Development of a Prediction Model for Short-Term Remission of Patients with Crohn’s Disease Treated with Anti-TNF Drugs
Abstract
:1. Introduction
2. Results
2.1. Descriptive Study and Clinical Predictors of Non-Short-Term Remission
2.2. Proteomic Markers of Short-Term Remission
2.3. Validation of ENOA and VINC as Potential Markers of Short-Term Remission
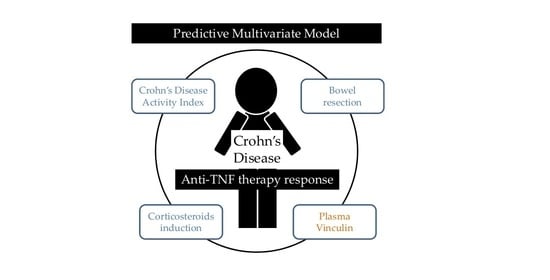
2.4. Prognostic Value of ENOA and VINC in CD Patients Undergoing Anti-TNF Therapy
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Study Population
4.3. Plasma Samples
4.4. Sample Preparation for LC-MS Analysis
4.5. Creation of the Spectral Library
4.6. Relative Quantification by SWATH Acquisition
4.7. SWATH Data Analysis
4.8. Functional Pathways Analysis
4.9. Enzyme-Linked Immunosorbent Assay
4.10. Statistical Analysis
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lungaro, L.; Costanzini, A.; Manza, F.; Barbalinardo, M.; Gentili, D.; Guarino, M.; Caputo, F.; Zoli, G.; De Giorgio, R.; Caio, G. Impact of Female Gender in Inflammatory Bowel Diseases: A Narrative Review. J. Pers. Med. 2023, 13, 165. [Google Scholar] [CrossRef]
- Caio, G.; Lungaro, L.; Caputo, F.; Zoli, E.; Giancola, F.; Chiarioni, G.; De Giorgio, R.; Zoli, G. Nutritional Treatment in Crohn’s Disease. Nutrients 2021, 13, 1628. [Google Scholar] [CrossRef]
- Danese, S.; Fiocchi, C. Etiopathogenesis of Inflammatory Bowel Diseases. World J. Gastroenterol. 2006, 12, 4807–4812. [Google Scholar] [CrossRef]
- Abraham, B.P.; Ahmed, T.; Ali, T. Inflammatory Bowel Disease: Pathophysiology and Current Therapeutic Approaches. In Gastrointestinal Pharmacology; Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2017; Volume 239, pp. 115–146. [Google Scholar] [CrossRef]
- Gomollón, F.; Dignass, A.; Annese, V.; Tilg, H.; Van Assche, G.; Lindsay, J.O.; Peyrin-Biroulet, L.; Cullen, G.J.; Daperno, M.; Kucharzik, T.; et al. 3rd European Evidence-Based Consensus on the Diagnosis and Management of Crohn’s Disease 2016: Part 1: Diagnosis and Medical Management. J. Crohn’s Colitis 2017, 11, 3–25. [Google Scholar] [CrossRef]
- Gisbert, J.P.; Panés, J. Loss of Response and Requirement of Infliximab Dose Intensification in Crohn’s Disease: A Review. Am. J. Gastroenterol. 2009, 104, 760–767. [Google Scholar] [CrossRef]
- Ben-Horin, S.; Kopylov, U.; Chowers, Y. Optimizing Anti-TNF Treatments in Inflammatory Bowel Disease. Autoimmun. Rev. 2014, 13, 24–30. [Google Scholar] [CrossRef]
- Stevens, T.W.; Matheeuwsen, M.; Lönnkvist, M.H.; Parker, C.E.; Wildenberg, M.E.; Gecse, K.B.; D’Haens, G.R. Systematic Review: Predictive Biomarkers of Therapeutic Response in Inflammatory Bowel Disease-Personalised Medicine in Its Infancy. Aliment. Pharm. 2018, 48, 1213–1231. [Google Scholar] [CrossRef]
- Na, S.-Y.; Moon, W. Perspectives on Current and Novel Treatments for Inflammatory Bowel Disease. Gut Liver 2019, 13, 604–616. [Google Scholar] [CrossRef]
- Gisbert, J.P.; Chaparro, M. Clinical Usefulness of Proteomics in Inflammatory Bowel Disease: A Comprehensive Review. J. Crohn’s Colitis 2019, 13, 374–384. [Google Scholar] [CrossRef]
- Gisbert, J.P.; Chaparro, M. Predictors of Primary Response to Biologic Treatment [Anti-TNF, Vedolizumab, and Ustekinumab] in Patients With Inflammatory Bowel Disease: From Basic Science to Clinical Practice. J. Crohn’s Colitis 2020, 14, 694–709. [Google Scholar] [CrossRef]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted Data Extraction of the MS/MS Spectra Generated by Data-Independent Acquisition: A New Concept for Consistent and Accurate Proteome Analysis. Mol. Cell Proteom. 2012, 11, O111.016717. [Google Scholar] [CrossRef]
- Kennedy, N.A.; Heap, G.A.; Green, H.D.; Hamilton, B.; Bewshea, C.; Walker, G.J.; Thomas, A.; Nice, R.; Perry, M.H.; Bouri, S.; et al. Predictors of Anti-TNF Treatment Failure in Anti-TNF-Naive Patients with Active Luminal Crohn’s Disease: A Prospective, Multicentre, Cohort Study. Lancet Gastroenterol. Hepatol. 2019, 4, 341–353. [Google Scholar] [CrossRef]
- Hirschmann, S.; Neurath, M.F. Top-down Approach to Biological Therapy of Crohn’s Disease. Expert. Opin. Biol. Ther. 2017, 17, 285–293. [Google Scholar] [CrossRef]
- Dvorakova, M.; Nenutil, R.; Bouchal, P. Transgelins, Cytoskeletal Proteins Implicated in Different Aspects of Cancer Development. Expert. Rev. Proteom. 2014, 11, 149–165. [Google Scholar] [CrossRef]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms Regulating Intestinal Barrier Integrity and Its Pathological Implications. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef]
- López-Posadas, R.; Stürzl, M.; Atreya, I.; Neurath, M.F.; Britzen-Laurent, N. Interplay of GTPases and Cytoskeleton in Cellular Barrier Defects during Gut Inflammation. Front. Immunol. 2017, 8, 1240. [Google Scholar] [CrossRef]
- Ma, T.Y.; Iwamoto, G.K.; Hoa, N.T.; Akotia, V.; Pedram, A.; Boivin, M.A.; Said, H.M. TNF-Alpha-Induced Increase in Intestinal Epithelial Tight Junction Permeability Requires NF-Kappa B Activation. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 286, G367–G376. [Google Scholar] [CrossRef]
- Maldonado-Contreras, A.; Birtley, J.R.; Boll, E.; Zhao, Y.; Mumy, K.L.; Toscano, J.; Ayehunie, S.; Reinecker, H.-C.; Stern, L.J.; McCormick, B.A. Shigella Depends on SepA to Destabilize the Intestinal Epithelial Integrity via Cofilin Activation. Gut. Microbes. 2017, 8, 544–560. [Google Scholar] [CrossRef]
- Choi, N.-R.; Lee, H.-J.; Jung, S.-H.; Hong, C.Y.; Vo, M.-C.; Hoang, M.-D.; Kim, H.-J.; Lee, J.-J. Generation of Potent Dendritic Cells with Improved Migration Ability through P-Cofilin and Sarco/Endoplasmic Reticulum Ca2+ Transport ATPase 2 Regulation. Cytotherapy 2015, 17, 1421–1433. [Google Scholar] [CrossRef]
- Ortega Moreno, L.; Fernández-Tomé, S.; Chaparro, M.; Marin, A.C.; Mora-Gutiérrez, I.; Santander, C.; Baldan-Martin, M.; Gisbert, J.P.; Bernardo, D. Profiling of Human Circulating Dendritic Cells and Monocyte Subsets Discriminates Between Type and Mucosal Status in Patients With Inflammatory Bowel Disease. Inflamm. Bowel. Dis. 2021, 27, 268–274. [Google Scholar] [CrossRef]
- Le Clainche, C.; Dwivedi, S.P.; Didry, D.; Carlier, M.-F. Vinculin Is a Dually Regulated Actin Filament Barbed End-Capping and Side-Binding Protein. J. Biol. Chem. 2010, 285, 23420–23432. [Google Scholar] [CrossRef]
- Dougherty, G.W.; Chopp, T.; Qi, S.-M.; Cutler, M.L. The Ras Suppressor Rsu-1 Binds to the LIM 5 Domain of the Adaptor Protein PINCH1 and Participates in Adhesion-Related Functions. Exp. Cell Res. 2005, 306, 168–179. [Google Scholar] [CrossRef]
- Moriggi, M.; Pastorelli, L.; Torretta, E.; Tontini, G.E.; Capitanio, D.; Bogetto, S.F.; Vecchi, M.; Gelfi, C. Contribution of Extracellular Matrix and Signal Mechanotransduction to Epithelial Cell Damage in Inflammatory Bowel Disease Patients: A Proteomic Study. Proteomics 2017, 17, 1700164. [Google Scholar] [CrossRef]
- Leffers, H.; Nielsen, M.S.; Andersen, A.H.; Honoré, B.; Madsen, P.; Vandekerckhove, J.; Celis, J.E. Identification of Two Human Rho GDP Dissociation Inhibitor Proteins Whose Overexpression Leads to Disruption of the Actin Cytoskeleton. Exp. Cell. Res. 1993, 209, 165–174. [Google Scholar] [CrossRef]
- Guo, Z.; Neilson, L.J.; Zhong, H.; Murray, P.S.; Zanivan, S.; Zaidel-Bar, R. E-Cadherin Interactome Complexity and Robustness Resolved by Quantitative Proteomics. Sci. Signal. 2014, 7, rs7. [Google Scholar] [CrossRef]
- Hirata, H.; Tatsumi, H.; Sokabe, M. Mechanical Forces Facilitate Actin Polymerization at Focal Adhesions in a Zyxin-Dependent Manner. J. Cell Sci. 2008, 121, 2795–2804. [Google Scholar] [CrossRef]
- Kotaka, M.; Kostin, S.; Ngai, S.; Chan, K.; Lau, Y.; Lee, S.M.Y.; Li, H.; Ng, E.K.O.; Schaper, J.; Tsui, S.K.W.; et al. Interaction of HCLIM1, an Enigma Family Protein, with α-Actinin 2. J. Cell Biochem. 2000, 78, 558–565. [Google Scholar] [CrossRef]
- Baig, A.; Bao, X.; Haslam, R.J. Proteomic Identification of Pleckstrin-Associated Proteins in Platelets: Possible Interactions with Actin. Proteomics 2009, 9, 4254–4258. [Google Scholar] [CrossRef]
- Shkoda, A.; Werner, T.; Daniel, H.; Gunckel, M.; Rogler, G.; Haller, D. Differential Protein Expression Profile in the Intestinal Epithelium from Patients with Inflammatory Bowel Disease. J. Proteome. Res. 2007, 6, 1114–1125. [Google Scholar] [CrossRef]
- Bearer, E.L.; Prakash, J.M.; Li, Z. Actin Dynamics in Platelets. Int. Rev. Cytol. 2002, 217, 137–182. [Google Scholar] [CrossRef]
- Meuwis, M.-A.; Fillet, M.; Lutteri, L.; Marée, R.; Geurts, P.; de Seny, D.; Malaise, M.; Chapelle, J.-P.; Wehenkel, L.; Belaiche, J.; et al. Proteomics for Prediction and Characterization of Response to Infliximab in Crohn’s Disease: A Pilot Study. Clin. Biochem. 2008, 41, 960–967. [Google Scholar] [CrossRef]
- Jenne, C.N.; Kubes, P. Platelets in Inflammation and Infection. Platelets 2015, 26, 286–292. [Google Scholar] [CrossRef]
- Nguyen, X.D.; Müller-Berghaus, J.; Kälsch, T.; Schadendorf, D.; Borggrefe, M.; Klüter, H. Differentiation of Monocyte-Derived Dendritic Cells under the Influence of Platelets. Cytotherapy 2008, 10, 720–729. [Google Scholar] [CrossRef]
- Nishat, S.; Wuescher, L.M.; Worth, R.G. Platelets Enhance Dendritic Cell Responses against Staphylococcus Aureus through CD40-CD40L. Infect. Immun. 2018, 86, e00186-18. [Google Scholar] [CrossRef]
- Tyers, M.; Haslam, R.J.; Rachubinski, R.A.; Harley, C.B. Molecular Analysis of Pleckstrin: The Major Protein Kinase C Substrate of Platelets. J. Cell Biochem. 1989, 40, 133–145. [Google Scholar] [CrossRef]
- Ma, A.D.; Abrams, C.S. Pleckstrin Induces Cytoskeletal Reorganization via a Rac-Dependent Pathway. J. Biol. Chem. 1999, 274, 28730–28735. [Google Scholar] [CrossRef]
- Roll, R.L.; Bauman, E.M.; Bennett, J.S.; Abrams, C.S. Phosphorylated Pleckstrin Induces Cell Spreading via an Integrin-Dependent Pathway. J. Cell Biol. 2000, 150, 1461–1466. [Google Scholar] [CrossRef]
- Hansen, C.G.; Bright, N.A.; Howard, G.; Nichols, B.J. SDPR Induces Membrane Curvature and Functions in the Formation of Caveolae. Nat. Cell Biol. 2009, 11, 807–814. [Google Scholar] [CrossRef]
- Iontcheva, I.; Amar, S.; Zawawi, K.H.; Kantarci, A.; Van Dyke, T.E. Role for Moesin in Lipopolysaccharide-Stimulated Signal Transduction. Infect. Immun. 2004, 72, 2312–2320. [Google Scholar] [CrossRef]
- Zhang, J.; Hou, S.; Gu, J.; Tian, T.; Yuan, Q.; Jia, J.; Qin, Z.; Chen, Z. S100A4 Promotes Colon Inflammation and Colitis-Associated Colon Tumorigenesis. Oncoimmunology 2018, 7, e1461301. [Google Scholar] [CrossRef]
- Morávková, P.; Kohoutová, D.; Vávrová, J.; Bureš, J. S100A4 Protein in Inflammatory Bowel Disease: Results of a Single Centre Prospective Study. Acta. Med. 2017, 60, 108–113. [Google Scholar] [CrossRef]
- Arijs, I.; Quintens, R.; Van Lommel, L.; Van Steen, K.; De Hertogh, G.; Lemaire, K.; Schraenen, A.; Perrier, C.; Van Assche, G.; Vermeire, S.; et al. Predictive Value of Epithelial Gene Expression Profiles for Response to Infliximab in Crohn’s Disease. Inflamm. Bowel. Dis. 2010, 16, 2090–2098. [Google Scholar] [CrossRef]
- Said, N.; Theodorescu, D. RhoGDI2 Suppresses Bladder Cancer Metastasis via Reduction of Inflammation in the Tumor Microenvironment. Oncoimmunology 2012, 1, 1175–1177. [Google Scholar] [CrossRef]
- Theodorescu, D.; Sapinoso, L.M.; Conaway, M.R.; Oxford, G.; Hampton, G.M.; Frierson, H.F. Reduced Expression of Metastasis Suppressor RhoGDI2 Is Associated with Decreased Survival for Patients with Bladder Cancer. Clin. Cancer. Res. 2004, 10, 3800–3806. [Google Scholar] [CrossRef]
- Fujita, A.; Shida, A.; Fujioka, S.; Kurihara, H.; Okamoto, T.; Yanaga, K. Clinical Significance of Rho GDP Dissociation Inhibitor 2 in Colorectal Carcinoma. Int. J. Clin. Oncol. 2012, 17, 137–142. [Google Scholar] [CrossRef]
- Kettritz, R.; Xu, Y.X.; Faass, B.; Klein, J.B.; Müller, E.C.; Otto, A.; Busjahn, A.; Luft, F.C.; Haller, H. TNF-Alpha-Mediated Neutrophil Apoptosis Involves Ly-GDI, a Rho GTPase Regulator. J. Leukoc. Biol. 2000, 68, 277–283. [Google Scholar]
- Menon, D.; Innes, A.; Oakley, A.J.; Dahlstrom, J.E.; Jensen, L.M.; Brüstle, A.; Tummala, P.; Rooke, M.; Casarotto, M.G.; Baell, J.B.; et al. GSTO1-1 Plays a pro-Inflammatory Role in Models of Inflammation, Colitis and Obesity. Sci. Rep. 2017, 7, 17832. [Google Scholar] [CrossRef]
- Kang, H.J.; Jung, S.-K.; Kim, S.J.; Chung, S.J. Structure of Human Alpha-Enolase (HENO1), a Multifunctional Glycolytic Enzyme. Acta Cryst. D. Biol. Cryst. 2008, 64, 651–657. [Google Scholar] [CrossRef]
- Petrak, J.; Ivanek, R.; Toman, O.; Cmejla, R.; Cmejlova, J.; Vyoral, D.; Zivny, J.; Vulpe, C.D. Déjà vu in Proteomics. A Hit Parade of Repeatedly Identified Differentially Expressed Proteins. Proteomics 2008, 8, 1744–1749. [Google Scholar] [CrossRef]
- Miles, L.A.; Dahlberg, C.M.; Plescia, J.; Felez, J.; Kato, K.; Plow, E.F. Role of Cell-Surface Lysines in Plasminogen Binding to Cells: Identification of Alpha-Enolase as a Candidate Plasminogen Receptor. Biochemistry 1991, 30, 1682–1691. [Google Scholar] [CrossRef]
- Gazouli, M.; Anagnostopoulos, A.K.; Papadopoulou, A.; Vaiopoulou, A.; Papamichael, K.; Mantzaris, G.; Theodoropoulos, G.E.; Anagnou, N.P.; Tsangaris, G.T. Serum Protein Profile of Crohn’s Disease Treated with Infliximab. J. Crohn’s Colitis 2013, 7, e461–e470. [Google Scholar] [CrossRef]
- Vermeulen, N.; Arijs, I.; Joossens, S.; Vermeire, S.; Clerens, S.; Van den Bergh, K.; Michiels, G.; Arckens, L.; Schuit, F.; Van Lommel, L.; et al. Anti-Alpha-Enolase Antibodies in Patients with Inflammatory Bowel Disease. Clin. Chem. 2008, 54, 534–541. [Google Scholar] [CrossRef]
- Roozendaal, C.; Zhao, M.H.; Horst, G.; Lockwood, C.M.; Kleibeuker, J.H.; Limburg, P.C.; Nelis, G.F.; Kallenberg, C.G. Catalase and Alpha-Enolase: Two Novel Granulocyte Autoantigens in Inflammatory Bowel Disease (IBD). Clin. Exp. Immunol. 1998, 112, 10–16. [Google Scholar] [CrossRef]
- Hu, H.; Juvekar, A.; Lyssiotis, C.A.; Lien, E.C.; Albeck, J.G.; Oh, D.; Varma, G.; Hung, Y.P.; Ullas, S.; Lauring, J.; et al. Phosphoinositide 3-Kinase Regulates Glycolysis through Mobilization of Aldolase from the Actin Cytoskeleton. Cell 2016, 164, 433–446. [Google Scholar] [CrossRef]
- Best, W.R.; Becktel, J.M.; Singleton, J.W.; Kern, F. Development of a Crohn’s Disease Activity Index. National Cooperative Crohn’s Disease Study. Gastroenterology 1976, 70, 439–444. [Google Scholar]
- Daperno, M.; D’Haens, G.; Van Assche, G.; Baert, F.; Bulois, P.; Maunoury, V.; Sostegni, R.; Rocca, R.; Pera, A.; Gevers, A.; et al. Development and Validation of a New, Simplified Endoscopic Activity Score for Crohn’s Disease: The SES-CD. Gastrointest. Endosc. 2004, 60, 505–512. [Google Scholar] [CrossRef]
- Vowinckel, J.; Zelezniak, A.; Bruderer, R.; Mülleder, M.; Reiter, L.; Ralser, M. Cost-Effective Generation of Precise Label-Free Quantitative Proteomes in High-Throughput by MicroLC and Data-Independent Acquisition. Sci. Rep. 2018, 8, 4346. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics Enrichment Tools: Paths toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic. Acids. Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]



| Missing Values | Univariate Analysis | ||||
|---|---|---|---|---|---|
| Variable | NSTR (16) | STR (97) | OR (95% CI) | p | |
| Gender (% male) | - | 11 (68.8%) | 53 (54.6%) | 0.5 (0.2–1.7) | 0.296 |
| Age (years, median, IQR) | - | 53.5 (43.0–59.3) | 39.0 (27.5–50.0) | 1.1 (1.0–1.1) | 0.009 * |
| Smoking habit (%) | 8 (7.1%) | ||||
| No | 9 (69.2%) | 46 (50.0%) | 1 (Ref.) | ||
| Yes | 4 (30.8%) | 32 (34.8%) | 0.6 (0.2–2.3) | 0.486 | |
| Former | 0 (0.0%) | 14 (15.2%) | 0.0 (0.0-) | 0.999 | |
| Anti-TNF | - | ||||
| Infliximab/biosimilar (%) | 9 (56.3%) | 48 (49.5%) | 1 (Ref.) | ||
| Adalimumab (%) | 7 (43.8%) | 49 (50.5%) | 0.8 (0.3–2.2) | 0.617 | |
| Treatment indication | - | ||||
| Luminal disease (%) | 15 (93.8%) | 84 (86.6%) | 1 (Ref.) | ||
| Perianal disease (%) | 1 (6.3%) | 8 (8.2%) | 0.7 (0.1–6.0) | 0.745 | |
| Both (%) | 0 (0.0%) | 5 (5.2%) | 0.0 (0.0-) | 0.999 | |
| Immunomodulatory therapy | - | ||||
| None (%) | 2 (12.5%) | 17 (17.5%) | 1 (Ref.) | ||
| Azathioprine (%) | 13 (81.3%) | 74 (76.3%) | 1.5 (0.3–7.2) | 0.619 | |
| Methotrexate (%) | 0 (0.0%) | 3 (3.1%) | 0.0 (0.0-) | 0.999 | |
| Other (%) | 1 (6.3%) | 3 (3.1%) | 2.8 (0.2–42.0) | 0.449 | |
| Corticosteroids induction (yes, %) | - | 8 (50.0%) | 16 (16.5%) | 5.1 (1.7–15.5) | 0.004 * |
| Basal BMI (Kg/m2) (median, IQR) | 10 (8.8%) | 23.0 (20.8–24.9) | 22.4 (20.5–26.0) | 1.0 (0.8–1.1) | 0.838 |
| Disease duration (years, median, IQR) | 8 (7.1%) | 22.0 (2.5–26.0) | 4.0 (1.0–13.0) | 1.1 (1.0–1.2) | 0.006 * |
| Montreal (age at diagnosis) | 11 (9.7%) | ||||
| <17 (%) | 0 (0.0%) | 7 (7.8%) | 1 (Ref.) | ||
| 17–40 (%) | 9 (75.0%) | 64 (71.1%) | 2.3 × 108 (0.0-) | 0.999 | |
| >40 (%) | 3 (25.0%) | 19 (21.1%) | 2.6 × 108 (0.0-) | 0.999 | |
| Montreal location | 3 (2.7%) | ||||
| Ileal (%) | 8 (53.3%) | 39 (41.1%) | 1 (Ref.) | ||
| Colonic (%) | 1 (6.7%) | 16 (16.8%) | 0.3 (0.0–2.6) | 0.281 | |
| Ileocolonic (%) | 6 (40.0%) | 37 (38.9%) | 0.8 (0.3–2.5) | 0.689 | |
| Isolated upper disease (%) | 0 (0.0%) | 3 (3.2%) | 0.0 (0.0-) | 0.999 | |
| Behavior | 8 (7.1%) | ||||
| Inflammatory (%) | 5 (38.5%) | 56 (60.9%) | 1 (Ref.) | ||
| Stricturing (%) | 1 (7.7%) | 16 (17.4%) | 0.7 (0.1–6.4) | 0.753 | |
| Fistulizing (%) | 7 (53.8%) | 18 (19.6%) | 4.4 (1.2–15.4) | 0.023 * | |
| Fistulizing and Stricturing (%) | 0 (0.0%) | 2 (2.2%) | 0.0 (0.0-) | 0.999 | |
| Perianal disease (yes, %) | 1 (0.9%) | 2 (12.5%) | 32 (33.3%) | 0.3 (0.1–1.3) | 0.111 |
| Extraintestinal manifestation (yes, %) | 8 (7.1%) | 2 (15.4%) | 14 (15.2%) | 1.0 (0.2–5.1) | 0.987 |
| Appendicectomy (yes, %) | 8 (7.1%) | 1 (7.7%) | 8 (8.7%) | 0.9 (0.1–7.6) | 0.904 |
| Bowel resection (yes, %) | - | 10 (62.5%) | 20 (20.6%) | 6.4 (2.1–19.8) | 0.001 * |
| Perianal surgery (yes, %) | - | 0 (0.0%) | 18 (18.6%) | 0.0 (0.0-) | 0.998 |
| Basal CDAI score (AU, median, IQR) | - | 235.5 (149.3–310.0) | 91.2 (54.9–166.0) | 1.0 (1.0–1.0) | 0.000 * |
| Basal hemoglobin (g/L, mean ± SD) | 9 (8.0%) | 12.7 ± 1.0 | 13.0 ± 1.5 | 0.8 (0.6–1.3) | 0.384 |
| Basal WBC (103/μL, median, IQR) | 9 (8.0%) | 7.5 (5.5–11.7) | 7.6 (5.6–10.4) | 1.1 (0.9–1.2) | 0.561 |
| Basal platelets (103/μL, median, IQR) | 10 (8.8%) | 304.0 (223.0–356.5) | 312.5 (263.5–363.5) | 1.0 (1.0–1.0) | 0.235 |
| Basal albumin (g/dL, mean ± SD) | 22 (19.5%) | 4.0 ± 0.4 | 4.0 ± 0.5 | 0.9 (0.2–3.8) | 0.940 |
| Basal ferritin (ng/mL, median, IQR) | 19 (16.8%) | 119.5 (51.2–187.3) | 55.0 (26.0–114.9) | 1.0 (1.0–1.0) | 0.449 |
| Basal CRP (mg/L, median, IQR) | 14 (12.4%) | 2.3 (0.6–3.4) | 4.4 (0.6–14.6) | 1.0 (0.9–1.0) | 0.253 |
| Basal ESR (mm/h, median, IQR) | 34 (30.1%) | 35.0 (10.8–85.8) | 27.0 (11.0–37.0) | 1.0 (1.0–1.0) | 0.121 |
| Basal ENOA (μg/mg protein) | 1 (0.9%) | 0.1 (0.0–0.2) | 0.1 (0.0–0.2) | 0.1 (0.0–4.7) | 0.199 |
| Basal VINC (pg/mg protein) | - | 0.8 (0.2–1.2) | 1.2 (0.6–2.0) | 0.7 (0.4–1.2) | 0.171 |
| Protein ID | Protein | p | Fold Change | Biological Process | Molecular Function |
|---|---|---|---|---|---|
| P06733 | ENOA | 0.0001 | 3.9 | Glycolysis/Plasminogen activation/Transcription regulation | DNA binding/Lyase/Repressor |
| P18206 | VINC | 0.0007 | 4.6 | Cell adhesion/Cytoskeleton; Hemostasis/Platelet function | Actin binding |
| O00151 | PDLI1 | 0.0013 | 2.4 | Cell adhesion/Cytoskeleton | Actin binding |
| P52566 | GDIR2 | 0.0013 | 4.9 | Cell adhesion/Cytoskeleton | GTPase activity |
| Q15942 | ZYX | 0.0014 | 5.5 | Cell adhesion/Cytoskeleton | Metal binding/RNA binding |
| P04075 | ALDOA | 0.0021 | 3.3 | Glycolysis; Cell adhesion/Cytoskeleton; Hemostasis/Platelet function | Actin binding/Fructose-bisphosphate aldolase activity |
| P26038 | MOES | 0.0023 | 2.5 | Cell adhesion/Cytoskeleton; Hemostasis/Platelet function | Actin binding |
| P78417 | GSTO1 | 0.0025 | 3.2 | Inflammatory response | Oxidoreductase/Transferase |
| P12814 | ACTN1 | 0.0025 | 3.2 | Cell adhesion/Cytoskeleton; Hemostasis/Platelet function | Actin binding |
| P37802 | TAGL2 | 0.0027 | 6.4 | Cell adhesion/Cytoskeleton; Hemostasis/Platelet function | Cadherin binding |
| O95810 | SDPR | 0.0027 | 4.2 | Cell adhesion/Cytoskeleton; Hemostasis/Platelet function | Lipid binding |
| P50395 | GDIB | 0.0028 | 3.0 | Inflammatory response | GTPase activation |
| P23528 | COF1 | 0.0029 | 3.0 | Cell adhesion/Cytoskeleton; Hemostasis/Platelet function | Actin binding |
| P26447 | S10A4 | 0.0031 | 3.3 | Cell adhesion/Cytoskeleton; Inflammatory response | Actin binding |
| Q15404 | RSU1 | 0.0037 | 3.9 | Cell adhesion/Cytoskeleton | Positive regulation of GTPase activity |
| P08567 | PLEK | 0.0048 | 5.7 | Cell adhesion/Cytoskeleton; Hemostasis/Platelet function | Protein binding |
| P63104 | 1433Z | 0.0078 | 4.2 | Cell adhesion/Cytoskeleton | Protein binding |
| P60174 | TPIS | 0.0093 | 3.6 | Glycolysis | Isomerase/Lyase |
| Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|
| w/o VINC/ENOA | w/ ENOA | w/ VINC | ||||
| Variable | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p |
| Age (years) | - | - | x | x | x | x |
| Corticosteroids induction (%) | 8.6 (1.7–43.5) | 0.009 * | 12.8 (2.4–68.8) | 0.003 * | 14.3 (2.6–77.7) | 0.002 * |
| Disease duration (years) | - | - | x | x | x | x |
| Bowel Resection (%) | 10.5 (2.1–52.0) | 0.004 * | 13.2 (2.5–68.9) | 0.002 * | 14.8 (2.8–78.0) | 0.001 * |
| Basal CDAI score (AU) | 1.0 (1.0–1.0) | 0.003 * | 1.0 (1.0–1.0) | 0.001 * | 1.0 (1.0–1.0) | 0.002 * |
| Basal ENOA (µg/mg protein) | x | x | 0.0 (0.0–1.7) | 0.067 | x | x |
| Basal VINC (pg/mg protein) | x | x | x | x | 0.5 (0.3–0.9) | 0.032 * |
| Variable | AUROC | p | 95% CI |
|---|---|---|---|
| Basal VINC (pg/mg) | 0.651 | 0.054 | (0.500–0.802) |
| Corticosteroids induction | 0.668 | 0.032 | (0.511–0.824) |
| Bowel resection | 0.709 | 0.007 | (0.562–0.857) |
| CDAI score | 0.829 | 0.000 | (0.718–0.940) |
| Adjusted model (w/o VINC) | 0.904 | 0.000 | (0.838–0.970) |
| Adjusted model (w/ VINC) | 0.919 | 0.000 | (0.862–0.977) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medina-Medina, R.; Iglesias-Flores, E.; Benítez, J.M.; Marín-Pedrosa, S.; Salgueiro-Rodríguez, I.; Linares, C.I.; González-Rubio, S.; Soto-Escribano, P.; Gros, B.; Rodríguez-Perálvarez, M.L.; et al. Development of a Prediction Model for Short-Term Remission of Patients with Crohn’s Disease Treated with Anti-TNF Drugs. Int. J. Mol. Sci. 2023, 24, 8695. https://doi.org/10.3390/ijms24108695
Medina-Medina R, Iglesias-Flores E, Benítez JM, Marín-Pedrosa S, Salgueiro-Rodríguez I, Linares CI, González-Rubio S, Soto-Escribano P, Gros B, Rodríguez-Perálvarez ML, et al. Development of a Prediction Model for Short-Term Remission of Patients with Crohn’s Disease Treated with Anti-TNF Drugs. International Journal of Molecular Sciences. 2023; 24(10):8695. https://doi.org/10.3390/ijms24108695
Chicago/Turabian StyleMedina-Medina, Rosario, Eva Iglesias-Flores, Jose M. Benítez, Sandra Marín-Pedrosa, Isabel Salgueiro-Rodríguez, Clara I. Linares, Sandra González-Rubio, Pilar Soto-Escribano, Beatriz Gros, Manuel L. Rodríguez-Perálvarez, and et al. 2023. "Development of a Prediction Model for Short-Term Remission of Patients with Crohn’s Disease Treated with Anti-TNF Drugs" International Journal of Molecular Sciences 24, no. 10: 8695. https://doi.org/10.3390/ijms24108695
APA StyleMedina-Medina, R., Iglesias-Flores, E., Benítez, J. M., Marín-Pedrosa, S., Salgueiro-Rodríguez, I., Linares, C. I., González-Rubio, S., Soto-Escribano, P., Gros, B., Rodríguez-Perálvarez, M. L., Cabriada, J. L., Chaparro, M., Gisbert, J. P., Chicano-Gálvez, E., Ortea, I., Ferrín, G., García-Sánchez, V., & Aguilar-Melero, P. (2023). Development of a Prediction Model for Short-Term Remission of Patients with Crohn’s Disease Treated with Anti-TNF Drugs. International Journal of Molecular Sciences, 24(10), 8695. https://doi.org/10.3390/ijms24108695