7-Ketocholesterol-Induced Micro-RNA-107-5p Increases Number and Activity of Osteoclasts by Targeting MKP1
Abstract
:1. Introduction
2. Results
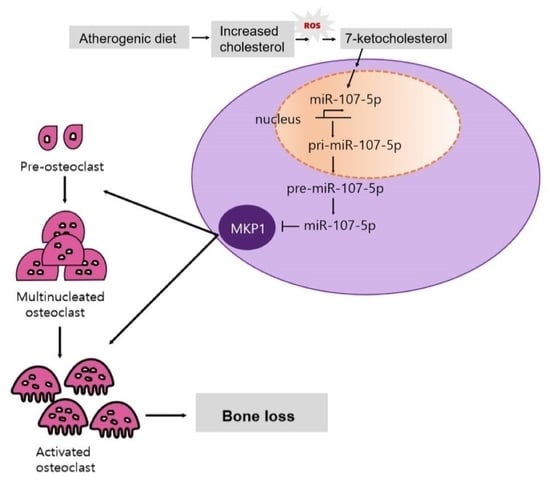
2.1. Atherogenic Diet Up-Regulates miR-107-5p
2.2. miR-107-5p Expression Regulates OC Differentiation
2.3. Identification of Target for 7-KC-Induced miR-107-5p in OCs
2.4. MiR-107-5p Specifically Targets MKP1 by Binding to 3′-UTRs of MKP1
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Reagents and Antibodies
4.3. Animals, Culture of Osteoblast and OC, and OC Formation
4.4. MiR and siRNA Transfection
4.5. RNA Isolation and Quantitative Polymerase Chain Reaction (qPCR)
4.6. Bone Resorption
4.7. Western Blot Analysis
4.8. Construction of 3′-UTR reporter of MKP1
4.9. Luciferase Assays
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Orozco, P. Atherogenic lipid profile and elevated lipoprotein (a) are associated with lower bone mineral density in early postmenopausal overweight women. Eur. J. Epidemiol. 2004, 19, 1105–1112. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Sugimoto, T.; Yano, S.; Yamauchi, M.; Sowa, H.; Chen, Q.; Chihara, K. Plasma Lipids and Osteoporosis in Postmenopausal Women. Endocr. J. 2002, 49, 211–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelton, K.; Krieder, J.; Joiner, D.; Freeman, M.R.; Goldstein, S.A.; Solomon, K.R. Hypercholesterolemia promotes an oste-oporotic phenotype. Am. J. Pathol. 2012, 181, 928–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, B.; Sundh, D.; Mellström, D.; Axelsson, K.; Nilsson, A.; Lorentzon, M. Association between cortical bone micro-structure and statin use in older women. J. Clin. Endocrinol. Metab. 2019, 104, 250–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oxlund, H.; Andreassen, T.T. Simvastatin treatment partially prevents ovariectomy-induced bone loss while increasing cortical bone formation. Bone 2004, 34, 609–618. [Google Scholar] [CrossRef]
- Sul, O.J.; Kim, J.E.; Ke, K.; Suh, J.H.; Choi, H.S. Atherogenic diet-induced bone loss is primarily due to increased osteo-clastogenesis in mice. J. Nutr. Biochem. 2020, 79, 108337. [Google Scholar] [CrossRef]
- Teitelbaum, S.L.; Ross, F.P. Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 2003, 4, 638–649. [Google Scholar] [CrossRef]
- Sul, O.-J.; Li, G.; Kim, J.-E.; Kim, E.-S.; Choi, H.-S. 7-ketocholesterol enhances autophagy via the ROS-TFEB signaling pathway in osteoclasts. J. Nutr. Biochem. 2021, 96, 108783. [Google Scholar] [CrossRef]
- Shalev, M.; Elson, A. The roles of protein tyrosine phosphatases in bone-resorbing osteoclasts. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 114–123. [Google Scholar] [CrossRef]
- Aoki, K.; Didomenico, E.; Sims, N.A.; Mukhopadhyay, K.; Neff, L.; Houghton, A.; Amling, M.; Levy, J.B.; Horne, W.C.; Baron, R. The tyrosine phosphatase SHP-1 is a negative regulator of osteoclastogenesis and osteoclast resorbing activity: Increased resorption and osteopenia in me(v)/me(v) mutant mice. Bone 1999, 25, 261–267. [Google Scholar] [CrossRef]
- Blüml, S.; Friedrich, M.; Lohmeyer, T.; Sahin, E.; Saferding, V.; Brunner, J.; Puchner, A.; Mandl, P.; Niederreiter, B.; Smolen, J.S.; et al. Loss of phosphatase and tensin homolog (PTEN) in myeloid cells controls inflammatory bone destruction by regulating the osteoclastogenic potential of myeloid cells. Ann. Rheum. Dis. 2013, 74, 227–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tonks, N.K. Protein tyrosine phosphatases—from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013, 280, 346–378. [Google Scholar] [CrossRef] [Green Version]
- Valerio, M.; Herbert, B.; Basilakos, D.; Browne, C.; Yu, H.; Kirkwood, K. Critical role of MKP-1 in lipopolysaccha-ride-induced osteoclast formation through CXCL1 and CXCL2. Cytokine 2015, 71, 71–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Valerio, M.S.; Kirkwood, K.L. MAPK Usage in Periodontal Disease Progression. J. Signal Transduct. 2012, 2012, 308943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, J.; Cui, W.; Zhang, Q.; Xu, X.; Mercan, F.; Bennett, A.; Vignery, A. Role of MKP-1 in osteoclasts and bone home-ostasis. Am. J. Pathol. 2009, 175, 1564–1573. [Google Scholar] [CrossRef] [Green Version]
- Ko, N.-Y.; Chen, L.-R.; Chen, K.-H. The Role of Micro RNA and Long-Non-Coding RNA in Osteoporosis. Int. J. Mol. Sci. 2020, 21, 4886. [Google Scholar] [CrossRef]
- Tang, P.; Xiong, Q.; Ge, W.; Zhang, L. The role of MicroRNAs in Osteoclasts and Osteoporosis. RNA Biol. 2014, 11, 1355–1363. [Google Scholar] [CrossRef] [Green Version]
- Mizoguchi, F.; Izu, Y.; Hayata, T.; Hemmi, H.; Nakashima, K.; Nakamura, T.; Kato, S.; Miyasaka, N.; Ezura, Y.; Noda, M. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J. Cell. Biochem. 2009, 109, 866–875. [Google Scholar] [CrossRef]
- Sugatani, T.; Hruska, K.A. Impaired Micro-RNA Pathways Diminish Osteoclast Differentiation and Function. J. Biol. Chem. 2009, 284, 4667–4678. [Google Scholar] [CrossRef] [Green Version]
- Ke, K.; Sul, O.-J.; Rajasekaran, M.; Choi, H.-S. MicroRNA-183 increases osteoclastogenesis by repressing heme oxygenase-1. Bone 2015, 81, 237–246. [Google Scholar] [CrossRef]
- Sugatani, T.; Vacher, J.; Hruska, K.A. A microRNA expression signature of osteoclastogenesis. Blood 2011, 117, 3648–3657. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cheng, P.; Xie, H.; Zhou, H.-D.; Wu, X.-P.; Liao, E.-Y.; Luo, X.-H. MiR-503 Regulates Osteoclastogenesis via Targeting RANK. J. Bone Miner. Res. 2014, 29, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Krzeszinskia, J.Y.; Wei, W.; Huynh, H.; Jin, Z.; Wang, X.; Chang, T.-C.; Xie, X.-J.; He, L.; Mangala, L.S.; Lopez-Berestein, G.; et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature 2014, 512, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Sul, O.-J.; Sung, Y.-B.; Rajasekaran, M.; Ke, K.; Yu, R.; Back, S.-H.; Choi, H.-S. MicroRNA-155 induces autophagy in osteoclasts by targeting transforming growth factor β-activated kinase 1-binding protein 2 upon lipopolysaccharide stimulation. Bone 2018, 116, 279–289. [Google Scholar] [CrossRef]
- Sul, O.-J.; Rajasekaran, M.; Park, H.-J.; Suh, J.-H.; Choi, H.-S. MicroRNA-29b Enhances Osteoclast Survival by Targeting BCL-2-Modifying Factor after Lipopolysaccharide Stimulation. Oxidative Med. Cell. Longev. 2019, 2019, 6018180. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.-h.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of Mammalian MicroRNA Targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; Da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Weigl, M.; Kocijan, R.; Ferguson, J.; Leinfellner, G.; Heimel, P.; Feichtinger, X.; Pietschmann, P.; Grillari, J.; Zwerina, J.; Redl, H.; et al. Longitudinal Changes of Circulating miRNAs During Bisphosphonate and Teriparatide Treatment in an Animal Model of Postmenopausal Osteoporosis. J. Bone Miner. Res. 2021, 36, 1131–1144. [Google Scholar] [CrossRef]
- Rottiers, V.; Näär, A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef]
- Kim, W.K.; Sul, O.J.; Choi, E.K.; Lee, M.H.; Jeong, C.S.; Kim, H.J.; Kim, S.Y.; Suh, J.H.; Yu, R.; Choi, H.S. Absence of Herpes virus entry mediator (HVEM) increases bone mass by attenuating receptor activator of nuclear factor-κB ligand (RANKL)-induced osteoclastogenesis. Endocrinology 2012, 153, 4808–4817. [Google Scholar] [CrossRef]
- Van Phan, T.; Sul, O.-J.; Ke, K.; Lee, M.-H.; Kim, W.-K.; Cho, Y.-S.; Kim, H.-J.; Kim, S.-Y.; Chung, H.-T.; Choi, H.-S. Carbon monoxide protects against ovariectomy-induced bone loss by inhibiting osteoclastogenesis. Biochem. Pharmacol. 2013, 85, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.-K.; Kim, W.-K.; Sul, O.-J.; Park, Y.-K.; Kim, E.-S.; Suh, J.-H.; Yu, R.; Choi, H.-S. TNFRSF14 deficiency protects against ovariectomy-induced adipose tissue inflammation. J. Endocrinol. 2014, 220, 25–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, E.-K.; Park, H.-J.; Sul, O.-J.; Rajasekaran, M.; Yu, R.; Choi, H.-S.; Rajesekaran, M. Carbon monoxide reverses adipose tissue inflammation and insulin resistance upon loss of ovarian function. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E621–E630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valerio, M.S.; Alexis, F.; Kirkwood, K.L. Functionalized nanoparticles containing MKP-1 agonists reduce periodontal bone loss. J. Periodontol. 2019, 90, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Ke, K.; Safder, M.; Sul, O.-J.; Kim, W.-K.; Suh, J.-H.; Joe, Y.; Chung, H.-T.; Choi, H.-S. Hemeoxygenase-1 maintains bone mass via attenuating a redox imbalance in osteoclast. Mol. Cell. Endocrinol. 2015, 409, 11–20. [Google Scholar] [CrossRef]
- Arai, F.; Miyamoto, T.; Ohneda, O.; Inada, T.; Sudo, T.; Brasel, K.; Miyata, T.; Anderson, D.M.; Suda, T. Commitment and differentiation of osteoclast precursor cells by the sequential expression of c-Fms and receptor activator of nuclear factor kappaB (RANK) receptors. J. Exp. Med. 1999, 190, 1741–1754. [Google Scholar] [CrossRef]
- Okayasu, M.; Nakayachi, M.; Hayashida, C.; Ito, J.; Kaneda, T.; Masuhara, M.; Suda, N.; Sato, T.; Hakeda, Y. Low-density lipoprotein receptor deficiency causes impaired osteoclastogenesis and increased bone mass in mice because of defect in oste-oclastic cell-cell fusion. J. Biol. Chem. 2012, 287, 19229–19241. [Google Scholar] [CrossRef] [Green Version]
- Park, H.J.; Son, H.-J.; Sul, O.-J.; Suh, J.-H.; Choi, H.-S. 4-Phenylbutyric acid protects against lipopolysaccharide-induced bone loss by modulating autophagy in osteoclasts. Biochem. Pharmacol. 2018, 151, 9–17. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Sul, O.-J.; Yu, R.; Choi, H.-S. 7-Ketocholesterol-Induced Micro-RNA-107-5p Increases Number and Activity of Osteoclasts by Targeting MKP1. Int. J. Mol. Sci. 2022, 23, 3697. https://doi.org/10.3390/ijms23073697
Li G, Sul O-J, Yu R, Choi H-S. 7-Ketocholesterol-Induced Micro-RNA-107-5p Increases Number and Activity of Osteoclasts by Targeting MKP1. International Journal of Molecular Sciences. 2022; 23(7):3697. https://doi.org/10.3390/ijms23073697
Chicago/Turabian StyleLi, Guoen, Ok-Joo Sul, Rina Yu, and Hye-Seon Choi. 2022. "7-Ketocholesterol-Induced Micro-RNA-107-5p Increases Number and Activity of Osteoclasts by Targeting MKP1" International Journal of Molecular Sciences 23, no. 7: 3697. https://doi.org/10.3390/ijms23073697
APA StyleLi, G., Sul, O. -J., Yu, R., & Choi, H. -S. (2022). 7-Ketocholesterol-Induced Micro-RNA-107-5p Increases Number and Activity of Osteoclasts by Targeting MKP1. International Journal of Molecular Sciences, 23(7), 3697. https://doi.org/10.3390/ijms23073697