, J.; Sistonen, L.; Sahlgren, C. Sumoylation of Notch1 represses its target gene expression during cell stress. Cell Death Differ. 2018, 25, 600–615. [Google Scholar] [CrossRef] [Green Version]
- Mo, J.S.; Kim, M.Y.; Han, S.O.; Kim, I.S.; Ann, E.J.; Lee, K.S.; Seo, M.S.; Kim, J.Y.; Lee, S.C.; Park, J.W.; et al. Integrin-linked kinase controls Notch1 signaling by down-regulation of protein stability through Fbw7 ubiquitin ligase. Mol. Cell Biol. 2007, 27, 5565–5574. [Google Scholar] [CrossRef] [Green Version]
- Yeh, C.H.; Bellon, M.; Nicot, C. FBXW7: A critical tumor suppressor of human cancers. Mol. Cancer 2018, 17, 115. [Google Scholar] [CrossRef]
- Lutz, D.; Wolters-Eisfeld, G.; Joshi, G.; Djogo, N.; Jakovcevski, I.; Schachner, M.; Kleen, R. Generation and nuclear translocation of sumoylated transmembrane fragment of cell adhesion molecule L1. J. Biol. Chem. 2012, 287, 17161–17175. [Google Scholar] [CrossRef] [Green Version]
- VanGenderen, C.; Harkness, T.A.A.; Arnason, T.G. The role of Anaphase Promoting Complex activation, inhibition and substrates in cancer development and progression. Aging 2020, 12, 15818–15855. [Google Scholar] [CrossRef] [PubMed]
- Thornton, B.R.; Toczyski, D.P. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell Biol. 2003, 5, 1090–1094. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Kang, X.; Zhang, G.; Fang, F.; Du, Y.; Lv, H. High expression of UBE2C is associated with the aggressive progression and poor outcome of malignant glioma. Oncol. Lett. 2016, 11, 2300–2304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.; Powell, C.; Yao, M.; Wu, J.; Dong, Q. Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int. J. Biochem. Cell Biol. 2014, 47, 113–117. [Google Scholar] [CrossRef]
- Van Ree, J.H.; Jeganathan, K.B.; Malureanu, L.; van Deursen, J.M. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J. Cell Biol. 2010, 188, 83–100. [Google Scholar] [CrossRef] [Green Version]
- Dastsooz, H.; Cereda, M.; Donna, D.; Oliviero, S. A Comprehensive Bioinformatics Analysis of UBE2C in Cancers. Int. J. Mol. Sci. 2019, 20, 2228. [Google Scholar] [CrossRef] [Green Version]
- Pallante, P.; Berlingieri, M.T.; Troncone, G.; Kruhoffer, M.; Orntoft, T.F.; Viglietto, G.; Caleo, A.; Migliaccio, I.; Decaussin-Petrucci, M.; Santoro, M.; et al. UbcH10 overexpression may represent a marker of anaplastic thyroid carcinomas. Br. J. Cancer 2005, 93, 464–471. [Google Scholar] [CrossRef] [Green Version]
- Fujita, T.; Ikeda, H.; Taira, N.; Hatoh, S.; Naito, M.; Doihara, H. Overexpression of UbcH10 alternates the cell cycle profile and accelerate the tumor proliferation in colon cancer. BMC Cancer 2009, 9, 87. [Google Scholar] [CrossRef] [Green Version]
- Berlingieri, M.T.; Pallante, P.; Sboner, A.; Barbareschi, M.; Bianco, M.; Ferraro, A.; Mansueto, G.; Borbone, E.; Guerriero, E.; Troncone, G.; et al. UbcH10 is overexpressed in malignant breast carcinomas. Eur. J. Cancer 2007, 43, 2729–2735. [Google Scholar] [CrossRef]
- Perrotta, I.; Bruno, L.; Maltese, L.; Russo, E.; Donato, A.; Donato, G. Immunohistochemical analysis of the ubiquitin-conjugating enzyme UbcH10 in lung cancer: A useful tool for diagnosis and therapy. J. Histochem. Cytochem. 2012, 60, 359–365. [Google Scholar] [CrossRef]
- Jiang, L.; Huang, C.G.; Lu, Y.C.; Luo, C.; Hu, G.H.; Liu, H.M.; Chen, J.X.; Han, H.X. Expression of ubiquitin-conjugating enzyme E2C/UbcH10 in astrocytic tumors. Brain Res. 2008, 1201, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Vriend, J.; Nachtigal, M.W. Ubiquitin Proteasome Pathway Transcriptome in Epithelial Ovarian Cancer. Cancers 2021, 13, 2659. [Google Scholar] [CrossRef] [PubMed]
- Psyrri, A.; Kalogeras, K.T.; Kronenwett, R.; Wirtz, R.M.; Batistatou, A.; Bournakis, E.; Timotheadou, E.; Gogas, H.; Aravantinos, G.; Christodoulou, C.; et al. Prognostic significance of UBE2C mRNA expression in high-risk early breast cancer. A Hellenic Cooperative Oncology Group (HeCOG) Study. Ann. Oncol. 2012, 23, 1422–1427. [Google Scholar] [CrossRef] [PubMed]
- Donato, G.; Iofrida, G.; Lavano, A.; Volpentesta, G.; Signorelli, F.; Pallante, P.L.; Berlingieri, M.T.; Pierantoni, M.G.; Palmieri, D.; Coforti, F.; et al. Analysis of UbcH10 expression represents a useful tool for the diagnosis and therapy of astrocytic tumors. Clin. Neuropathol. 2008, 27, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.P.; Huang, N.C.; Jhuang, S.J.; Pan, H.B.; Peng, N.J.; Cheng, J.T.; Chen, C.F.; Chen, J.J.; Chan, T.H. Ubiquitin-conjugating enzyme UBE2C is highly expressed in breast microcalcification lesions. PLoS ONE 2014, 9, e93934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, H.; Luo, Y.; Ding, G.; Fu, Z. A Pan-Cancer Analysis of UBE2S in Tumorigenesis, Prognosis, Pathway, Immune Infiltration and Evasion, and Therapy Response from an Immune-Oncology Perspective. J. Oncol. 2022, 2022, 3982539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shi, C.C.; Zhang, H.P.; Li, G.Q.; Li, S.S. MLN4924 suppresses neddylation and induces cell cycle arrest, senescence, and apoptosis in human osteosarcoma. Oncotarget 2016, 7, 45263–45274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machida, Y.J.; Machida, Y.; Chen, Y.; Gurtan, A.M.; Kupfer, G.M.; D’Andrea, A.D.; Dutta, A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell 2006, 23, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Tan, Y.; Deng, H.; Shen, Z.; Liu, Y.; Wu, P.; Tan, C.; Jiang, Y. UBE2J2 promotes hepatocellular carcinoma cell epithelial-mesenchymal transition and invasion in vitro. Oncotarget 2017, 8, 71736–71749. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Lv, Y.; Zhang, Y.; Gao, H. Regulation of p53 level by UBE4B in breast cancer. PLoS ONE 2014, 9, e90154. [Google Scholar] [CrossRef]
- Topno, R.; Singh, I.; Kumar, M.; Agarwal, P. Integrated bioinformatic analysis identifies UBE2Q1 as a potential prognostic marker for high grade serous ovarian cancer. BMC Cancer 2021, 21, 220. [Google Scholar] [CrossRef] [PubMed]
- Essers, J.; Theil, A.F.; Baldeyron, C.; van Cappellen, W.A.; Houtsmuller, A.B.; Kanaar, R.; Vermeulen, W. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell Biol. 2005, 25, 9350–9359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehm, E.M.; Gildenberg, M.S.; Washington, M.T. The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes 2016, 39, 231–254. [Google Scholar]
- Du, X.; Song, H.; Shen, N.; Hua, R.; Yang, G. The Molecular Basis of Ubiquitin-Conjugating Enzymes (E2s) as a Potential Target for Cancer Therapy. Int. J. Mol. Sci. 2021, 22, 3440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Qin, Z.; Zhang, X.; Xiao, W. Roles of sequential ubiquitination of PCNA in DNA-damage tolerance. FEBS Lett. 2011, 585, 2786–2794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseini, S.M.; Okoye, I.; Chaleshtari, M.G.; Hazhirkarzar, B.; Mohamadnejad, J.; Azizi, G.; Hojjat-Farsangi, M.; Mohammadi, H.; Shotorbani, S.S.; Jadidi-Niar, F. E2 ubiquitin-conjugating enzymes in cancer: Implications for immunotherapeutic interventions. Clin. Chim. Acta 2019, 498, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Bhat, A.; Xiao, W. Regulation of nucleotide excision repair through ubiquitination. Acta Biochim. Biophys. Sin. 2011, 43, 919–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Jenkins, L.M.M.; Su, Y.P.; Nitiss, K.C.; Nitiss, J.L.; Pommier, Y. A conserved SUMO pathway repairs topoisomerase DNA-protein cross-links by engaging ubiquitin-mediated proteasomal degradation. Sci. Adv. 2020, 6, eaba6290. [Google Scholar] [CrossRef]
- Morris, J.R. SUMO in the mammalian response to DNA damage. Biochem. Soc. Trans. 2010, 38 Pt 1, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Galanty, Y.; Belotserkovskaya, R.; Coates, J.; Polo, S.; Miller, K.M.; Jackson, S.P. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 2009, 462, 935–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, R.T. SUMO: A history of modification. Mol. Cell 2005, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Agnihotri, S.; Wolf, A.; Munoz, D.M.; Smith, C.J.; Gajadhar, A.; Restrepo, A.; Clark, I.D.; Fuller, G.N.; Kesari, G.N.; Dirks, P.B.; et al. A GATA4-regulated tumor suppressor network represses formation of malignant human astrocytomas. J. Exp. Med. 2011, 208, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Schelpe, J.; Monte, D.; Dewitte, F.; Sixma, T.K.; Rucktooa, P. Structure of UBE2Z Enzyme Provides Functional Insight into Specificity in the FAT10 Protein Conjugation Machinery. J. Biol. Chem. 2016, 291, 630–639. [Google Scholar] [CrossRef] [Green Version]
- Morrow, J.K.; Lin, H.K.; Sun, S.C.; Zhang, S. Targeting ubiquitination for cancer therapies. Future Med. Chem. 2015, 7, 2333–2350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Jia, L. Neddylation Pathway as a Novel Anti-cancer Target: Mechanistic Investigation and Therapeutic Implication. Anticancer Agents Med. Chem. 2015, 15, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Song, Y. Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J. Hematol. Oncol. 2020, 13, 50. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, M.; Crews, C.M. PROteolysis TArgeting Chimeras (PROTACs)—Past, present and future. Drug Discov. Today Technol. 2019, 31, 15–27. [Google Scholar] [CrossRef]
- Mullard, A. Targeted protein degraders crowd into the clinic. Nat. Rev. Drug Discov. 2021, 20, 247–250. [Google Scholar] [CrossRef]
- Chi, J.J.; Li, H.; Zhou, Z.; Izquierdo-Ferrer, J.; Xue, Y.; Wavelet, C.M.; Schiltz, G.E.; Zhang, B.; Cristofanilli, M.; Lu, X.; et al. A novel strategy to block mitotic progression for targeted therapy. EBioMedicine 2019, 49, 40–54. [Google Scholar] [CrossRef]





















| Age/Subtype | 0–10 | 11–20 | 21–30 | 31–40 | 41–50 | 51–60 | 61–70 | Total |
|---|---|---|---|---|---|---|---|---|
| PF-EPN-A | 54 | 9 | 0 | 1 | 0 | 1 | 0 | 65 |
| PF-EPN-B | 0 | 9 | 12 | 7 | 5 | 3 | 0 | 36 |
| PF-SE | 1 | 0 | 0 | 1 | 1 | 5 | 3 | 11 |
| SP-EPN | 1 | 1 | 1 | 2 | 4 | 1 | 0 | 10 |
| SP-MPE | 1 | 2 | 2 | 1 | 0 | 2 | 0 | 8 |
| ST-EPN-RELA | 30 | 7 | 2 | 0 | 1 | 0 | 0 | 40 |
| ST-EPN-YAP1 | 5 | 0 | 1 | 0 | 0 | 1 | 0 | 7 |
| ST-SE | 0 | 1 | 2 | 2 | 2 | 0 | 1 | 8 |
| Gene Classification | Number of Reactome Pathways | Number of E2 Genes Associated with This Pathway | Genes | p Value for Group |
|---|---|---|---|---|
| Synthesis of active ubiquitin: roles of E1 and E2 enzymes | 1 | 17 | UBE2A, UBE2B, UBE2C, UBE2D1, UBE2D2, UBE2E1, UBE2E3, UBE2G1, UBE2G2, UBE2H, UBE2K, UBE2L3, UBE2Q2, UBE2S, UBE2T, UBE2W, UBE2Z | 2.84 × 10−39 |
| Protein ubiqutination | 3 | 20 | UBE2A, UBE2B, UBE2C, UBE2D1, UBE2D2, UBE2E1, UBE2E3, UBE2G1, UBE2G2, UBE2H, UBE2J2, UBE2K, UBE2L3, UBE2N, UBE2Q2, UBE2S, UBE2T, UBE2V2, UBE2W, UBE2Z | 2.84 × 10−39 |
| Antigen processing: ubiquitination and proteasome degradation | 2 | 25 | UBE2E1, UBE2E2, UBE2G1, UBE2A, UBE2G2, UBE2C, UBE2B, UBE2E3, UBE2H, UBE2K, UBE2Q1, UBE2Q2, UBE2D1, UBE2L6, UBE3B, UBE2D4, UBE2D2, UBE2V2, UBE2N, UBE2S, UBE2W, UBE2J2, UBE2Z, UBE2J1, UBE2L3 | 1.40 × 10−33 |
| DDX58/IFIH1 and induction of interferon-alpha/beta | 2 | 4 | UBE2D1, UBE2D2, UBE2K, UBE2L6 | 2.63 × 10−6 |
| IKK complex and RIP1 | 2 | 3 | UBE2D1, UBE2D2, UBE2N | 2.63 × 10−6 |
| APC/C:Cdc20 conversion | 22 | 4 | UBE2C, UBE2S, UBE2D1, UBE2E1 | 3.27 × 10−4 |
| Formation of incision complex in Global Genome nucleotide excision repair (GG-NER) | 1 | 3 | UBE2I, UBE2N, UBE2V2 | 1.41 × 10−4 |
| ISG15 antiviral mechanism | 1 | 3 | UBE2E1, UBE2L6, UBE2N | 6.52 × 10−4 |
| Gene Classification | Reactome Reactions | Number of Genes | Genes | p for Group |
|---|---|---|---|---|
| Transfer of ubiquitin | 4 | 25 | UBE2A, UBE2B, UBE2C, UBE2D1, UBE2D2, UBE2D4, UBE2E1, UBE2E2, UBE2E3, UBE2G1, UBE2G2, UBE2H, UBE2J1, UBE2J2, UBE2K, UBE2L3, UBE2L6, UBE2N, UBE2Q1, UBE2Q2, UBE2S, UBE2V2, UBE2W, UBE2Z, UBE3B | 2.89 × 10−39 |
| UBA1 conjugates ubiquitin to cytosolic E2 | 1 | 11 | UBE2C, UBE2D1, UBE2D2, UBE2E3, UBE2G1, UBE2G2, UBE2H, UBE2K, UBE2L3, UBE2Q2, UBE2S | 3.90 × 10−28 |
| UBA6 conjugates ubiquitin to cytosolic E2 | 1 | 7 | UBE2D1, UBE2D2, UBE2E3, UBE2G2, UBE2L3, UBE2S, UBE2Z | 3.90 × 10−28 |
| UBA1 conjugates ubiquitin to nuclear E2 | 1 | 10 | UBE2A, UBE2B, UBE2C, UBE2D2, UBE2E1, UBE2E3, UBE2L3, UBE2S, UBE2T, UBE2W | 3.22 × 10−23 |
| APC/c * related reactions | 37 | 4 | UBE2C, UBE2S, UBE2D1, UBE2E1 | 7.02 × 10−10 |
| PCNA related reactions | 5 | 3 | UBE2B (aka RAD6B), UBE2N, UBE2V2 | 2.09 × 10−5 |
| Reactome Term | No. of Reactome Pathways | No. of E3 Ligase Genes in Pathway | Genes | p for Pathway |
|---|---|---|---|---|
| Antigen processing: Ubiquitination & proteasome degradation | 1 | 13 | AREL1, DZIP3, HACE1, HECW2, HERC6, PJA1, RNF114, RNF19A, RNF34, SMURF2, TRIM71, TRIM9, UBE3D | 4.94 × 10−10 |
| Interferon gamma signaling | 1 | 5 | MID1, PIAS1, TRIM2, TRIM22, TRIM45 | 4.78 × 10−5 |
| SUMOlyation of intracellular receptors | 1 | 3 | HDAC4, PIAS1, PPARG | 3.03 × 10−4 |
| SUMOlyation of Ubiquitinylation proteins | 1 | 3 | MDM2, PIAS1, TRIM27 | 6.14 × 10−4 |
| Signaling by Notch | 1 | 3 | DTX1, DTX4, HDAC4 | 4.22 × 10−3 |
| Reaction Term | No. of Reactome Pathways | No. of E3 Ligase Genes in Pathway | Genes | Group p Value |
|---|---|---|---|---|
| 1.Interaction of E3, E2-Ub complex and substrate 2.Transfer of UB from E2 to substrate 3.Polyubiquitination of substrate 4. Release of E3 from substrate | 4 | 13 | AREL1, DZIP3, HACE1, HECW2, HERC6, PJA1, RNF114, RNF19A, RNF34, SMURF2, TRIM71, TRIM9, UBE3D | 4.32 × 10−11 |
| SUMOylation of MDM2 with SUMO1 | 1 | 3 | MDM2, PIAS1, TRIM27 | 2.81 × 10−6 |
| Expression of IFNG-stimulated genes | 1 | 4 | MID1, TRIM2, TRIM22, TRIM45 | 2.93 × 10−4 |
| Group | No. of Reactome Pathways | No. of E3 Adpator Genes in Pathway | Genes | p for Group |
|---|---|---|---|---|
| Neddylation | 1 | 12 | CISH, DCAF10, DCAF16, DCAF7, DDB2, FBXL13, FBXL14, FBXO15, FBXO31, GAN, KCTD6, KLHL42 | 3.46 × 10−11 |
| APC/c related pathways | 17 | 3 | ANAPC1, ANAPC5, ANAPC7 | 1.31 × 10−4 |
| Group | No. of Reactome Reactions | No. of Genes in Pathway | Genes | p for Group |
|---|---|---|---|---|
| HCMV nuclear pore docking | 1 | 4 | DYNC1I1, DYNC1I2, NUP37, RAE1 | 4.49 × 10−5 |
| Neddylation–CUL 4 adaptors | 3 | 4 | DCAF10, DCAF16, DCAF7, DDB2 | 3.56 × 10−5 |
| Neddylation–CUL1,CUL3,CUL5 adaptors | 8 | 8 | FBXL13, FBXL14, FBXO15, FBXO31, GAN, KCTD6, KLHL42, CISH | 1.71 × 10−9 |
| APC/c related pathways | 37 | 3 | ANAPC1, ANAPC5, ANAPC7 | 5.28 × 10−3 |
| UPS Component | Pathways | UPS Genes |
|---|---|---|
| Ubiquitin activators | Neddylation, | UBA3 |
| Activation of UBE2Z | UBA6 | |
| Ubiquitin conjugases | Antigen processing: ubiquitination and proteasome degradation | Many (Table 2) |
| APC/c complex and cell cycle | UBE2C, UBE2S | |
| DNA stability and repair | UBE2T | |
| SUMOlyation | UBE2I | |
| Neddylation | UBE2M, UBE2F | |
| * Fatylation | UBE2Z | |
| Ubiquitin ligases | Antigen processing: ubiquitination and proteasome degradation | Many (Table 4) |
| Cell cycle regulation | APC/c, CDC20 | |
| Induction (or inhibition) of interferon | HDAC4, PIAS1 | |
| SUMOlyation | PIAS1 | |
| Notch signaling | HDAC4, DTX1, DTX4 | |
| Regulation of Hox genes | RNF 20, RNF40 | |
| Ubiquitin ligase Adaptors | Neddylation | Many (Table 6) |
| APC/c related pathways | ANAPC1, ANAPC5, ANAPC7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vriend, J.; Thanasupawat, T.; Sinha, N.; Klonisch, T. Ubiquitin Proteasome Gene Signatures in Ependymoma Molecular Subtypes. Int. J. Mol. Sci. 2022, 23, 12330. https://doi.org/10.3390/ijms232012330
Vriend J, Thanasupawat T, Sinha N, Klonisch T. Ubiquitin Proteasome Gene Signatures in Ependymoma Molecular Subtypes. International Journal of Molecular Sciences. 2022; 23(20):12330. https://doi.org/10.3390/ijms232012330
Chicago/Turabian StyleVriend, Jerry, Thatchawan Thanasupawat, Namita Sinha, and Thomas Klonisch. 2022. "Ubiquitin Proteasome Gene Signatures in Ependymoma Molecular Subtypes" International Journal of Molecular Sciences 23, no. 20: 12330. https://doi.org/10.3390/ijms232012330
APA StyleVriend, J., Thanasupawat, T., Sinha, N., & Klonisch, T. (2022). Ubiquitin Proteasome Gene Signatures in Ependymoma Molecular Subtypes. International Journal of Molecular Sciences, 23(20), 12330. https://doi.org/10.3390/ijms232012330