Proteome Landscape during Ripening of Solid Endosperm from Two Different Coconut Cultivars Reveals Contrasting Carbohydrate and Fatty Acid Metabolic Pathway Modulation
Abstract
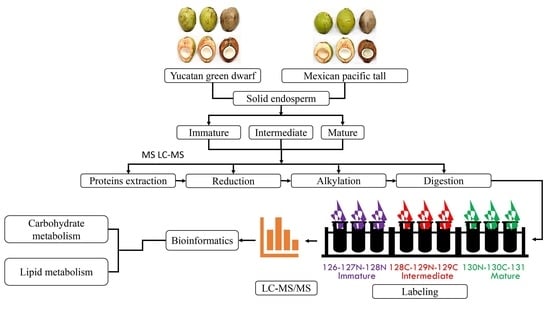
:1. Introduction
2. Results
2.1. Physiological and Physicochemical Parameters of the Coconut Fruit
2.2. Proteome Landscape of Coconut Solid Endosperm during Ripening
2.3. Dynamics of the Coconut Solid Endosperm Proteome during Ripening
2.4. Detoxification of ROS and Methylglyoxal Pathways in MPT and YGD Cultivars
2.5. DAPs Involved in Carbohydrate Metabolism in the YGD and MPT Cultivars
2.5.1. Glycolysis and Gluconeogenesis
2.5.2. The Tricarboxylic Acid Cycle (TCA)
2.6. DAPs Involved in Lipid Metabolism in the YGD and MPT Cultivars
2.7. Other Proteins
3. Discussion
4. Materials and Methods
4.1. Plant Material Collection
4.2. Proteome Extraction from Coconut Solid Endosperm
4.3. Protein Quantification and Analysis by SDS-PAGE
4.4. Sample Preparation for Proteomics
4.5. TMT Labeling and Peptide Fractionation
4.6. Nano-LC-MS/MS and Synchronous Precursor Selection (SPS)-MS3
4.7. Bioinformatics Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Niral, V.; Jerard, B.A.; Rajesh, M.K. Germplasm Resources: Diversity and Conservation. In The Coconut Genome; Rajesh, M.K., Ramesh, S.V., Perera, L., Kole, C., Eds.; Compendium of Plant Genomes; Springer International Publishing: Cham, Switzerland, 2021; pp. 27–46. ISBN 978-3-030-76648-1. [Google Scholar]
- Burns, D.T.; Johnston, E.-L.; Walker, M.J. Authenticity and the Potability of Coconut Water—A Critical Review. J. AOAC Int. 2020, 103, 800–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, E.Y.Y.; Biddle, J.; Foale, M.; Panis, B.; Adkins, S.W. The Potential to Propagate Coconut Clones through Direct Shoot Organogenesis: A Review. Sci. Hortic. 2021, 289, 110400. [Google Scholar] [CrossRef]
- Nartvaranant, P. Genetic Variations for “Nam Hom” Coconut (Cocos nucifera L.) Grownin the Western Region of Thailand Using AFLP Markers. J. Thai Interdiscip. Res. 2019, 14, 12. [Google Scholar] [CrossRef]
- Ignacio, I.-F.; Miguel, T.-S. Research Opportunities on the Coconut (Cocos nucifera L.) Using New Technologies. S. Afr. J. Bot. 2021, 141, 414–420. [Google Scholar] [CrossRef]
- Sudha, R.; Niral, V.; Samsudeen, K. Botanical Study and Cytology. In The Coconut Genome; Rajesh, M.K., Ramesh, S.V., Perera, L., Kole, C., Eds.; Compendium of Plant Genomes; Springer International Publishing: Cham, Switzerland, 2021; pp. 13–25. ISBN 978-3-030-76648-1. [Google Scholar]
- Lantican, D.V.; Strickler, S.R.; Canama, A.O.; Gardoce, R.R.; Mueller, L.A.; Galvez, H.F. De Novo Genome Sequence Assembly of Dwarf Coconut (Cocos nucifera L. ‘Catigan Green Dwarf’) Provides Insights into Genomic Variation between Coconut Types and Related Palm Species. G3 Genes Genomes Genet. 2019, 9, 2377–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boonkaew, T.; Mongkolsiriwatana, C.; Vongvanrungruang, A.; Srikulnath, K.; Peyachoknagul, S. Characterization of GA20ox Genes in Tall and Dwarf Types Coconut (Cocos nucifera L.). Genes Genom. 2018, 40, 735–745. [Google Scholar] [CrossRef]
- Naik, M.; Sunil, C.M.; Rawson, A.; Venkatachalapathy, N. Tender Coconut Water: A Review on Recent Advances in Processing and Preservation. Food Rev. Int. 2022, 38, 1215–1236. [Google Scholar] [CrossRef]
- Zheng, Y.; Jin, Y.; Yuan, Y.; Feng, D.; Chen, L.; Li, D.; Zhou, P. Identification and Function Analysis of a Type 2 Diacylglycerol Acyltransferase (DGAT2) from the Endosperm of Coconut (Cocos nucifera L.). Gene 2019, 702, 75–82. [Google Scholar] [CrossRef]
- Tenda, E.; Miftahorrachman; Kumaunang, J. Profile of Amino Acids and Fatty Acids of Some Indonesia Tall Coconut Varieties. IOP Conf. Ser. Earth Environ. Sci. 2022, 974, 012133. [Google Scholar] [CrossRef]
- Kumar, S.N. Variability in Coconut (Cocos nucifera L.) Germplasm and Hybrids for Fatty Acid Profile of Oil. J. Agric. Food Chem. 2011, 59, 13050–13058. [Google Scholar] [CrossRef]
- Reynolds, K.B.; Cullerne, D.P.; El Tahchy, A.; Rolland, V.; Blanchard, C.L.; Wood, C.C.; Singh, S.P.; Petrie, J.R. Identification of Genes Involved in Lipid Biosynthesis through de Novo Transcriptome Assembly from Cocos nucifera Developing Endosperm. Plant Cell Physiol. 2019, 60, 945–960. [Google Scholar] [CrossRef] [PubMed]
- Angeles, J.; Lado, J.; Pascual, E.; Cueto, C.; Laurena, A.; Laude, R. Towards the Understanding of Important Coconut Endosperm Phenotypes: Is There an Epigenetic Control? Agronomy 2018, 8, 225. [Google Scholar] [CrossRef] [Green Version]
- Bourgis, F.; Kilaru, A.; Cao, X.; Ngando-Ebongue, G.-F.; Drira, N.; Ohlrogge, J.B.; Arondel, V. Comparative Transcriptome and Metabolite Analysis of Oil Palm and Date Palm Mesocarp That Differ Dramatically in Carbon Partitioning. Proc. Natl. Acad. Sci. USA 2011, 108, 12527–12532. [Google Scholar] [CrossRef] [Green Version]
- Naganeeswaran, S.; Fayas, T.P.; Rajesh, M.K. Dataset of Transcriptome Assembly of Date Palm Embryogenic Calli and Functional Annotation. Data Brief 2020, 31, 105760. [Google Scholar] [CrossRef]
- D’Amato, A.; Fasoli, E.; Righetti, P.G. Harry Belafonte and the Secret Proteome of Coconut Milk. J. Proteom. 2012, 75, 914–920. [Google Scholar] [CrossRef]
- Huang, J.; Liu, X.; Lan, Q.; Lai, X.; Luo, Z.; Yang, G. Proteomic Profile of Coconuts. Eur. Food Res. Technol. 2016, 242, 449–455. [Google Scholar] [CrossRef]
- Ma, J.; Pan, C.; Chen, H.; Chen, W.; Chen, W.; Zhang, M.; Zhong, Q. Insight of the Functional and Biological Activities of Coconut (Cocos nucifera L.) Protein by Proteomics Analysis and Protein-Based Bioinformatics. Molecules 2022, 27, 2987. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Palma, J.M. Nitric Oxide on/off in Fruit Ripening. Plant Biol. 2018, 20, 805–807. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, X.; Li, L.; Tang, Y.; Qi, W.; Liu, X.; Qiao, L.; Wang, W.; Jia, X. A Label-Free Quantitative Proteomic Investigation Reveals Stage-Responsive Ripening Genes in Apricot Fruits. J. Hortic. Sci. Biotechnol. 2017, 92, 261–269. [Google Scholar] [CrossRef]
- Williams, R.S.; Benkeblia, N. Biochemical and Physiological Changes of Star Apple Fruit (Chrysophyllum cainito) during Different “on Plant” Maturation and Ripening Stages. Sci. Hortic. 2018, 236, 36–42. [Google Scholar] [CrossRef]
- Juarez-Escobar, J.; Guerrero-Analco, J.A.; Zamora-Briseño, J.A.; Elizalde-Contreras, J.M.; Bautista-Valle, M.V.; Bojórquez-Velázquez, E.; Loyola-Vargas, V.M.; Mata-Rosas, M.; Ruíz-May, E. Tissue-Specific Proteome Characterization of Avocado Seed during Postharvest Shelf Life. J. Proteom. 2021, 235, 104112. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Lin, B.; Tan, J.; Hao, P.; Hua, S.; Deng, Z. Tandem Mass Tag-Based Quantitative Proteomics Reveals Implication of a Late Embryogenesis Abundant Protein (BnLEA57) in Seed Oil Accumulation in Brassica napus L. Front. Plant Sci. 2022, 13, 907244. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.C.; Gordon, A.; Wizzard, G.; McCook, K.; Rolle, R. Changes in Chemical Composition of Coconut (Cocos nucifera) Water during Maturation of the Fruit. J. Sci. Food Agric. 2004, 84, 1049–1052. [Google Scholar] [CrossRef]
- Farooq, M.A.; Zhang, X.; Zafar, M.M.; Ma, W.; Zhao, J. Roles of Reactive Oxygen Species and Mitochondria in Seed Germination. Front. Plant Sci. 2021, 12, 781734. [Google Scholar] [CrossRef] [PubMed]
- Miray, R.; Kazaz, S.; To, A.; Baud, S. Molecular Control of Oil Metabolism in the Endosperm of Seeds. Int. J. Mol. Sci. 2021, 22, 1621. [Google Scholar] [CrossRef]
- Nair, K.P. The Coconut Palm (Cocos nucifera L.). In Tree Crops; Springer International Publishing: Cham, Switzerland, 2021; pp. 79–128. ISBN 978-3-030-62139-1. [Google Scholar]
- Baslam, M.; Mitsui, T.; Sueyoshi, K.; Ohyama, T. Recent Advances in Carbon and Nitrogen Metabolism in C3 Plants. Int. J. Mol. Sci. 2020, 22, 318. [Google Scholar] [CrossRef]
- Ramzan, A.; Shah, M.; Ullah, N.; Sheheryar; Nascimento, J.R.S.; Campos, F.A.P.; Domont, G.B.; Nogueira, F.C.S.; Abdellattif, M.H. Proteomic Analysis of Embryo Isolated from Mature Jatropha curcas L. Seeds. Front. Plant Sci. 2022, 13, 843764. [Google Scholar] [CrossRef]
- Allaman, I.; Bélanger, M.; Magistretti, P.J. Methylglyoxal, the Dark Side of Glycolysis. Front. Neurosci. 2015, 9, 23. [Google Scholar] [CrossRef] [Green Version]
- Hoque, T.S.; Hossain, M.A.; Mostofa, M.G.; Burritt, D.J.; Fujita, M.; Tran, L.-S.P. Methylglyoxal: An Emerging Signaling Molecule in Plant Abiotic Stress Responses and Tolerance. Front. Plant Sci. 2016, 7, 1341. [Google Scholar] [CrossRef] [Green Version]
- Kalapos, M.P. The Tandem of Free Radicals and Methylglyoxal. Chem. Biol. Interact. 2008, 171, 251–271. [Google Scholar] [CrossRef]
- Xu, Z.; Ma, J.; Qu, C.; Hu, Y.; Hao, B.; Sun, Y.; Liu, Z.; Yang, H.; Yang, C.; Wang, H.; et al. Identification and Expression Analyses of the Alanine Aminotransferase (AlaAT) Gene Family in Poplar Seedlings. Sci. Rep. 2017, 7, 45933. [Google Scholar] [CrossRef] [Green Version]
- Fushinobu, S. Molecular Evolution and Functional Divergence of UDP-Hexose 4-Epimerases. Curr. Opin. Chem. Biol. 2021, 61, 53–62. [Google Scholar] [CrossRef]
- Mortimer, J.C.; Laohavisit, A.; Macpherson, N.; Webb, A.; Brownlee, C.; Battey, N.H.; Davies, J.M. Annexins: Multifunctional Components of Growth and Adaptation. J. Exp. Bot. 2008, 59, 533–544. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Xiu, J.; Li, Y.; Ma, H.; Wu, J.; Wang, B.; Guo, G. Characterization and Expression Pattern Analysis of the T-Complex Protein-1 Zeta Subunit in Musca domestica L. (Diptera). J. Insect Sci. 2017, 17, 90. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Wu, X.; Liu, X. Temperature Stress Response of Heat Shock Protein 90 (Hsp90) in the Clam Paphia Undulata. Aquac. Fish. 2018, 3, 106–114. [Google Scholar] [CrossRef]
- Keereetaweep, J.; Liu, H.; Zhai, Z.; Shanklin, J. Biotin Attachment Domain-Containing Proteins Irreversibly Inhibit Acetyl CoA Carboxylase. Plant Physiol. 2018, 177, 208–215. [Google Scholar] [CrossRef] [Green Version]
- Kundu, P.; Nehra, A.; Gill, R.; Tuteja, N.; Gill, S.S. Unraveling the Importance of EF-Hand-Mediated Calcium Signaling in Plants. S. Afr. J. Bot. 2022, 148, 615–633. [Google Scholar] [CrossRef]
- Denancé, N.; Szurek, B.; Noël, L.D. Emerging Functions of Nodulin-Like Proteins in Non-Nodulating Plant Species. Plant Cell Physiol. 2014, 55, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.; Ge, Q.; Zhang, S.; Zhang, Z.; Liu, A.; Fan, S.; Jiang, X.; Feng, Y.; Zhang, L.; Niu, D.; et al. UDP-Glucose Dehydrogenases: Identification, Expression, and Function Analyses in Upland Cotton (Gossypium hirsutum). Front. Genet. 2021, 11, 597890. [Google Scholar] [CrossRef] [PubMed]
- Stein, O.; Granot, D. An Overview of Sucrose Synthases in Plants. Front. Plant Sci. 2019, 10, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, L. Structure and Function of Biotin-Dependent Carboxylases. Cell. Mol. Life Sci. 2013, 70, 863–891. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Liu, M.; He, L.; Li, X.; Wang, F.; Yan, B.; Wei, J.; Zhao, C.; Li, Z.; Xu, J. A Cytosolic NAD+-Dependent GPDH from Maize (ZmGPDH1) Is Involved in Conferring Salt and Osmotic Stress Tolerance. BMC Plant Biol. 2019, 19, 16. [Google Scholar] [CrossRef]
- Yao, Y.; Geng, M.-T.; Wu, X.-H.; Sun, C.; Wang, Y.-L.; Chen, X.; Shang, L.; Lu, X.-H.; Li, Z.; Li, R.-M.; et al. Identification, Expression, and Functional Analysis of the Fructokinase Gene Family in Cassava. Int. J. Mol. Sci. 2017, 18, 2398. [Google Scholar] [CrossRef] [Green Version]
- Ueda, Y.; Zhao, W.; Ihara, H.; Imahori, Y.; Tsantili, E.; Wendakoon, S.; Chambers, A.; Bai, J. Functional Characteristics of Aldehyde Dehydrogenase and Its Involvement in Aromatic Volatile Biosynthesis in Postharvest Banana Ripening. Foods 2022, 11, 347. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Yu, J.; Yan, W.; Zhang, J.; Yang, D.; Yao, G.; Liu, Z.; Wu, Y.; Hou, X. Integrative ITRAQ-Based Proteomic and Transcriptomic Analysis Reveals the Accumulation Patterns of Key Metabolites Associated with Oil Quality during Seed Ripening of Camellia oleifera. Hortic. Res. 2021, 8, 157. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wang, B. Comparative Analysis of Ascorbate Peroxidases (APXs) from Selected Plants with a Special Focus on Oryza sativa Employing Public Databases. PLoS ONE 2019, 14, e0226543. [Google Scholar] [CrossRef] [PubMed]
- Abid, G.; Silue, S.; Muhovski, Y.; Jacquemin, J.-M.; Toussaint, A.; Baudoin, J.-P. Role of Myo-Inositol Phosphate Synthase and Sucrose Synthase Genes in Plant Seed Development. Gene 2009, 439, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rosa-Téllez, S.; Anoman, A.D.; Flores-Tornero, M.; Toujani, W.; Alseek, S.; Fernie, A.R.; Nebauer, S.G.; Muñoz-Bertomeu, J.; Segura, J.; Ros, R. Phosphoglycerate Kinases Are Co-Regulated to Adjust Metabolism and to Optimize Growth. Plant Physiol. 2018, 176, 1182–1198. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Saand, M.A.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Fan, H.; Wang, F. ITRAQ-Based Comparative Proteomic Analysis of Two Coconut Varieties Reveals Aromatic Coconut Cold-Sensitive in Response to Low Temperature. J. Proteom. 2020, 220, 103766. [Google Scholar] [CrossRef]
- Hassan, H.; Amiruddin, M.D.; Weckwerth, W.; Ramli, U.S. Deciphering Key Proteins of Oil Palm (Elaeis Guineensis Jacq.) Fruit Mesocarp Development by Proteomics and Chemometrics. Electrophoresis 2019, 40, 254–265. [Google Scholar] [CrossRef]
- Chen, Y.-Y.; Zhang, Z.-H.; Zhong, C.-Y.; Song, X.-M.; Lin, Q.-H.; Huang, C.-M.; Huang, R.-H.; Chen, W. Functional Analysis of Differentially Expressed Proteins in Chinese Bayberry (Myrica rubra Sieb. et Zucc.) Fruits during Ripening. Food Chem. 2016, 190, 763–770. [Google Scholar] [CrossRef]
- D’Ambrosio, C.; Arena, S.; Rocco, M.; Verrillo, F.; Novi, G.; Viscosi, V.; Marra, M.; Scaloni, A. Proteomic Analysis of Apricot Fruit during Ripening. J. Proteom. 2013, 78, 39–57. [Google Scholar] [CrossRef] [Green Version]
- Correa, S.M.; Fernie, A.R.; Nikoloski, Z.; Brotman, Y. Towards Model-Driven Characterization and Manipulation of Plant Lipid Metabolism. Prog. Lipid Res. 2020, 80, 101051. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Hou, D.; Li, Y.; Chao, H.; Zhang, K.; Wang, H.; Xiang, J.; Raboanatahiry, N.; Wang, B.; Li, M. Integration of Proteomic and Genomic Approaches to Dissect Seed Germination Vigor in Brassica napus Seeds Differing in Oil Content. BMC Plant Biol. 2019, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; Li, F.; Liu, A. Comparative Proteomic and Transcriptomic Analyses Provide New Insight into the Formation of Seed Size in Castor Bean. BMC Plant Biol. 2020, 20, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, J.; Yu, L.; Xu, C. A Central Role for Triacylglycerol in Membrane Lipid Breakdown, Fatty Acid β-Oxidation, and Plant Survival under Extended Darkness. Plant Physiol. 2017, 174, 1517–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zulu, N.N.; Zienkiewicz, K.; Vollheyde, K.; Feussner, I. Current Trends to Comprehend Lipid Metabolism in Diatoms. Prog. Lipid Res. 2018, 70, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Teng, M.; Zhao, Y.J.; Khoo, A.L.; Yeo, T.C.; Yong, Q.W.; Lim, B.P. Impact of Coconut Oil Consumption on Cardiovascular Health: A Systematic Review and Meta-Analysis. Nutr. Rev. 2020, 78, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Lamade, E.; Tcherkez, G. Seed Germination in Oil Palm (Elaeis guineensis Jacq.): A Review of Metabolic Pathways and Control Mechanisms. Int. J. Mol. Sci. 2020, 21, 4227. [Google Scholar] [CrossRef]
- Liu, H.; Li, H.; Gu, J.; Deng, L.; Ren, L.; Hong, Y.; Lu, Q.; Chen, X.; Liang, X. Identification of the Candidate Proteins Related to Oleic Acid Accumulation during Peanut (Arachis hypogaea L.) Seed Development through Comparative Proteome Analysis. Int. J. Mol. Sci. 2018, 19, 1235. [Google Scholar] [CrossRef] [Green Version]
- Pedreschi, R.; Uarrota, V.; Fuentealba, C.; Alvaro, J.E.; Olmedo, P.; Defilippi, B.G.; Meneses, C.; Campos-Vargas, R. Primary Metabolism in Avocado Fruit. Front. Plant Sci. 2019, 10, 795. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Chen, C.; Wang, X.; Zhang, Z.; Yu, F.; Guy, R.D. Proteomic Analysis of Metabolic Mechanisms Associated with Fatty Acid Biosynthesis during Styrax tonkinensis Kernel Development. J. Sci. Food Agric. 2021, 101, 6053–6063. [Google Scholar] [CrossRef]
- Maskromo, I.; Karouw, S.; Pandin, D.S.; Mahayu, W.M.; Santosa, B.; Alouw, J.C. Physichochemical Properties of Kebumen Entog Dwarf Coconut. IOP Conf. Ser. Earth Environ. Sci. 2020, 418, 012037. [Google Scholar] [CrossRef]
- Yu, L.; Fan, J.; Zhou, C.; Xu, C. Chloroplast Lipid Biosynthesis Is Fine-Tuned to Thylakoid Membrane Remodeling during Light Acclimation. Plant Physiol. 2021, 185, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Benning, C. Functions of Triacylglycerols during Plant Development and Stress. Curr. Opin. Biotechnol. 2018, 49, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Kilaru, A.; Cao, X.; Dabbs, P.B.; Sung, H.-J.; Rahman, M.M.; Thrower, N.; Zynda, G.; Podicheti, R.; Ibarra-Laclette, E.; Herrera-Estrella, L.; et al. Oil Biosynthesis in a Basal Angiosperm: Transcriptome Analysis of Persea americana Mesocarp. BMC Plant Biol. 2015, 15, 203. [Google Scholar] [CrossRef] [Green Version]
- Antonets, K.S.; Belousov, M.V.; Sulatskaya, A.I.; Belousova, M.E.; Kosolapova, A.O.; Sulatsky, M.I.; Andreeva, E.A.; Zykin, P.A.; Malovichko, Y.V.; Shtark, O.Y.; et al. Accumulation of Storage Proteins in Plant Seeds Is Mediated by Amyloid Formation. PLoS Biol. 2020, 18, e3000564. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.N.; Arocena, R.V.; Laurena, A.C.; Tecson-Mendoza, E.M. 11S and 7S Globulins of Coconut (Cocos nucifera L.): Purification and Characterization. J. Agric. Food Chem. 2005, 53, 1734–1739. [Google Scholar] [CrossRef] [PubMed]
- Patil, U.; Benjakul, S. Coconut Milk and Coconut Oil: Their Manufacture Associated with Protein Functionality. J. Food Sci. 2018, 83, 2019–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dave, A.C.; Ye, A.; Singh, H. Structural and Interfacial Characteristics of Oil Bodies in Coconuts (Cocos nucifera L.). Food Chem. 2019, 276, 129–139. [Google Scholar] [CrossRef]
- Nguyen, T.-P.; Cueff, G.; Hegedus, D.D.; Rajjou, L.; Bentsink, L. A Role for Seed Storage Proteins in Arabidopsis Seed Longevity. J. Exp. Bot. 2015, 66, 6399–6413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oropeza, C.; Cordova, I.; Chumba, A.; Narváez, M.; Sáenz, L.; Ashburner, R.; Harrison, N. Phytoplasma Distribution in Coconut Palms Affected by Lethal Yellowing Disease. Ann. Appl. Biol. 2011, 159, 109–117. [Google Scholar] [CrossRef]
- Perera, P.I.P.; Hocher, V.; Weerakoon, L.K.; Yakandawala, D.M.D.; Fernando, S.C.; Verdeil, J.-L. Early Inflorescence and Floral Development in Cocos nucifera L. (Arecaceae: Arecoideae). S. Afr. J. Bot. 2010, 76, 482–492. [Google Scholar] [CrossRef] [Green Version]
- Islas-Flores, I.; Oropeza, C.; Hernández-Sotomayor, S.M.T. Protein Phosphorylation during Coconut Zygotic Embryo Development1. Plant Physiol. 1998, 118, 257–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Burgos-Canul, Y.Y.; Canto-Canché, B.; Berezovski, M.V.; Mironov, G.; Loyola-Vargas, V.M.; Barba de Rosa, A.P.; Tzec-Simá, M.; Brito-Argáez, L.; Carrillo-Pech, M.; Grijalva-Arango, R.; et al. The Cell Wall Proteome from Two Strains of Pseudocercospora fijiensis with Differences in Virulence. World J. Microbiol. Biotechnol. 2019, 35, 105. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Pérez, A.; Zamora-Briseño, J.A.; Ruiz-May, E.; Pereira-Santana, A.; Elizalde-Contreras, J.M.; Pozos-González, S.; Torres-Irineo, E.; Hernández-López, J.; Gaxiola-Cortés, M.G.; Rodríguez-Canul, R. Proteomic Profiling of the White Shrimp Litopenaeus vannamei (Boone, 1931) Hemocytes Infected with White Spot Syndrome Virus Reveals the Induction of Allergy-Related Proteins. Dev. Comp. Immunol. 2019, 91, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-Supervised Learning for Peptide Identification from Shotgun Proteomics Datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Félix, J.W.; Granados-Alegría, M.I.; Gómez-Tah, R.; Tzec-Simá, M.; Ruíz-May, E.; Canto-Canché, B.; Zamora-Briseño, J.A.; Bojórquez-Velázquez, E.; Oropeza-Salín, C.; Islas-Flores, I. Proteome Landscape during Ripening of Solid Endosperm from Two Different Coconut Cultivars Reveals Contrasting Carbohydrate and Fatty Acid Metabolic Pathway Modulation. Int. J. Mol. Sci. 2023, 24, 10431. https://doi.org/10.3390/ijms241310431
Félix JW, Granados-Alegría MI, Gómez-Tah R, Tzec-Simá M, Ruíz-May E, Canto-Canché B, Zamora-Briseño JA, Bojórquez-Velázquez E, Oropeza-Salín C, Islas-Flores I. Proteome Landscape during Ripening of Solid Endosperm from Two Different Coconut Cultivars Reveals Contrasting Carbohydrate and Fatty Acid Metabolic Pathway Modulation. International Journal of Molecular Sciences. 2023; 24(13):10431. https://doi.org/10.3390/ijms241310431
Chicago/Turabian StyleFélix, Jean Wildort, María Inés Granados-Alegría, Rufino Gómez-Tah, Miguel Tzec-Simá, Eliel Ruíz-May, Blondy Canto-Canché, Jesús Alejandro Zamora-Briseño, Esaú Bojórquez-Velázquez, Carlos Oropeza-Salín, and Ignacio Islas-Flores. 2023. "Proteome Landscape during Ripening of Solid Endosperm from Two Different Coconut Cultivars Reveals Contrasting Carbohydrate and Fatty Acid Metabolic Pathway Modulation" International Journal of Molecular Sciences 24, no. 13: 10431. https://doi.org/10.3390/ijms241310431
APA StyleFélix, J. W., Granados-Alegría, M. I., Gómez-Tah, R., Tzec-Simá, M., Ruíz-May, E., Canto-Canché, B., Zamora-Briseño, J. A., Bojórquez-Velázquez, E., Oropeza-Salín, C., & Islas-Flores, I. (2023). Proteome Landscape during Ripening of Solid Endosperm from Two Different Coconut Cultivars Reveals Contrasting Carbohydrate and Fatty Acid Metabolic Pathway Modulation. International Journal of Molecular Sciences, 24(13), 10431. https://doi.org/10.3390/ijms241310431