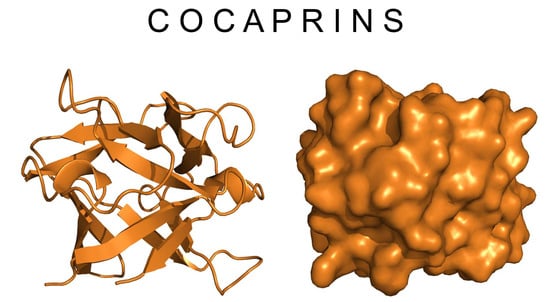Cocaprins, β-Trefoil Fold Inhibitors of Cysteine and Aspartic Proteases from Coprinopsis cinerea
Abstract
:1. Introduction
2. Results
2.1. Identification, Expression and Homologs of Cocaprins
2.2. Biochemical Characterization
2.3. Crystal Structure of Cocaprin 1
2.4. Cocaprins Are Cysteine Protease Inhibitors
2.5. Cocaprins Are Aspartic Protease Inhibitors
2.6. Aspartic and Cysteine Proteases Are Not Inhibited through the Same Inhibitory Reactive Site
2.7. Cocaprins Inhibit the Activity of Peptidases from C. cinerea Fruiting Bodies
2.8. Are Cocaprins Lectins?
3. Discussion
4. Materials and Methods
4.1. Enzymes, Substrates and Inhibitors
4.2. Cloning, Heterologous Expression and Purification of Recombinant Cocaprins
4.3. Mutagenesis
4.4. SDS-PAGE, Native-PAGE, and Isoelectric Focusing
4.5. Structure Solution and Refinement
4.6. Inhibition Assays and Determination of Kinetic Constants
4.7. Glycan Microarray Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sabotič, J.; Kos, J. Fungal Protease Inhibitors. In Fungal Metabolites; Mérillon, J.-M., Ramawat, K.G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 853–885. [Google Scholar]
- Sabotič, J.; Renko, M.; Kos, J. β-Trefoil Protease Inhibitors Unique to Higher Fungi. Acta Chim. Slov. 2019, 66, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Murzin, A.G.; Lesk, A.M.; Chothia, C. beta-Trefoil fold. Patterns of structure and sequence in the Kunitz inhibitors interleukins-1 beta and 1 alpha and fibroblast growth factors. J. Mol. Biol. 1992, 223, 531–543. [Google Scholar] [CrossRef]
- Renko, M.; Sabotič, J.; Turk, D. β-Trefoil inhibitors—From the work of Kunitz onward. Biol. Chem. 2012, 393, 1043. [Google Scholar] [CrossRef] [PubMed]
- Avanzo Caglič, P.; Renko, M.; Turk, D.; Kos, J.; Sabotič, J. Fungal beta-trefoil trypsin inhibitors cnispin and cospin demonstrate the plasticity of the beta-trefoil fold. Biochim. Biophys. Acta 2014, 1844, 1749–1756. [Google Scholar] [CrossRef]
- Sabotič, J.; Bleuler-Martinez, S.; Renko, M.; Avanzo Caglič, P.; Kallert, S.; Štrukelj, B.; Turk, D.; Aebi, M.; Kos, J.; Künzler, M. Structural basis of trypsin inhibition and entomotoxicity of cospin, serine protease inhibitor involved in defense of Coprinopsis cinerea fruiting bodies. J. Biol. Chem. 2012, 287, 3898–3907. [Google Scholar] [CrossRef] [Green Version]
- Renko, M.; Sabotič, J.; Mihelič, M.; Brzin, J.; Kos, J.; Turk, D. Versatile loops in mycocypins inhibit three protease families. J. Biol. Chem. 2010, 285, 308–316. [Google Scholar] [CrossRef] [Green Version]
- Sabotič, J.; Galeša, K.; Popovič, T.; Leonardi, A.; Brzin, J. Comparison of natural and recombinant clitocypins, the fungal cysteine protease inhibitors. Protein Expr. Purif. 2007, 53, 104–111. [Google Scholar] [CrossRef]
- Sabotič, J.; Popovič, T.; Puizdar, V.; Brzin, J. Macrocypins, a family of cysteine protease inhibitors from the basidiomycete Macrolepiota procera. FEBS J. 2009, 276, 4334–4345. [Google Scholar] [CrossRef]
- Plett, J.; Sabotič, J.; Vogt, E.; Snijders, F.; Kohler, A.; Nielsen, U.; Künzler, M.; Martin, F.; Veneault-Fourrey, C. Mycorrhiza-induced mycocypins of Laccaria bicolor are potent protease inhibitors with nematotoxic and collembola antifeedant activity. New Phytol. 2022; submitted. [Google Scholar]
- Guo, J.; Erskine, P.T.; Coker, A.R.; Wood, S.P.; Cooper, J.B. Structure of a Kunitz-type potato cathepsin D inhibitor. J. Struct. Biol. 2015, 192, 554–560. [Google Scholar] [CrossRef]
- Cater, S.A.; Lees, W.E.; Hill, J.; Brzin, J.; Kay, J.; Phylip, L.H. Aspartic proteinase inhibitors from tomato and potato are more potent against yeast proteinase A than cathepsin D. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 2002, 1596, 76–82. [Google Scholar] [CrossRef]
- Guerra, Y.; Valiente, P.A.; Berry, C.; Pons, T. Predicting functional residues of the Solanum lycopersicum aspartic protease inhibitor (SLAPI) by combining sequence and structural analysis with molecular docking. J. Mol. Model. 2012, 18, 2673–2687. [Google Scholar] [CrossRef] [PubMed]
- Kues, U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. MMBR 2000, 64, 316–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Künzler, M. How fungi defend themselves against microbial competitors and animal predators. PLoS Pathog. 2018, 14, e1007184. [Google Scholar] [CrossRef]
- Kombrink, A.; Tayyrov, A.; Essig, A.; Stockli, M.; Micheller, S.; Hintze, J.; van Heuvel, Y.; Durig, N.; Lin, C.W.; Kallio, P.T.; et al. Induction of antibacterial proteins and peptides in the coprophilous mushroom Coprinopsis cinerea in response to bacteria. ISME J. 2019, 13, 588–602. [Google Scholar] [CrossRef] [Green Version]
- Muraguchi, H.; Umezawa, K.; Niikura, M.; Yoshida, M.; Kozaki, T.; Ishii, K.; Sakai, K.; Shimizu, M.; Nakahori, K.; Sakamoto, Y.; et al. Strand-Specific RNA-Seq Analyses of Fruiting Body Development in Coprinopsis cinerea. PLoS ONE 2015, 10, e0141586. [Google Scholar] [CrossRef] [Green Version]
- Plaza, D.F.; Lin, C.W.; van der Velden, N.S.; Aebi, M.; Künzler, M. Comparative transcriptomics of the model mushroom Coprinopsis cinerea reveals tissue-specific armories and a conserved circuitry for sexual development. BMC Genom. 2014, 15, 492. [Google Scholar] [CrossRef] [Green Version]
- Tayyrov, A.; Stanley, C.E.; Azevedo, S.; Kunzler, M. Combining microfluidics and RNA-sequencing to assess the inducible defensome of a mushroom against nematodes. BMC Genom. 2019, 20, 243. [Google Scholar] [CrossRef] [Green Version]
- Bleuler-Martinez, S.; Stutz, K.; Sieber, R.; Collot, M.; Mallet, J.-M.; Hengartner, M.; Schubert, M.; Varrot, A.; Künzler, M. Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes. Glycobiology 2016, 27, 486–500. [Google Scholar] [CrossRef] [Green Version]
- Schubert, M.; Bleuler-Martinez, S.; Butschi, A.; Wälti, M.A.; Egloff, P.; Stutz, K.; Yan, S.; Wilson, I.B.H.; Hengartner, M.O.; Aebi, M.; et al. Plasticity of the β-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System. PLoS Pathog. 2012, 8, e1002706. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhong, Y.; Chang, J.; Kwan, H.S. Chromosome-level de novo assembly of Coprinopsis cinerea A43mut B43mut pab1–1 #326 and genetic variant identification of mutants using Nanopore MinION sequencing. Fungal Genet. Biol. FG B 2021, 146, 103485. [Google Scholar] [PubMed]
- Pohleven, J.; Renko, M.; Magister, Š.; Smith, D.F.; Künzler, M.; Štrukelj, B.; Turk, D.; Kos, J.; Sabotič, J. Bivalent Carbohydrate Binding Is Required for Biological Activity of Clitocybe nebularis Lectin (CNL), the N, N’-Diacetyllactosediamine (GalNAc beta 1–4GlcNAc, LacdiNAc)-specific Lectin from Basidiomycete C. nebularis. J. Biol. Chem. 2012, 287, 10602–10612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Žurga, S.; Pohleven, J.; Renko, M.; Bleuler-Martinez, S.; Sosnowski, P.; Turk, D.; Künzler, M.; Kos, J.; Sabotič, J. A novel β-trefoil lectin from the parasol mushroom (Macrolepiota procera) is nematotoxic. FEBS J. 2014, 281, 3489–3506. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Henderson, P.J.F. Linear Equation That Describes Steady-State Kinetics of Enzymes and Subcellular Particles Interacting with Tightly Bound Inhibitors. Biochem. J. 1972, 127, 321–333. [Google Scholar] [CrossRef] [Green Version]
- Sabotič, J.; Trček, T.; Popovič, T.; Brzin, J. Basidiomycetes harbour a hidden treasure of proteolytic diversity. J. Biotechnol. 2007, 128, 297–307. [Google Scholar] [CrossRef]
- Sabotič, J.; Ohm, R.A.; Künzler, M. Entomotoxic and nematotoxic lectins and protease inhibitors from fungal fruiting bodies. Appl. Microbiol. Biotechnol. 2016, 100, 91–111. [Google Scholar] [CrossRef] [Green Version]
- Gosavi, S.; Whitford, P.C.; Jennings, P.A.; Onuchic, J.N. Extracting function from a beta-trefoil folding motif. Proc. Natl. Acad. Sci. USA 2008, 105, 10384–10389. [Google Scholar] [CrossRef] [Green Version]
- Langstein-Skora, I.; Schmid, A.; Emenecker, R.J.; Richardson, M.O.G.; Götz, M.J.; Payer, S.K.; Korber, P.; Holehouse, A.S. Sequence- and chemical specificity define the functional landscape of intrinsically disordered regions. bioRxiv 2022. [Google Scholar] [CrossRef]
- Bleuler-Martínez, S.; Butschi, A.; Garbani, M.; Wälti, M.A.; Wohlschlager, T.; Potthoff, E.; Sabotič, J.; Pohleven, J.; Lüthy, P.; Hengartner, M.O.; et al. A lectin-mediated resistance of higher fungi against predators and parasites. Mol. Ecol. 2011, 20, 3056–3070. [Google Scholar] [CrossRef] [PubMed]
- Sabotič, J.; Koruza, K.; Gabor, B.; Peterka, M.; Barut, M.; Kos, J.; Brzin, J. The value of fungal protease inhibitors in affinity chromatography. In Affinity Chromatography; Magdeldin, S., Ed.; Intech: Rijeka, Croatia, 2012. [Google Scholar]
- Fleurkens, M.S. Carbohydrate-Based Vaccines against the Sheep Parasite Haemonchus Contortus. Ph.D. Thesis, ETH Zürich, Zurich, Switzerland, 2017. [Google Scholar]
- Künzler, M.; Bleuler-Martinez, S.; Butschi, A.; Garbani, M.; Lüthy, P.; Hengartner, M.O.; Aebi, M. Biotoxicity assays for fruiting body lectins and other cytoplasmic proteins. Methods Enzymol. 2010, 480, 141–150. [Google Scholar] [PubMed]
- Weiner, M.P.; Costa, G.L. Rapid PCR site-directed mutagenesis. PCR Methods Appl. 1994, 4, S131–S136. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Cryst. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grahn, E.; Askarieh, G.; Holmner, A.; Tateno, H.; Winter, H.C.; Goldstein, I.J.; Krengel, U. Crystal structure of the Marasmius oreades mushroom lectin in complex with a xenotransplantation epitope. J. Mol. Biol. 2007, 369, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Langer, G.G.; Hazledine, S.; Wiegels, T.; Carolan, C.; Lamzin, V.S. Visual automated macromolecular model building. Acta Cryst. D Biol. Cryst. 2013, 69, 635–641. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Cryst. D Biol. Cryst. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Cryst. D Biol. Cryst. 2011, 67, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.F.; Song, X.; Cummings, R.D. Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods Enzymol. 2010, 480, 417–444. [Google Scholar]
- Sabotič, J.; Kos, J. Microbial and fungal protease inhibitors—Current and potential applications. Appl. Microbiol. Biotechnol. 2012, 93, 1351–1375. [Google Scholar] [CrossRef]
- Garcia-Mesa, Y.; Jay, T.R.; Checkley, M.A.; Luttge, B.; Dobrowolski, C.; Valadkhan, S.; Landreth, G.E.; Karn, J.; Alvarez-Carbonell, D. Immortalization of primary microglia: A new platform to study HIV regulation in the central nervous system. J. Neurovirology 2017, 23, 47–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perišić Nanut, M.; Žurga, S.; Konjar, Š.; Prunk, M.; Kos, J.; Sabotič, J. The fungal Clitocybe nebularis lectin binds distinct cell surface glycoprotein receptors to induce cell death selectively in Jurkat cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2022, 36, e22215. [Google Scholar] [CrossRef] [PubMed]





| CCP1 | |
|---|---|
| Resolution range | 34.6–1.60 (1.66–1.60) |
| Space group | P 1 21 1 |
| Unit cell | 34.7 94.4 52.2 90 103.1 90 |
| Total reflections | 310,224 (19,426) |
| Unique reflections | 41,795 (3485) |
| Multiplicity | 7.4 (5.6) |
| Completeness (%) | 97.2 (81.7) |
| Mean I/sigma(I) | 26.4 (2.8) |
| Wilson B-factor | 12.22 |
| R-merge | 0.062 (0.58) |
| R-meas | 0.066 (0.64) |
| R-pim | 0.024 (0.2642) |
| CC1/2 | 0.999 (0.831) |
| CC* | 1 (0.953) |
| Reflections used in refinement | 39,704 (3484) |
| Reflections used for R-free | 2090 (174) |
| R-work | 0.163 (0.220) |
| R-free | 0.201 (0.222) |
| CC(work) | 0.965 (0.891) |
| CC(free) | 0.950 (0.902) |
| Number of non-hydrogen atoms | 2494 |
| macromolecules | 2111 |
| ligands | 0 |
| solvent | 383 |
| Protein residues | 272 |
| RMS(bonds) | 0.014 |
| RMS(angles) | 1.76 |
| Ramachandran favored (%) | 98.88 |
| Ramachandran allowed (%) | 1.12 |
| Ramachandran outliers (%) | 0.00 |
| Rotamer outliers (%) | 1.40 |
| Clashscore | 1.74 |
| Average B-factor | 15.59 |
| macromolecules | 13.45 |
| solvent | 27.40 |
| Protease | Protease Family | Source Organism | Inhibition (Ki [µM] or *IC50* [µM]) | |
|---|---|---|---|---|
| Cocaprin 1 | Cocaprin 2 | |||
| papain | C1 | Carica papaya | 5.63 ± 2.62 | 16.25 ± 2.49 |
| ficain | C1 | Ficus glabrata | 2.09 ± 0.18 | 1.19 ± 0.12 |
| cathepsin L | C1 | Homo sapiens | NI | NI |
| cathepsin H | C1 | Homo sapiens | NI | NI |
| legumain | C13 | Phaseolus vulgaris | NI | NI |
| pepsin | A1 | Sus scrofa | 0.86 ± 0.20 | 0.34 ± 0.11 |
| rennin | A1 | Bos taurus | *44.5* | *20.5* |
| rhizopuspepsin | A1 | Rhizopus sp. | NI | NI |
| PEP1 | H. contortus | NI | NI | |
| APR1 | H. contortus | *30.7* | *25.0* | |
| trypsin | S1 | Bos taurus | NI | NI |
| chymotrypsin | S1 | Bos taurus | NI | NI |
| thrombin | S1 | Bos taurus | NI | NI |
| elastase | S1 | Sus scrofa | NI | NI |
| kallikrein | S1 | Sus scrofa | NI | NI |
| subtilisin | S8 | Bacillus subtilis | NI | NI |
| Protease | Inhibition (Ki [µM]) | ||||
|---|---|---|---|---|---|
| WT | G13E | N22R | FH32EE | D47R | |
| Papain (C1) | 5.63 ± 2.62 | 5.22 ± 0.40 | 48.04 ± 11.99 | 7.90 ± 4.04 | 1.22 ± 0.32 |
| Pepsin (A1) | 0.86 ± 0.20 | 0.61 ± 0.37 | 0.33 ± 0.16 | 0.45 ± 0.22 | 1.83 ± 0.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renko, M.; Zupan, T.; Plaza, D.F.; Schmieder, S.S.; Perišić Nanut, M.; Kos, J.; Turk, D.; Künzler, M.; Sabotič, J. Cocaprins, β-Trefoil Fold Inhibitors of Cysteine and Aspartic Proteases from Coprinopsis cinerea. Int. J. Mol. Sci. 2022, 23, 4916. https://doi.org/10.3390/ijms23094916
Renko M, Zupan T, Plaza DF, Schmieder SS, Perišić Nanut M, Kos J, Turk D, Künzler M, Sabotič J. Cocaprins, β-Trefoil Fold Inhibitors of Cysteine and Aspartic Proteases from Coprinopsis cinerea. International Journal of Molecular Sciences. 2022; 23(9):4916. https://doi.org/10.3390/ijms23094916
Chicago/Turabian StyleRenko, Miha, Tanja Zupan, David F. Plaza, Stefanie S. Schmieder, Milica Perišić Nanut, Janko Kos, Dušan Turk, Markus Künzler, and Jerica Sabotič. 2022. "Cocaprins, β-Trefoil Fold Inhibitors of Cysteine and Aspartic Proteases from Coprinopsis cinerea" International Journal of Molecular Sciences 23, no. 9: 4916. https://doi.org/10.3390/ijms23094916
APA StyleRenko, M., Zupan, T., Plaza, D. F., Schmieder, S. S., Perišić Nanut, M., Kos, J., Turk, D., Künzler, M., & Sabotič, J. (2022). Cocaprins, β-Trefoil Fold Inhibitors of Cysteine and Aspartic Proteases from Coprinopsis cinerea. International Journal of Molecular Sciences, 23(9), 4916. https://doi.org/10.3390/ijms23094916