The Price of Human Evolution: Cancer-Testis Antigens, the Decline in Male Fertility and the Increase in Cancer
Abstract
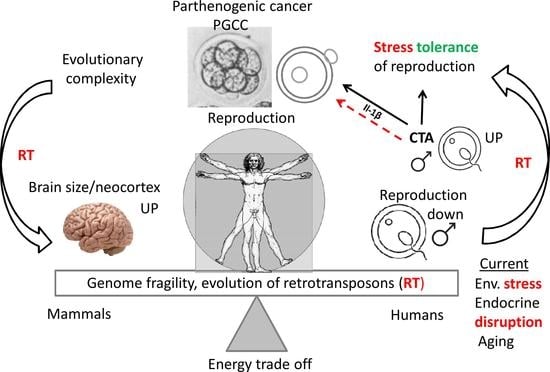
:1. Introduction
2. Evolution of the Human Genome via Segmental Duplications, Adaptation via Transposition and Fragility, CTA Origin, Reactivation of the X-Chromosome in Spermatogenesis, and X-Doubling in Male Cancer
3. The MAGE Protein Oncogene Family Functions in Gametogenesis and the Adaptive Stress Response
- -
- -
- MAGE-B2 (locus Xp21.2) coincides with the position of NORB1, whose doubling or deletion leads to dosage-sensitive sex reversal [81].
- -
- MAGE-C1/C2—involved in p53 suppression and cancer invasion [70].
- -
- -
- The antigenic PRAME is a very important master gene connecting the meiotic giant nucleus of the cancer cell genome network with the MAGE-A cluster (Figure 3A). As a nuclear receptor transcriptional regulator, it activates the embryonal stemness (through OCT4A) and PGC program (through SOX17) [84] and it also downregulates cell differentiation as a dominant retinoic acid receptor signalling gene [85]. So, PRAME can be crucial for cell fate change and soma-germ transition. PRAME is overexpressed not only in many solid tumours but also in myeloid leukaemia [86].
4. STRING Network Analysis of GG Genes in the Human Genome along the Evolutionary Phylostratigraphic Axis
5. Mammalian Placenta Evolution, Immunity, and the Enrichment of the “Female Pregnancy” GO Module in Genotoxically Challenged PGCCs
6. String Network Analysis (Continuation) Strata 10–16
7. The Causal Link between the GG Genes of the 8th Phylogenetic Stratum and CTA-Enriched Strata 12 and 14 in Spermatogenesis and in Cancer Progression
8. The Global Decline of Male Fertility, the Link to Cancer Risk, and the Consequence of Endocrine Disruption
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmad, A.S.; Ormiston-Smith, N.; Sasieni, P.D. Trends in the Lifetime Risk of Developing Cancer in Great Britain: Comparison of Risk for Those Born from 1930 to 1960. Br. J. Cancer 2015, 112, 943–947. [Google Scholar] [CrossRef]
- Weinberg, B.A.; Marshall, J.L. Colon Cancer in Young Adults: Trends and Their Implications. Curr. Oncol. Rep. 2019, 21, 3. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-I.; Lim, H.; Moon, A. Sex Differences in Cancer: Epidemiology, Genetics and Therapy. Biomol. Ther. 2018, 26, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Levine, H.; Jørgensen, N.; Martino-Andrade, A.; Mendiola, J.; Weksler-Derri, D.; Jolles, M.; Pinotti, R.; Swan, S.H. Temporal Trends in Sperm Count: A Systematic Review and Meta-Regression Analysis of Samples Collected Globally in the 20th and 21st Centuries. Hum. Reprod. Update 2023, 29, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Kalejs, M.; Erenpreisa, J. Cancer/testis Antigens and Gametogenesis: A Review and “Brain-Storming” Session. Cancer Cell Int. 2005, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.C.; Münger, K. Role and Clinical Utility of Cancer/Testis Antigens in Head and Neck Squamous Cell Carcinoma. Cancers 2021, 13, 5690. [Google Scholar] [CrossRef]
- Pjanova, D.M.; Vainshelbaum, N.; Salmina, K.; Erenpreisa, J. The Role of the Meiotic Component in Reproduction of B-RAF-Mutated Melanoma: A Review and “brainstorming” Session. In Melanoma; IntechOpen: Rijeka, Croatia, 2021; ISBN 9781838808785. [Google Scholar]
- Vainshelbaum, N.M.; Giuliani, A.; Salmina, K.; Pjanova, D.; Erenpreisa, J. The Transcriptome and Proteome Networks of Malignant Tumours Reveal Atavistic Attractors of Polyploidy-Related Asexual Reproduction. Int. J. Mol. Sci. 2022, 23, 14930. [Google Scholar] [CrossRef]
- Zhang, Y.E.; Long, M. New Genes Contribute to Genetic and Phenotypic Novelties in Human Evolution. Curr. Opin. Genet. Dev. 2014, 29, 90–96. [Google Scholar] [CrossRef] [Green Version]
- Domazet-Loso, T.; Tautz, D. Phylostratigraphic Tracking of Cancer Genes Suggests a Link to the Emergence of Multicellularity in Metazoa. BMC Biol. 2010, 8, 66. [Google Scholar] [CrossRef] [Green Version]
- Trigos, A.S.; Pearson, R.B.; Papenfuss, A.T.; Goode, D.L. Altered Interactions between Unicellular and Multicellular Genes Drive Hallmarks of Transformation in a Diverse Range of Solid Tumors. Proc. Natl. Acad. Sci. USA 2017, 114, 6406–6411. [Google Scholar] [CrossRef]
- Niculescu, V.F. Developmental and Non Developmental Polyploidy in Xenic and Axenic Cultured Stem Cell Lines of Entamoeba Invadens and E. Histolytica. Insights Stem Cells 2016, 2, 14. [Google Scholar]
- Niculescu, V.F. Carcinogenesis: Recent Insights in Protist Stem Cell Biology Lead to a Better Understanding of Atavistic Mechanisms Implied in Cancer Development. MOJ Tumor Res. 2018, 1, 18–29. [Google Scholar] [CrossRef]
- Niculescu, V.F. Cancer Exploits a Late Premetazoan Gene Module Conserved in the Human Genome. Genes Dis. 2023, 10, 1136–1138. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Salmina, K.; Anatskaya, O.; Cragg, M.S. Paradoxes of Cancer: Survival at the Brink. Semin. Cancer Biol. 2022, 81, 119–131. [Google Scholar] [CrossRef]
- Vinogradov, A.E.; Anatskaya, O.V. Systemic Alterations of Cancer Cells and Their Boost by Polyploidization: Unicellular Attractor (UCA) Model. Int. J. Mol. Sci. 2023, 24, 6196. [Google Scholar] [CrossRef] [PubMed]
- Kasperski, A. Life Entrapped in a Network of Atavistic Attractors: How to Find a Rescue. Int. J. Mol. Sci. 2022, 23, 4017. [Google Scholar] [CrossRef] [PubMed]
- Trigos, A.S.; Pearson, R.B.; Papenfuss, A.T.; Goode, D.L. How the Evolution of Multicellularity Set the Stage for Cancer. Br. J. Cancer 2018, 118, 145–152. [Google Scholar] [CrossRef]
- Lineweaver, C.H.; Bussey, K.J.; Blackburn, A.C.; Davies, P.C.W. Cancer Progression as a Sequence of Atavistic Reversions. Bioessays 2021, 43, e2000305. [Google Scholar] [CrossRef]
- Salmina, K.; Vainshelbaum, N.M.; Kreishmane, M.; Inashkina, I.; Cragg, M.S.; Pjanova, D.; Erenpreisa, J. The Role of Mitotic Slippage in Creating a “Female Pregnancy-like System” in a Single Polyploid Giant Cancer Cell. Int. J. Mol. Sci. 2023, 24, 3237. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Vinogradov, A.E. Polyploidy and Myc Proto-Oncogenes Promote Stress Adaptation via Epigenetic Plasticity and Gene Regulatory Network Rewiring. Int. J. Mol. Sci. 2022, 23, 9691. [Google Scholar] [CrossRef]
- Vainshelbaum, N.M.; Zayakin, P.; Kleina, R.; Giuliani, A.; Erenpreisa, J. Meta-Analysis of Cancer Triploidy: Rearrangements of Genome Complements in Male Human Tumors Are Characterized by XXY Karyotypes. Genes 2019, 10, 613. [Google Scholar] [CrossRef] [Green Version]
- Bruggeman, J.W.; Koster, J.; Lodder, P.; Repping, S.; Hamer, G. Massive Expression of Germ Cell-Specific Genes Is a Hallmark of Cancer and a Potential Target for Novel Treatment Development. Oncogene 2018, 37, 5694–5700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinnitsky, V.B. Oncogerminative Hypothesis of Tumor Formation. Med. Hypotheses 1993, 40, 19–27. [Google Scholar] [CrossRef]
- Vinnitsky, V. The Development of a Malignant Tumor Is due to a Desperate Asexual Self-Cloning Process in Which Cancer Stem Cells Develop the Ability to Mimic the Genetic Program of Germline Cells. Intrinsically Disord. Proteins 2014, 2, e29997. [Google Scholar] [CrossRef] [Green Version]
- Erenpreiss Current Concepts of Malignant Growth. In Part A: From a Normal Cell to Cancer; Zvaigzne: Riga, Latvia, 1993.
- Erenpreisa, J.; Salmina, K.; Huna, A.; Jackson, T.R.; Vazquez-Martin, A.; Cragg, M.S. The “Virgin Birth”, Polyploidy, and the Origin of Cancer. Oncoscience 2015, 2, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Mercado-Uribe, I.; Liu, J. Dedifferentiation into Blastomere-like Cancer Stem Cells via Formation of Polyploid Giant Cancer Cells. Oncogene 2017, 36, 4887–4900. [Google Scholar] [CrossRef] [Green Version]
- Moein, S.; Adibi, R.; da Silva Meirelles, L.; Nardi, N.B.; Gheisari, Y. Cancer Regeneration: Polyploid Cells Are the Key Drivers of Tumor Progression. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188408. [Google Scholar] [CrossRef]
- Zhang, S.; Mercado-Uribe, I.; Xing, Z.; Sun, B.; Kuang, J.; Liu, J. Generation of Cancer Stem-like Cells through the Formation of Polyploid Giant Cancer Cells. Oncogene 2014, 33, 116–128. [Google Scholar] [CrossRef]
- Díaz-Carballo, D.; Gustmann, S.; Jastrow, H.; Acikelli, A.H.; Dammann, P.; Klein, J.; Dembinski, U.; Bardenheuer, W.; Malak, S.; Araúzo-Bravo, M.J.; et al. Atypical Cell Populations Associated with Acquired Resistance to Cytostatics and Cancer Stem Cell Features: The Role of Mitochondria in Nuclear Encapsulation. DNA Cell Biol. 2014, 33, 749–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirzayans, R.; Andrais, B.; Murray, D. Roles of Polyploid/Multinucleated Giant Cancer Cells in Metastasis and Disease Relapse Following Anticancer Treatment. Cancers 2018, 10, 118. [Google Scholar] [CrossRef]
- Salmina, K.; Gerashchenko, B.I.; Hausmann, M.; Vainshelbaum, N.M.; Zayakin, P.; Erenpreiss, J.; Freivalds, T.; Cragg, M.S.; Erenpreisa, J. When Three Isn’t a Crowd: A Digyny Concept for Treatment-Resistant, Near-Triploid Human Cancers. Genes 2019, 10, 551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmina, K.; Bojko, A.; Inashkina, I.; Staniak, K.; Dudkowska, M.; Podlesniy, P.; Rumnieks, F.; Vainshelbaum, N.M.; Pjanova, D.; Sikora, E.; et al. “Mitotic Slippage” and Extranuclear DNA in Cancer Chemoresistance: A Focus on Telomeres. Int. J. Mol. Sci. 2020, 21, 2779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weihua, Z.; Lin, Q.; Ramoth, A.J.; Fan, D.; Fidler, I.J. Formation of Solid Tumors by a Single Multinucleated Cancer Cell. Cancer 2011, 117, 4092–4099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundaram, M.; Guernsey, D.L.; Rajaraman, M.M.; Rajaraman, R. Neosis: A Novel Type of Cell Division in Cancer. Cancer Biol. Ther. 2004, 3, 207–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erenpreisa, J.A.; Cragg, M.S.; Fringes, B.; Sharakhov, I.; Illidge, T.M. Release of Mitotic Descendants by Giant Cells from Irradiated Burkitt’s Lymphoma Cell Lines. Cell Biol. Int. 2000, 24, 635–648. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Cragg, M.S. Life-Cycle Features of Tumour Cells. In Evolutionary Biology from Concept to Application; Pontarotti, P., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 61–71. ISBN 9783540789932. [Google Scholar]
- Puig, P.-E.; Guilly, M.-N.; Bouchot, A.; Droin, N.; Cathelin, D.; Bouyer, F.; Favier, L.; Ghiringhelli, F.; Kroemer, G.; Solary, E.; et al. Tumor Cells Can Escape DNA-Damaging Cisplatin through DNA Endoreduplication and Reversible Polyploidy. Cell Biol. Int. 2008, 32, 1031–1043. [Google Scholar] [CrossRef]
- Green, K.J.; Kirk, D.L. Cleavage Patterns, Cell Lineages, and Development of a Cytoplasmic Bridge System in Volvox Embryos. J. Cell Biol. 1981, 91, 743–755. [Google Scholar] [CrossRef]
- Bailey, J.A.; Eichler, E.E. Primate Segmental Duplications: Crucibles of Evolution, Diversity and Disease. Nat. Rev. Genet. 2006, 7, 552–564. [Google Scholar] [CrossRef]
- Kojima, K.K. Structural and Sequence Diversity of Eukaryotic Transposable Elements. Genes Genet. Syst. 2020, 94, 233–252. [Google Scholar] [CrossRef] [Green Version]
- Ravichandran, S.; Subramani, V.K.; Kim, K.K. Z-DNA in the Genome: From Structure to Disease. Biophys. Rev. 2019, 11, 383–387. [Google Scholar] [CrossRef]
- Gu, S.; Yuan, B.; Campbell, I.M.; Beck, C.R.; Carvalho, C.M.B.; Nagamani, S.C.S.; Erez, A.; Patel, A.; Bacino, C.A.; Shaw, C.A.; et al. Alu-Mediated Diverse and Complex Pathogenic Copy-Number Variants within Human Chromosome 17 at p13.3. Hum. Mol. Genet. 2015, 24, 4061–4077. [Google Scholar] [CrossRef] [Green Version]
- Walen, K.H. Epigenetic Enabled Normal Human Cells, Lead to First Cell’s Unique Division System, Driving Tumorigenesis Evolution. J. Cancer Ther. 2021, 13, 48–69. [Google Scholar] [CrossRef]
- Ji, F.; Zhu, X.; Liao, H.; Ouyang, L.; Huang, Y.; Syeda, M.Z.; Ying, S. New Era of Mapping and Understanding Common Fragile Sites: An Updated Review on Origin of Chromosome Fragility. Front. Genet. 2022, 13, 906957. [Google Scholar] [CrossRef] [PubMed]
- Prieler, S.; Chen, D.; Huang, L.; Mayrhofer, E.; Zsótér, S.; Vesely, M.; Mbogning, J.; Klein, F. Spo11 Generates Gaps through Concerted Cuts at Sites of Topological Stress. Nature 2021, 594, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Su, B. Evolutionary Origin and Human-Specific Expansion of a Cancer/testis Antigen Gene Family. Mol. Biol. Evol. 2014, 31, 2365–2375. [Google Scholar] [CrossRef] [Green Version]
- Bailey, J.A.; Carrel, L.; Chakravarti, A.; Eichler, E.E. Molecular Evidence for a Relationship between LINE-1 Elements and X Chromosome Inactivation: The Lyon Repeat Hypothesis. Proc. Natl. Acad. Sci. USA 2000, 97, 6634–6639. [Google Scholar] [CrossRef]
- Belyayev, A. Bursts of Transposable Elements as an Evolutionary Driving Force. J. Evol. Biol. 2014, 27, 2573–2584. [Google Scholar] [CrossRef] [PubMed]
- Roy-Engel, A.M.; Batzer, M.A.; Deininger, P.L. Evolution of Human Retrosequences: Alu. eLS 2008. [Google Scholar] [CrossRef]
- Depienne, C.; Mandel, J.-L. 30 Years of Repeat Expansion Disorders: What Have We Learned and What Are the Remaining Challenges? Am. J. Hum. Genet. 2021, 108, 764–785. [Google Scholar] [CrossRef]
- Plant, T.M.; Zeleznik, A.J. Knobil and Neill’s Physiology of Reproduction; Academic Press: New York, NY, USA, 2014; ISBN 9780123977694. [Google Scholar]
- Cui, X.; Jing, X.; Wu, X.; Yan, M.; Li, Q.; Shen, Y.; Wang, Z. DNA Methylation in Spermatogenesis and Male Infertility. Exp. Ther. Med. 2016, 12, 1973–1979. [Google Scholar] [CrossRef] [Green Version]
- Kohlrausch, F.B.; Berteli, T.S.; Wang, F.; Navarro, P.A.; Keefe, D.L. Control of LINE-1 Expression Maintains Genome Integrity in Germline and Early Embryo Development. Reprod. Sci. 2022, 29, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Blythe, M.J.; Kocer, A.; Rubio-Roldan, A.; Giles, T.; Abakir, A.; Ialy-Radio, C.; Wheldon, L.M.; Bereshchenko, O.; Bruscoli, S.; Kondrashov, A.; et al. LINE-1 Transcription in Round Spermatids Is Associated with Accretion of 5-Carboxylcytosine in Their Open Reading Frames. Commun. Biol. 2021, 4, 691. [Google Scholar] [CrossRef] [PubMed]
- Pittoggi, C.; Renzi, L.; Zaccagnini, G.; Cimini, D.; Degrassi, F.; Giordano, R.; Magnano, A.R.; Lorenzini, R.; Lavia, P.; Spadafora, C. A Fraction of Mouse Sperm Chromatin Is Organized in Nucleosomal Hypersensitive Domains Enriched in Retroposon DNA. J. Cell Sci. 1999, 112, 3537–3548. [Google Scholar] [CrossRef]
- Lazaros, L.; Kitsou, C.; Kostoulas, C.; Bellou, S.; Hatzi, E.; Ladias, P.; Stefos, T.; Markoula, S.; Galani, V.; Vartholomatos, G.; et al. Retrotransposon Expression and Incorporation of Cloned Human and Mouse Retroelements in Human Spermatozoa. Fertil. Steril. 2017, 107, 821–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazazian, H.H., Jr.; Wong, C.; Youssoufian, H.; Scott, A.F.; Phillips, D.G.; Antonarakis, S.E. Haemophilia A Resulting from de Novo Insertion of L1 Sequences Represents a Novel Mechanism for Mutation in Man. Nature 1988, 332, 164–166. [Google Scholar] [CrossRef]
- Boissinot, S.; Chevret, P.; Furano, A.V. L1 (LINE-1) Retrotransposon Evolution and Amplification in Recent Human History. Mol. Biol. Evol. 2000, 17, 915–928. [Google Scholar] [CrossRef] [Green Version]
- Batzer, M.A.; Deininger, P.L. Alu Repeats and Human Genomic Diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef]
- Skowronski, J.; Fanning, T.G.; Singer, M.F. Unit-Length Line-1 Transcripts in Human Teratocarcinoma Cells. Mol. Cell. Biol. 1988, 8, 1385–1397. [Google Scholar]
- Siebenkäs, C.; Chiappinelli, K.B.; Guzzetta, A.A.; Sharma, A.; Jeschke, J.; Vatapalli, R.; Baylin, S.B.; Ahuja, N. Inhibiting DNA Methylation Activates Cancer Testis Antigens and Expression of the Antigen Processing and Presentation Machinery in Colon and Ovarian Cancer Cells. PLoS ONE 2017, 12, e0179501. [Google Scholar] [CrossRef]
- Talon, I.; Janiszewski, A.; Chappell, J.; Vanheer, L.; Pasque, V. Recent Advances in Understanding the Reversal of Gene Silencing During X Chromosome Reactivation. Front. Cell Dev. Biol. 2019, 7, 169. [Google Scholar] [CrossRef]
- Sun, Z.; Prodduturi, N.; Sun, S.Y.; Thompson, E.A.; Kocher, J.-P.A. Chromosome X Genomic and Epigenomic Aberrations and Clinical Implications in Breast Cancer by Base Resolution Profiling. Epigenomics 2015, 7, 1099–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Luo, B.; Xie, X.-X.; Liao, X.-S.; Fu, J.; Ge, Y.-Y.; Li, X.-S.; Guo, G.-S.; Shen, N.; Xiao, S.-W.; et al. Involvement of X-Chromosome Reactivation in Augmenting Cancer Testis Antigens Expression: A Hypothesis. Curr. Med. Sci. 2018, 38, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Foschini, M.P.; Morandi, L.; Sanchez, A.M.; Santoro, A.; Mulè, A.; Zannoni, G.F.; Varga, Z.; Moskovszky, L.; Cucchi, M.C.; Moelans, C.B.; et al. Methylation Profile of X-Chromosome-Related Genes in Male Breast Cancer. Front. Oncol. 2020, 10, 784. [Google Scholar] [CrossRef]
- Isler, K.; van Schaik, C.P. The Expensive Brain: A Framework for Explaining Evolutionary Changes in Brain Size. J. Hum. Evol. 2009, 57, 392–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heldstab, S.A.; Isler, K.; Graber, S.M.; Schuppli, C.; van Schaik, C.P. The Economics of Brain Size Evolution in Vertebrates. Curr. Biol. 2022, 32, R697–R708. [Google Scholar] [CrossRef] [PubMed]
- Florke Gee, R.R.; Chen, H.; Lee, A.K.; Daly, C.A.; Wilander, B.A.; Fon Tacer, K.; Potts, P.R. Emerging Roles of the MAGE Protein Family in Stress Response Pathways. J. Biol. Chem. 2020, 295, 16121–16155. [Google Scholar] [CrossRef]
- Fon Tacer, K.; Montoya, M.C.; Oatley, M.J.; Lord, T.; Oatley, J.M.; Klein, J.; Ravichandran, R.; Tillman, H.; Kim, M.; Connelly, J.P.; et al. MAGE Cancer-Testis Antigens Protect the Mammalian Germline under Environmental Stress. Sci. Adv. 2019, 5, eaav4832. [Google Scholar] [CrossRef] [Green Version]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A Majority of the Cancer/testis Antigens Are Intrinsically Disordered Proteins. J. Cell. Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, P.; Bhattacharya, S.; Achuthan, S.; Behal, A.; Jolly, M.K.; Kotnala, S.; Mohanty, A.; Rangarajan, G.; Salgia, R.; Uversky, V. Intrinsically Disordered Proteins: Critical Components of the Wetware. Chem. Rev. 2022, 122, 6614–6633. [Google Scholar] [CrossRef]
- Uversky, V.N. Introduction to Intrinsically Disordered Proteins (IDPs). Chem. Rev. 2014, 114, 6557–6560. [Google Scholar] [CrossRef]
- Marcar, L.; Maclaine, N.J.; Hupp, T.R.; Meek, D.W. Mage-A Cancer/testis Antigens Inhibit p53 Function by Blocking Its Interaction with Chromatin. Cancer Res. 2010, 70, 10362–10370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Cai, Z.; Jin, G.; Peng, D.; Pan, B.-S.; Zhang, X.; Han, F.; Xu, X.; Lin, H.-K. Abnormal Gametogenesis Induced by p53 Deficiency Promotes Tumor Progression and Drug Resistance. Cell Discov. 2018, 4, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gjerstorff, M.F.; Kock, K.; Nielsen, O.; Ditzel, H.J. MAGE-A1, GAGE and NY-ESO-1 Cancer/testis Antigen Expression during Human Gonadal Development. Hum. Reprod. 2007, 22, 953–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravichandran, R.; Kodali, K.; Peng, J.; Potts, P.R. Regulation of MAGE-A3/6 by the CRL4-DCAF12 Ubiquitin Ligase and Nutrient Availability. EMBO Rep. 2019, 20, e47352. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Chitiprolu, M.; Gagnon, D.; Meng, L.; Perez-Iratxeta, C.; Lagace, D.; Gibbings, D. Autophagy Supports Genomic Stability by Degrading Retrotransposon RNA. Nat. Commun. 2014, 5, 5276. [Google Scholar] [CrossRef] [Green Version]
- Bai, S.; Grossman, G.; Yuan, L.; Lessey, B.A.; French, F.S.; Young, S.L.; Wilson, E.M. Hormone Control and Expression of Androgen Receptor Coregulator MAGE-11 in Human Endometrium during the Window of Receptivity to Embryo Implantation. Mol. Hum. Reprod. 2008, 14, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Bardoni, B.; Zanaria, E.; Guioli, S.; Floridia, G.; Worley, K.C.; Tonini, G.; Ferrante, E.; Chiumello, G.; McCabe, E.R.; Fraccaro, M. A Dosage Sensitive Locus at Chromosome Xp21 Is Involved in Male to Female Sex Reversal. Nat. Genet. 1994, 7, 497–501. [Google Scholar] [CrossRef]
- Urizar-Arenaza, I.; Osinalde, N.; Akimov, V.; Puglia, M.; Muñoa-Hoyos, I.; Gianzo, M.; Rodriguez, J.A.; Ganzabal, T.; Blagoev, B.; Kratchmarova, I.; et al. SPANX-A/D Protein Subfamily Plays a Key Role in Nuclear Organisation, Metabolism and Flagellar Motility of Human Spermatozoa. Sci. Rep. 2020, 10, 5625. [Google Scholar] [CrossRef] [Green Version]
- Lazar, I.; Fabre, B.; Feng, Y.; Khateb, A.; Turko, P.; Martinez Gomez, J.M.; Frederick, D.T.; Levesque, M.P.; Feld, L.; Zhang, G.; et al. SPANX Control of Lamin A/C Modulates Nuclear Architecture and Promotes Melanoma Growth. Mol. Cancer Res. 2020, 18, 1560–1573. [Google Scholar] [CrossRef]
- Nettersheim, D.; Arndt, I.; Sharma, R.; Riesenberg, S.; Jostes, S.; Schneider, S.; Hölzel, M.; Kristiansen, G.; Schorle, H. The Cancer/testis-Antigen PRAME Supports the Pluripotency Network and Represses Somatic and Germ Cell Differentiation Programs in Seminomas. Br. J. Cancer 2016, 115, 454–464. [Google Scholar] [CrossRef] [Green Version]
- Epping, M.T.; Wang, L.; Edel, M.J.; Carlée, L.; Hernandez, M.; Bernards, R. The Human Tumor Antigen PRAME Is a Dominant Repressor of Retinoic Acid Receptor Signaling. Cell 2005, 122, 835–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellat-Deceunynck, C.; Mellerin, M.P.; Labarrière, N.; Jego, G.; Moreau-Aubry, A.; Harousseau, J.L.; Jotereau, F.; Bataille, R. The Cancer Germ-Line Genes MAGE-1, MAGE-3 and PRAME Are Commonly Expressed by Human Myeloma Cells. Eur. J. Immunol. 2000, 30, 803–809. [Google Scholar] [CrossRef]
- Wu, D.-D.; Irwin, D.M.; Zhang, Y.-P. De Novo Origin of Human Protein-Coding Genes. PLoS Genet. 2011, 7, e1002379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verkerk, A.J.; Pieretti, M.; Sutcliffe, J.S.; Fu, Y.H.; Kuhl, D.P.; Pizzuti, A.; Reiner, O.; Richards, S.; Victoria, M.F.; Zhang, F.P. Identification of a Gene (FMR-1) Containing a CGG Repeat Coincident with a Breakpoint Cluster Region Exhibiting Length Variation in Fragile X Syndrome. Cell 1991, 65, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Kidd, S.A.; Lachiewicz, A.; Barbouth, D.; Blitz, R.K.; Delahunty, C.; McBrien, D.; Visootsak, J.; Berry-Kravis, E. Fragile X Syndrome: A Review of Associated Medical Problems. Pediatrics 2014, 134, 995–1005. [Google Scholar] [CrossRef]
- Murray, A.; Webb, J.; Grimley, S.; Conway, G.; Jacobs, P. Studies of FRAXA and FRAXE in Women with Premature Ovarian Failure. J. Med. Genet. 1998, 35, 637–640. [Google Scholar] [CrossRef] [Green Version]
- Hagerman, R.J.; Hagerman, P. Fragile X-Associated Tremor/ataxia Syndrome—Features, Mechanisms and Management. Nat. Rev. Neurol. 2016, 12, 403–412. [Google Scholar] [CrossRef]
- Almeida, L.G.; Sakabe, N.J.; deOliveira, A.R.; Silva, M.C.C.; Mundstein, A.S.; Cohen, T.; Chen, Y.-T.; Chua, R.; Gurung, S.; Gnjatic, S.; et al. CTdatabase: A Knowledge-Base of High-Throughput and Curated Data on Cancer-Testis Antigens. Nucleic Acids Res. 2009, 37, D816–D819. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein–protein Networks, and Functional Characterization of User-Uploaded Gene/measurement Sets. Nucleic Acids Res. 2020, 49, D605–D612. [Google Scholar] [CrossRef]
- Pedersen, T.L. Ggraph: An Implementation of Grammar of Graphics for Graphs and Networks; R Package Version 1.0. 2 2018; RStudio: Boston, MA, USA, 2018. [Google Scholar]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis (2nd Ed.). Measurement 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Cleveland, L.R. The Origin and Evolution of Meiosis. Science 1947, 105, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Raĭkov, I.B. The Protozoan Nucleus, Morphology and Evolution; Springer: New York, NY, USA, 1982; ISBN 9780387816784. [Google Scholar]
- Grishaeva, T.M.; Bogdanov, Y.F. Conservation and Variability of Synaptonemal Complex Proteins in Phylogenesis of Eukaryotes. Int. J. Evol. Biol. 2014, 2014, 856230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finsterbusch, F.; Ravindranathan, R.; Dereli, I.; Stanzione, M.; Tränkner, D.; Tóth, A. Alignment of Homologous Chromosomes and Effective Repair of Programmed DNA Double-Strand Breaks during Mouse Meiosis Require the Minichromosome Maintenance Domain Containing 2 (MCMDC2) Protein. PLoS Genet. 2016, 12, e1006393. [Google Scholar] [CrossRef] [Green Version]
- Verrotti, A.C.; Wharton, R.P. Nanos Interacts with Cup in the Female Germline of Drosophila. Development 2000, 127, 5225–5232. [Google Scholar] [CrossRef]
- De Keuckelaere, E.; Hulpiau, P.; Saeys, Y.; Berx, G.; van Roy, F. Nanos Genes and Their Role in Development and beyond. Cell. Mol. Life Sci. 2018, 75, 1929–1946. [Google Scholar] [CrossRef] [PubMed]
- Cauffman, G.; Van de Velde, H.; Liebaers, I.; Van Steirteghem, A. DAZL Expression in Human Oocytes, Preimplantation Embryos and Embryonic Stem Cells. Mol. Hum. Reprod. 2005, 11, 405–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, M.; Yamaji, M. Primordial Germ Cells in Mice. Cold Spring Harb. Perspect. Biol. 2012, 4, a008375. [Google Scholar] [CrossRef] [Green Version]
- Erenpreisa, J.; Cragg, M.S. Three Steps to the Immortality of Cancer Cells: Senescence, Polyploidy and Self-Renewal. Cancer Cell Int. 2013, 13, 92. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, H.; Bernstein, C. Evolutionary Origin and Adaptive Function of Meiosis. In Meiosis; InTech: Vienna, Austria, 2013; ISBN 9789535111979. [Google Scholar]
- Kumar, M.; Pushpa, K.; Mylavarapu, S.V.S. Splitting the Cell, Building the Organism: Mechanisms of Cell Division in Metazoan Embryos. IUBMB Life 2015, 67, 575–587. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Rosewell, K.L.; Brännström, M.; Akin, J.W.; Curry, T.E., Jr.; Jo, M. FOS, a Critical Downstream Mediator of PGR and EGF Signaling Necessary for Ovulatory Prostaglandins in the Human Ovary. J. Clin. Endocrinol. Metab. 2018, 103, 4241–4252. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.-M.; Tian, H.-F.; Hu, Q.-M.; Meng, Y.; Xiao, H.-B. Evolution and Multiple Origins of Zona Pellucida Genes in Vertebrates. Biol. Open 2018, 7, bio036137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, W.; Chen, C.; De Felici, M.; Shen, W. In Vitro Differentiation of Germ Cells from Stem Cells: A Comparison between Primordial Germ Cells and in Vitro Derived Primordial Germ Cell-like Cells. Cell Death Dis. 2015, 6, e1906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, N.; Hornbaker, K.I.; Rice, D.A.; Karpova, T.; Agbor, V.A.; Heckert, L.L. Sex-Specific Differences in Mouse DMRT1 Expression Are Both Cell Type- and Stage-Dependent during Gonad Development. Biol. Reprod. 2007, 77, 466–475. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Ye, L.; Chen, H. Sex Determination and Maintenance: The Role of DMRT1 and FOXL2. Asian J. Androl. 2017, 19, 619–624. [Google Scholar] [PubMed]
- Domingos, J.A.; Budd, A.M.; Banh, Q.Q.; Goldsbury, J.A.; Zenger, K.R.; Jerry, D.R. Sex-Specific dmrt1 and cyp19a1 Methylation and Alternative Splicing in Gonads of the Protandrous Hermaphrodite Barramundi. PLoS ONE 2018, 13, e0204182. [Google Scholar] [CrossRef]
- Kensler, K.H.; Regan, M.M.; Heng, Y.J.; Baker, G.M.; Pyle, M.E.; Schnitt, S.J.; Hazra, A.; Kammler, R.; Thürlimann, B.; Colleoni, M.; et al. Prognostic and Predictive Value of Androgen Receptor Expression in Postmenopausal Women with Estrogen Receptor-Positive Breast Cancer: Results from the Breast International Group Trial 1-98. Breast Cancer Res. 2019, 21, 30. [Google Scholar] [CrossRef] [Green Version]
- Iacopetta, D.; Rechoum, Y.; Fuqua, S.A. The Role of Androgen Receptor in Breast Cancer. Drug Discov. Today Dis. Mech. 2012, 9, e19–e27. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.; Nonomura, N. Role of Androgen Receptor in Prostate Cancer: A Review. World J. Mens. Health 2019, 37, 288–295. [Google Scholar] [CrossRef]
- Davey, R.A.; Grossmann, M. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. Clin. Biochem. Rev. 2016, 37, 3–15. [Google Scholar]
- van de Lagemaat, L.N.; Landry, J.-R.; Mager, D.L.; Medstrand, P. Transposable Elements in Mammals Promote Regulatory Variation and Diversification of Genes with Specialized Functions. Trends Genet. 2003, 19, 530–536. [Google Scholar] [CrossRef]
- Kwan, L.; Fris, M.; Rodd, F.H.; Rowe, L.; Tuhela, L.; Panhuis, T.M. An Examination of the Variation in Maternal Placentae across the Genus Poeciliopsis (Poeciliidae). J. Morphol. 2015, 276, 707–720. [Google Scholar] [CrossRef]
- Jedrusik, A.; Cox, A.; Wicher, K.B.; Glover, D.M.; Zernicka-Goetz, M. Maternal-Zygotic Knockout Reveals a Critical Role of Cdx2 in the Morula to Blastocyst Transition. Dev. Biol. 2015, 398, 147–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, R.M.; Green, J.A.; Schulz, L.C. The Evolution of the Placenta. Reproduction 2016, 152, R179–R189. [Google Scholar] [CrossRef] [Green Version]
- Zybina, T. Genome Modifications Involved in Developmental Programs of the Placental Trophoblast. In Cytogenetics—Classical and Molecular Strategies for Analysing Heredity Material; IntechOpen: London, UK, 2021; ISBN 9781839689413. [Google Scholar]
- Cross, J.C.; Baczyk, D.; Dobric, N.; Hemberger, M.; Hughes, M.; Simmons, D.G.; Yamamoto, H.; Kingdom, J.C.P. Genes, Development and Evolution of the Placenta. Placenta 2003, 24, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.; Heidmann, O.; Degrelle, S.A.; Vernochet, C.; Lavialle, C.; Letzelter, C.; Bernard-Stoecklin, S.; Hassanin, A.; Mulot, B.; Guillomot, M.; et al. Captured Retroviral Envelope Syncytin Gene Associated with the Unique Placental Structure of Higher Ruminants. Proc. Natl. Acad. Sci. USA 2013, 110, E828–E837. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B. The Placenta Goes Viral: Retroviruses Control Gene Expression in Pregnancy. PLoS Biol. 2018, 16, e3000028. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Ma, H.; Yang, X.; Fan, L.; Tian, S.; Niu, R.; Yan, M.; Zheng, M.; Zhang, S. Cell Fusion-Related Proteins and Signaling Pathways, and Their Roles in the Development and Progression of Cancer. Front. Cell Dev. Biol. 2021, 9, 809668. [Google Scholar] [CrossRef]
- Motwani, M.; Pesiridis, S.; Fitzgerald, K.A. DNA Sensing by the cGAS-STING Pathway in Health and Disease. Nat. Rev. Genet. 2019, 20, 657–674. [Google Scholar] [CrossRef]
- Maxfield, K.E.; Taus, P.J.; Corcoran, K.; Wooten, J.; Macion, J.; Zhou, Y.; Borromeo, M.; Kollipara, R.K.; Yan, J.; Xie, Y.; et al. Comprehensive Functional Characterization of Cancer-Testis Antigens Defines Obligate Participation in Multiple Hallmarks of Cancer. Nat. Commun. 2015, 6, 8840. [Google Scholar] [CrossRef] [Green Version]
- Doghman-Bouguerra, M.; Granatiero, V.; Sbiera, S.; Sbiera, I.; Lacas-Gervais, S.; Brau, F.; Fassnacht, M.; Rizzuto, R.; Lalli, E. FATE1 Antagonizes Calcium- and Drug-Induced Apoptosis by Uncoupling ER and Mitochondria. EMBO Rep. 2016, 17, 1264–1280. [Google Scholar] [CrossRef] [Green Version]
- Kouprina, N.; Noskov, V.N.; Pavlicek, A.; Collins, N.K.; Schoppee Bortz, P.D.; Ottolenghi, C.; Loukinov, D.; Goldsmith, P.; Risinger, J.I.; Kim, J.-H.; et al. Evolutionary Diversification of SPANX-N Sperm Protein Gene Structure and Expression. PLoS ONE 2007, 2, e359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urizar-Arenaza, I.; Benedicto, A.; Perez-Valle, A.; Osinalde, N.; Akimov, V.; Muñoa-Hoyos, I.; Rodriguez, J.A.; Asumendi, A.; Boyano, M.D.; Blagoev, B.; et al. The Multifunctional Role of SPANX-A/D Protein Subfamily in the Promotion of pro-Tumoural Processes in Human Melanoma. Sci. Rep. 2021, 11, 3583. [Google Scholar] [CrossRef] [PubMed]
- Malcher, A.; Stokowy, T.; Berman, A.; Olszewska, M.; Jedrzejczak, P.; Sielski, D.; Nowakowski, A.; Rozwadowska, N.; Yatsenko, A.N.; Kurpisz, M.K. Whole-Genome Sequencing Identifies New Candidate Genes for Nonobstructive Azoospermia. Andrology 2022, 10, 1605–1624. [Google Scholar] [CrossRef]
- Shuvalov, O.; Kizenko, A.; Petukhov, A.; Fedorova, O.; Daks, A.; Barlev, N. Emerging Roles of Cancer-Testis Antigenes, Semenogelin 1 and 2, in Neoplastic Cells. Cell Death Discov. 2021, 7, 97. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Iseli, C.; Venditti, C.A.; Old, L.J.; Simpson, A.J.G.; Jongeneel, C.V. Identification of a New Cancer/testis Gene Family, CT47, among Expressed Multicopy Genes on the Human X Chromosome. Genes Chromosomes Cancer 2006, 45, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Wadelin, F.R.; Fulton, J.; Collins, H.M.; Tertipis, N.; Bottley, A.; Spriggs, K.A.; Falcone, F.H.; Heery, D.M. PRAME Is a Golgi-Targeted Protein That Associates with the Elongin BC Complex and Is Upregulated by Interferon-Gamma and Bacterial PAMPs. PLoS ONE 2013, 8, e58052. [Google Scholar] [CrossRef] [Green Version]
- Larson, E.D.; Marsh, A.J.; Harrison, M.M. Pioneering the Developmental Frontier. Mol. Cell 2021, 81, 1640–1650. [Google Scholar] [CrossRef]
- Quinton, R.J.; DiDomizio, A.; Vittoria, M.A.; Kotýnková, K.; Ticas, C.J.; Patel, S.; Koga, Y.; Vakhshoorzadeh, J.; Hermance, N.; Kuroda, T.S.; et al. Whole-Genome Doubling Confers Unique Genetic Vulnerabilities on Tumour Cells. Nature 2021, 590, 492–497. [Google Scholar] [CrossRef]
- Kristensen, D.M.; Sonne, S.B.; Ottesen, A.M.; Perrett, R.M.; Nielsen, J.E.; Almstrup, K.; Skakkebaek, N.E.; Leffers, H.; Rajpert-De Meyts, E. Origin of Pluripotent Germ Cell Tumours: The Role of Microenvironment during Embryonic Development. Mol. Cell. Endocrinol. 2008, 288, 111–118. [Google Scholar] [CrossRef] [Green Version]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Cooper, O.; Vlotides, G.; Fukuoka, H.; Greene, M.I.; Melmed, S. Expression and Function of ErbB Receptors and Ligands in the Pituitary. Endocr. Relat. Cancer 2011, 18, R197–R211. [Google Scholar] [CrossRef] [Green Version]
- Carlsen, E.; Giwercman, A.; Keiding, N.; Skakkebaek, N.E. Evidence for Decreasing Quality of Semen during Past 50 Years. BMJ 1992, 305, 609–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, H.; Jørgensen, N.; Martino-Andrade, A.; Mendiola, J.; Weksler-Derri, D.; Mindlis, I.; Pinotti, R.; Swan, S.H. Temporal Trends in Sperm Count: A Systematic Review and Meta-Regression Analysis. Hum. Reprod. Update 2017, 23, 646–659. [Google Scholar] [CrossRef] [Green Version]
- Vierula, M.; Niemi, M.; Keiski, A.; Saaranen, M.; Saarikoski, S.; Suominen, J. High and Unchanged Sperm Counts of Finnish Men. Int. J. Androl. 1996, 19, 11–17. [Google Scholar] [CrossRef]
- Fisch, H.; Goluboff, E.T.; Olson, J.H.; Feldshuh, J.; Broder, S.J.; Barad, D.H. Semen Analyses in 1,283 Men from the United States over a 25-Year Period: No Decline in Quality. Fertil. Steril. 1996, 65, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Auger, J.; Kunstmann, J.M.; Czyglik, F.; Jouannet, P. Decline in Semen Quality among Fertile Men in Paris during the Past 20 Years. N. Engl. J. Med. 1995, 332, 281–285. [Google Scholar] [CrossRef]
- Irvine, S.; Cawood, E.; Richardson, D.; MacDonald, E.; Aitken, J. Evidence of Deteriorating Semen Quality in the United Kingdom: Birth Cohort Study in 577 Men in Scotland over 11 Years. BMJ 1996, 312, 467–471. [Google Scholar] [CrossRef] [Green Version]
- Van Waeleghem, K.; De Clercq, N.; Vermeulen, L.; Schoonjans, F.; Comhaire, F. Deterioration of Sperm Quality in Young Healthy Belgian Men. Hum. Reprod. 1996, 11, 325–329. [Google Scholar] [CrossRef] [Green Version]
- Swan, S.H.; Elkin, E.P.; Fenster, L. The Question of Declining Sperm Density Revisited: An Analysis of 101 Studies Published 1934-1996. Environ. Health Perspect. 2000, 108, 961–966. [Google Scholar] [CrossRef]
- Lokeshwar, S.D.; Patel, P.; Fantus, R.J.; Halpern, J.; Chang, C.; Kargi, A.Y.; Ramasamy, R. Decline in Serum Testosterone Levels Among Adolescent and Young Adult Men in the USA. Eur. Urol. Focus 2021, 7, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Bloom, M.S.; Whitcomb, B.W.; Chen, Z.; Ye, A.; Kannan, K.; Buck Louis, G.M. Associations between Urinary Phthalate Concentrations and Semen Quality Parameters in a General Population. Hum. Reprod. 2015, 30, 2645–2657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gore, A.C.; Chappell, V.A.; Fenton, S.E.; Flaws, J.A.; Nadal, A.; Prins, G.S.; Toppari, J.; Zoeller, R.T. EDC-2: The Endocrine Society’s Second Scientific Statement on Endocrine-Disrupting Chemicals. Endocr. Rev. 2015, 36, E1–E150. [Google Scholar] [CrossRef]
- Chiu, Y.-H.; Gaskins, A.J.; Williams, P.L.; Mendiola, J.; Jørgensen, N.; Levine, H.; Hauser, R.; Swan, S.H.; Chavarro, J.E. Intake of Fruits and Vegetables with Low-to-Moderate Pesticide Residues Is Positively Associated with Semen-Quality Parameters among Young Healthy Men. J. Nutr. 2016, 146, 1084–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.-H.; Shi, Z.-D.; Yu, J.-C.; Zhang, Y.-P.; Wang, L.-G.; Qiu, Y. Scrotal Heat Stress Causes Sperm Chromatin Damage and Cysteinyl Aspartate-Spicific Proteinases 3 Changes in Fertile Men. J. Assist. Reprod. Genet. 2015, 32, 747–755. [Google Scholar] [CrossRef] [Green Version]
- Afeiche, M.; Williams, P.L.; Mendiola, J.; Gaskins, A.J.; Jørgensen, N.; Swan, S.H.; Chavarro, J.E. Dairy Food Intake in Relation to Semen Quality and Reproductive Hormone Levels among Physically Active Young Men. Hum. Reprod. 2013, 28, 2265–2275. [Google Scholar] [CrossRef]
- Jensen, T.K.; Heitmann, B.L.; Blomberg Jensen, M.; Halldorsson, T.I.; Andersson, A.-M.; Skakkebæk, N.E.; Joensen, U.N.; Lauritsen, M.P.; Christiansen, P.; Dalgård, C.; et al. High Dietary Intake of Saturated Fat Is Associated with Reduced Semen Quality among 701 Young Danish Men from the General Population. Am. J. Clin. Nutr. 2013, 97, 411–418. [Google Scholar] [CrossRef] [Green Version]
- Gollenberg, A.L.; Liu, F.; Brazil, C.; Drobnis, E.Z.; Guzick, D.; Overstreet, J.W.; Redmon, J.B.; Sparks, A.; Wang, C.; Swan, S.H. Semen Quality in Fertile Men in Relation to Psychosocial Stress. Fertil. Steril. 2010, 93, 1104–1111. [Google Scholar] [CrossRef]
- Nordkap, L.; Jensen, T.K.; Hansen, Å.M.; Lassen, T.H.; Bang, A.K.; Joensen, U.N.; Blomberg Jensen, M.; Skakkebæk, N.E.; Jørgensen, N. Psychological Stress and Testicular Function: A Cross-Sectional Study of 1,215 Danish Men. Fertil. Steril. 2016, 105, 174–187.e1–e2. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Harlev, A.; Agarwal, A.; Esteves, S.C. Cigarette Smoking and Semen Quality: A New Meta-Analysis Examining the Effect of the 2010 World Health Organization Laboratory Methods for the Examination of Human Semen. Eur. Urol. 2016, 70, 635–645. [Google Scholar] [CrossRef]
- Sermondade, N.; Faure, C.; Fezeu, L.; Shayeb, A.G.; Bonde, J.P.; Jensen, T.K.; Van Wely, M.; Cao, J.; Martini, A.C.; Eskandar, M.; et al. BMI in Relation to Sperm Count: An Updated Systematic Review and Collaborative Meta-Analysis. Hum. Reprod. Update 2013, 19, 221–231. [Google Scholar] [CrossRef]
- Eisenberg, M.L.; Kim, S.; Chen, Z.; Sundaram, R.; Schisterman, E.F.; Buck Louis, G.M. The Relationship between Male BMI and Waist Circumference on Semen Quality: Data from the LIFE Study. Hum. Reprod. 2014, 29, 193–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordkap, L.; Joensen, U.N.; Blomberg Jensen, M.; Jørgensen, N. Regional Differences and Temporal Trends in Male Reproductive Health Disorders: Semen Quality May Be a Sensitive Marker of Environmental Exposures. Mol. Cell. Endocrinol. 2012, 355, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Puzuka, A.; Alksere, B.; Gailite, L.; Erenpreiss, J. Idiopathic Infertility as a Feature of Genome Instability. Life 2021, 11, 628. [Google Scholar] [CrossRef] [PubMed]
- Salonia, A.; Matloob, R.; Gallina, A.; Abdollah, F.; Saccà, A.; Briganti, A.; Suardi, N.; Colombo, R.; Rocchini, L.; Guazzoni, G.; et al. Are Infertile Men Less Healthy than Fertile Men? Results of a Prospective Case-Control Survey. Eur. Urol. 2009, 56, 1025–1031. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, M.L.; Li, S.; Cullen, M.R.; Baker, L.C. Increased Risk of Incident Chronic Medical Conditions in Infertile Men: Analysis of United States Claims Data. Fertil. Steril. 2016, 105, 629–636. [Google Scholar] [CrossRef] [Green Version]
- Glazer, C.H.; Bonde, J.P.; Eisenberg, M.L.; Giwercman, A.; Hærvig, K.K.; Rimborg, S.; Vassard, D.; Pinborg, A.; Schmidt, L.; Bräuner, E.V. Male Infertility and Risk of Nonmalignant Chronic Diseases: A Systematic Review of the Epidemiological Evidence. Semin. Reprod. Med. 2017, 35, 282–290. [Google Scholar]
- Brubaker, W.D.; Li, S.; Baker, L.C.; Eisenberg, M.L. Increased Risk of Autoimmune Disorders in Infertile Men: Analysis of US Claims Data. Andrology 2018, 6, 94–98. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, M.L.; Li, S.; Brooks, J.D.; Cullen, M.R.; Baker, L.C. Increased Risk of Cancer in Infertile Men: Analysis of U.S. Claims Data. J. Urol. 2015, 193, 1596–1601. [Google Scholar] [CrossRef]
- Ringbäck Weitoft, G.; Burström, B.; Rosén, M. Premature Mortality among Lone Fathers and Childless Men. Soc. Sci. Med. 2004, 59, 1449–1459. [Google Scholar] [CrossRef]
- Glazer, C.H.; Eisenberg, M.L.; Tøttenborg, S.S.; Giwercman, A.; Flachs, E.M.; Bräuner, E.V.; Vassard, D.; Pinborg, A.; Schmidt, L.; Bonde, J.P. Male Factor Infertility and Risk of Death: A Nationwide Record-Linkage Study. Hum. Reprod. 2019, 34, 2266–2273. [Google Scholar] [CrossRef] [PubMed]
- Skakkebaek, N.E.; Rajpert-De Meyts, E.; Main, K.M. Testicular Dysgenesis Syndrome: An Increasingly Common Developmental Disorder with Environmental Aspects. Hum. Reprod. 2001, 16, 972–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho Henriques, M.; Loureiro, S.; Fardilha, M.; Teresa Herdeiro, M. The Role of Endocrine-Disrupting Chemicals in Male Fertility Decline. In Male Reproductive Health; IntechOpen: Rijeka, Croatia, 2020; ISBN 9781789856477. [Google Scholar]
- Tsuchiya, M.; Giuliani, A.; Zimatore, G.; Erenpreisa, J.; Yoshikawa, K. A Unified Genomic Mechanism of Cell-Fate Change. Results Probl. Cell Differ. 2022, 70, 35–69. [Google Scholar]
- Erenpreisa, J.; Giuliani, A. Resolution of Complex Issues in Genome Regulation and Cancer Requires Non-Linear and Network-Based Thermodynamics. Int. J. Mol. Sci. 2019, 21, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavia, P.; Sciamanna, I.; Spadafora, C. An Epigenetic LINE-1-Based Mechanism in Cancer. Int. J. Mol. Sci. 2022, 23, 14610. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. |














Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erenpreisa, J.; Vainshelbaum, N.M.; Lazovska, M.; Karklins, R.; Salmina, K.; Zayakin, P.; Rumnieks, F.; Inashkina, I.; Pjanova, D.; Erenpreiss, J. The Price of Human Evolution: Cancer-Testis Antigens, the Decline in Male Fertility and the Increase in Cancer. Int. J. Mol. Sci. 2023, 24, 11660. https://doi.org/10.3390/ijms241411660
Erenpreisa J, Vainshelbaum NM, Lazovska M, Karklins R, Salmina K, Zayakin P, Rumnieks F, Inashkina I, Pjanova D, Erenpreiss J. The Price of Human Evolution: Cancer-Testis Antigens, the Decline in Male Fertility and the Increase in Cancer. International Journal of Molecular Sciences. 2023; 24(14):11660. https://doi.org/10.3390/ijms241411660
Chicago/Turabian StyleErenpreisa, Jekaterina, Ninel Miriam Vainshelbaum, Marija Lazovska, Roberts Karklins, Kristine Salmina, Pawel Zayakin, Felikss Rumnieks, Inna Inashkina, Dace Pjanova, and Juris Erenpreiss. 2023. "The Price of Human Evolution: Cancer-Testis Antigens, the Decline in Male Fertility and the Increase in Cancer" International Journal of Molecular Sciences 24, no. 14: 11660. https://doi.org/10.3390/ijms241411660
APA StyleErenpreisa, J., Vainshelbaum, N. M., Lazovska, M., Karklins, R., Salmina, K., Zayakin, P., Rumnieks, F., Inashkina, I., Pjanova, D., & Erenpreiss, J. (2023). The Price of Human Evolution: Cancer-Testis Antigens, the Decline in Male Fertility and the Increase in Cancer. International Journal of Molecular Sciences, 24(14), 11660. https://doi.org/10.3390/ijms241411660