New Insights into Hemopexin-Binding to Hemin and Hemoglobin
Abstract
:1. Introduction
2. Results
2.1. Identification of the Hemopexin-Hemin and Hemopexin-Hemoglobin Interaction Regions
2.2. Frequency of Amino Acids in Hemopexin-Hemin/Hemoglobin Binding Sites
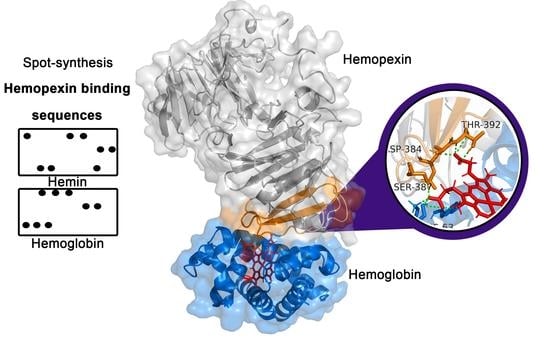
2.3. Molecular Docking Analysis of Hemopexin and Hemoglobin
3. Discussion
4. Materials and Methods
4.1. Spot-Synthesis
4.2. In Silico Analysis
4.3. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tolosano, E.; Altruda, F. Hemopexin: Structure, function, and regulation. DNA Cell Biol. 2002, 21, 297–306. [Google Scholar] [CrossRef]
- Delanghe, J.R.; Langlois, M.R. Hemopexin: A review of biological aspects and the role in laboratory medicine. Clin. Chim. Acta 2001, 312, 13–23. [Google Scholar] [CrossRef]
- Lim, S.K.; Ferraro, B.; Moore, K.; Halliwell, B. Role of haptoglobin in free hemoglobin metabolism. Redox Rep. 2001, 6, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Tolosano, E.; Fagoonee, S.; Morello, N.; Vinchi, F.; Fiorito, V. Heme scavenging and the other facets of hemopexin. Antioxid. Redox Signal. 2010, 12, 305–320. [Google Scholar] [CrossRef]
- Kumar, S.; Bandyopadhyay, U. Free heme toxicity and its detoxification systems in humans. Toxicol. Lett. 2005, 157, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Eldor, A.; Rachmilewitz, E.A. The hypercoagulable state in thalassemia. Blood 2002, 99, 36–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiabrando, D.; Vinchi, F.; Fiorito, V.; Mercurio, S.; Tolosano, E. Heme in pathophysiology: A matter of scavenging, metabolism, and trafficking across cell membranes. Front. Pharmacol. 2014, 5, 61. [Google Scholar] [CrossRef] [Green Version]
- Hahl, P.; Davis, T.; Washburn, C.; Rogers, J.T.; Smith, A. Mechanisms of neuroprotection by hemopexin: Modeling the control of heme and iron homeostasis in brain neurons in inflammatory states. J. Neurochem. 2013, 125, 89–101. [Google Scholar] [CrossRef] [Green Version]
- Vinchi, F.; Costa da Silva, M.; Ingoglia, G.; Petrillo, S.; Brinkman, N.; Zuercher, A.; Cerwenka, A.; Tolosano, E.; Muckenthaler, M.U. Hemopexin therapy reverts heme-induced proinflammatory phenotypic switching of macrophages in a mouse model of sickle cell disease. Blood 2016, 127, 473–486. [Google Scholar] [CrossRef]
- Ashraf, A.; Ashton, N.J.; Chatterjee, P.; Goozee, K.; Shen, K.; Fripp, J.; Ames, D.; Rowe, C.; Masters, C.L.; Villemagne, V.; et al. Plasma transferrin and hemopexin are associated with altered Aβ uptake and cognitive decline in Alzheimer’s disease pathology. Alzheimer’s Res. Ther. 2020, 12, 72. [Google Scholar] [CrossRef]
- Detzel, M.S.; Schmalohr, B.F.; Steinbock, F.; Hopp, M.T.; Ramoji, A.; Paul George, A.A.; Neugebauer, U.; Imhof, D. Revisiting the interaction of heme with hemopexin. Biol. Chem. 2021, 402, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Kassa, T.; Jana, S.; Meng, F.; Alayash, A.I. Differential heme release from various hemoglobin redox states and the upregulation of cellular heme oxygenase-1. FEBS Open Biol. 2016, 6, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Hrkal, Z.; Vodrázka, Z.; Kalousek, I. Transfer of heme from ferrihemoglobin and ferrihemoglobin isolated chains to hemopexin. Eur. J. Biochem. 1974, 43, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Immenschuh, S.; Vijayan, V.; Janciauskiene, S.; Gueler, F. Heme as a target for therapeutic interventions. Front. Pharmacol. 2017, 8, 146. [Google Scholar] [CrossRef] [Green Version]
- Belcher, J.D.; Chen, C.; Nguyen, J.; Abdulla, F.; Zhang, P.; Nguyen, H.; Nguyen, P.; Killeen, T.; Miescher, S.M.; Brinkman, N.; et al. Haptoglobin and hemopexin inhibit vaso-occlusion and inflammation in murine sickle cell disease: Role of heme oxygenase-1 induction. PLoS ONE 2018, 13, e0196455. [Google Scholar] [CrossRef] [Green Version]
- Paoli, M.; Anderson, B.F.; Baker, H.M.; Morgan, W.T.; Smith, A.; Baker, E.N. Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two beta-propeller domains. Nat. Struct. Biol. 1999, 6, 926–931. [Google Scholar] [CrossRef]
- Li, T.; Bonkovsky, H.L.; Guo, J. Structural analysis of heme proteins: Implications for design and prediction. BMC Struct. Biol. 2011, 11, 13. [Google Scholar] [CrossRef] [Green Version]
- Wißbrock, A.; George, A.A.P.; Brewitz, H.H.; Kühl, T.; Imhof, D. The molecular basis of transient heme-protein interactions: Analysis, concept, and implementation. Biosci. Rep. 2019, 39, BSR20181940. [Google Scholar] [CrossRef]
- Ruiz, E.F.; Cervantes, M.A. Diagnostic approach to hemolytic anemias in the adult. Rev. Bras. Hematol. Hemoter. 2015, 37, 423–425. [Google Scholar] [CrossRef] [Green Version]
- Jeong, W.J.; Bu, J.; Kubiatowicz, L.J.; Chen, S.S.; Kim, Y.; Hong, S. Peptide-nanoparticle conjugates: A next generation of diagnostic and therapeutic platforms? Nano Converg. 2018, 5, 38. [Google Scholar] [CrossRef]
- Canesin, G.; Di Ruscio, A.; Li, M.; Ummarino, S.; Hedblom, A.; Choudhury, R.; Krzyzanowska, A.; Csizmadia, E.; Palominos, M.; Stiehm, A.; et al. Scavenging of labile heme by hemopexin is a key checkpoint in cancer growth and metastases. Cell Rep. 2020, 32, 108181. [Google Scholar] [CrossRef] [PubMed]
- Larsen, R.; Gozzelino, R.; Jeney, V.; Tokaji, L.; Bozza, F.A.; Japiassú, A.M.; Bonaparte, D.; Cavalcante, M.M.; Chora, Â.; Ferreira, A.; et al. A central role for free heme in the pathogenesis of severe sepsis. Sci. Transl. Med. 2010, 2, 51ra71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graw, J.A.; Mayeur, C.; Rosales, I.; Liu, Y.; Sabbisetti, V.S.; Riley, F.E.; Rechester, O.; Malhotra, R.; Warren, H.S.; Colvin, R.B.; et al. Haptoglobin or hemopexin therapy prevents acute adverse effects of resuscitation after prolonged storage of red cells. Circulation 2016, 134, 945–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pukajło-Marczyk, A.; Zwolińska, D. Involvement of hemopexin in the pathogenesis of proteinuria in children with idiopathic nephrotic syndrome. J. Clin. Med. 2021, 10, 3160. [Google Scholar] [CrossRef]
- Hernández, C.; Garcia-Ramírez, M.; Simó, R. Overexpression of hemopexin in the diabetic eye: A new pathogenic candidate for diabetic macular edema. Diabetes Care 2013, 36, 2815–2821. [Google Scholar] [CrossRef] [Green Version]
- Spiller, F.; Costa, C.; Souto, F.O.; Vinchi, F.; Mestriner, F.L.; Laure, H.J.; Alves-Filho, J.C.; Freitas, A.; Rosa, J.C.; Ferreira, S.H.; et al. Inhibition of neutrophil migration by hemopexin leads to increased mortality due to sepsis in mice. Am. J. Respir. Crit. Care Med. 2011, 183, 922–931. [Google Scholar] [CrossRef]
- Chen-Roetling, J.; Ma, S.K.; Cao, Y.; Shah, A.; Regan, R.F. Hemopexin increases the neurotoxicity of hemoglobin when haptoglobin is absent. J. Neurochem. 2018, 145, 464–473. [Google Scholar] [CrossRef]
- Mikkelsen, J.H.; Runager, K.; Andersen, C.B.F. The human protein haptoglobin inhibits IsdH-mediated heme-sequestering by Staphylococcus aureus. J. Biol. Chem. 2020, 295, 1781–1791. [Google Scholar] [CrossRef]
- Watanabe, M.; Tanaka, Y.; Suenaga, A.; Kuroda, M.; Yao, M.; Watanabe, N.; Arisaka, F.; Tanaka, I.; Tsumoto, K. Structural basis for multimeric heme complexation through a specific protein-heme interaction: The case of the third neat domain of IsdH from Staphylococcus aureus. J. Biol. Chem. 2008, 283, 28649–28659. [Google Scholar] [CrossRef] [Green Version]
- Sakipov, S.; Rafikova, O.; Kurnikova, M.G.; Rafikov, R. Molecular mechanisms of bio-catalysis of heme extraction from hemoglobin. Redox. Biol. 2017, 11, 516–523. [Google Scholar] [CrossRef] [Green Version]
- Ponka, P. Cell biology of heme. Am. J. Med. Sci. 1999, 318, 241–256. [Google Scholar] [CrossRef]
- Lin, T.; Sammy, F.; Yang, H.; Thundivalappil, S.; Hellman, J.; Tracey, K.J.; Warren, H.S. Identification of hemopexin as an anti-inflammatory factor that inhibits synergy of hemoglobin with HMGB1 in sterile and infectious inflammation. J. Immunol. 2012, 189, 2017–2022. [Google Scholar] [CrossRef] [Green Version]
- De-Simone, S.G.; Gomes, L.R.; Napoleão-Pêgo, P.; Lechuga, G.C.; Pina, J.C.; Silva, F.R. Identification of linear B epitopes liable for the protective immunity of diphtheria toxin. Vaccines 2021, 9, 313. [Google Scholar] [CrossRef]
- Lechuga, G.C.; Souza-Silva, F.; Sacramento, C.Q.; Trugilho, M.R.O.; Valente, R.H.; Napoleão-Pêgo, P.; Dias, S.S.G.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Carels, N.; et al. SARS-CoV-2 proteins bind to hemoglobin and its metabolites. Int. J. Mol. Sci. 2021, 22, 9035. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [Green Version]




| Code | Interaction Site | Molecule | Position | Secondary Structure |
|---|---|---|---|---|
| H1 | KNFPSPVDAAFRQGH | Hemin | 91–105 | C + S |
| H2 | FRQGHNSVFL | Hemin | 101–110 | S |
| H3 | GWHSWPIAHQ | Hemin | 291–300 | C |
| H4 | CPGSSRLHIMAGRRL | Hemin | 366–380 | C + S |
| H5 | KSGAQATWTELPWPH | Hemin | 386–400 | C |
| H6 | NSCSANGPGLYLIHG | Hemin | 416–430 | C + S |
| H7 | LPQPQNVTSLLGCTH | Hemin | 448–462 | C |
| Hb1 | MARVLGAPVALGLWSLCWSL | Hemoglobin | 1–20 | H |
| Hb2 | FFQGDREWFWDLATG | Hemoglobin | 161–175 | C |
| Hb3 | LVLSALTSDNHGATYAFSGTHYWRLDTSRD | Hemoglobin | 261–290 | C |
| Hb4 | AFSWEEKLYLVQGTQ | Hemoglobin | 311–324 | C |
| Hb5 | RLHIMAGRRLWWLDLKSGAQATWTE | Hemoglobin | 371–395 | C |
| Atom | Donor | Residue | Chain | Atom | Acceptor | Residue | Chain | Distance (Å) |
|---|---|---|---|---|---|---|---|---|
| OG1 | THR | 87 | B-Hb | O | TRP | 393 | A-Hx | 2.986 |
| NZ | LYS | 95 | B-Hb | O | ALA | 391 | A-Hx | 2.767 |
| NZ | LYS | 95 | B-Hb | O | ALA | 389 | A-Hx | 2.897 |
| NZ | LYS | 59 | B-Hb | O | LYS | 386 | A-Hx | 2.781 |
| NE1 | TRP | 382 | A-Hx | OD2 | ASP | 73 | B-Hb | 2.885 |
| NH2 | ARG | 371 | A-Hx | OD1 | ASP | 73 | B-Hb | 2.643 |
| NH2 | ARG | 371 | A-Hx | O | GLY | 69 | B-Hb | 3.095 |
| NH1 | ARG | 371 | A-Hx | O | GLY | 69 | B-Hb | 3.008 |
| NZ | LYS | 66 | B-Hb | OG | SER | 370 | A-Hx | 2.785 |
| NZ | LYS | 65 | B-Hb | OG | SER | 369 | A-Hx | 2.850 |
| NZ | LYS | 65 | B-Hb | O | GLY | 368 | A-Hx | 2.914 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lechuga, G.C.; Napoleão-Pêgo, P.; Morel, C.M.; Provance, D.W.; De-Simone, S.G. New Insights into Hemopexin-Binding to Hemin and Hemoglobin. Int. J. Mol. Sci. 2022, 23, 3789. https://doi.org/10.3390/ijms23073789
Lechuga GC, Napoleão-Pêgo P, Morel CM, Provance DW, De-Simone SG. New Insights into Hemopexin-Binding to Hemin and Hemoglobin. International Journal of Molecular Sciences. 2022; 23(7):3789. https://doi.org/10.3390/ijms23073789
Chicago/Turabian StyleLechuga, Guilherme C., Paloma Napoleão-Pêgo, Carlos M. Morel, David W. Provance, and Salvatore G. De-Simone. 2022. "New Insights into Hemopexin-Binding to Hemin and Hemoglobin" International Journal of Molecular Sciences 23, no. 7: 3789. https://doi.org/10.3390/ijms23073789
APA StyleLechuga, G. C., Napoleão-Pêgo, P., Morel, C. M., Provance, D. W., & De-Simone, S. G. (2022). New Insights into Hemopexin-Binding to Hemin and Hemoglobin. International Journal of Molecular Sciences, 23(7), 3789. https://doi.org/10.3390/ijms23073789