The Lung Microbiome in COPD and Lung Cancer: Exploring the Potential of Metal-Based Drugs
Abstract
:1. Introduction
2. The Healthy Lung Microbiome
3. The Lung Microbiome in Lung Disease
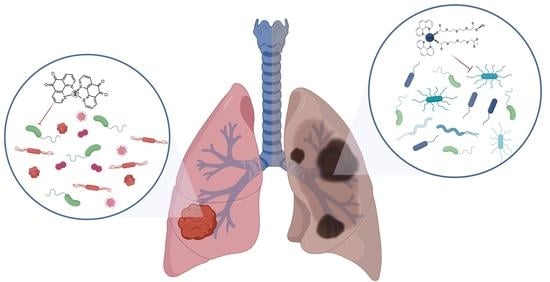
3.1. Chronic Obstructive Pulmonary Disease (COPD)
3.2. Lung Cancer
4. Novel Therapeutic Strategies: Metal-Based Drugs
5. Metal Drugs as Microbiome Modulators
5.1. Bismuth (Bi)
5.2. Gold (Au)
5.3. Silver (Ag)
6. 1,10-Phenanthroline and Its Metal Complexes
7. Mechanisms of Metal-Phen Complexes
7.1. The Bacterial Cell Envelope and Activity of Metal-Phen Complexes
7.2. DNA as an Antibacterial Target for Metal-Phen Complexes
7.3. The Activity of Metal-Phen Complexes on Biofilms
8. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aggarwal, N.; Kitano, S.; Puah, G.R.Y.; Kittelmann, S.; Hwang, I.Y.; Chang, M.W. Microbiome and Human Health: Current Understanding, Engineering, and Enabling Technologies. Chem. Rev. 2023, 123, 31–72. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Dickson, R.P.; Erb-Downward, J.R.; Martinez, F.J.; Huffnagle, G.B. The Microbiome and the Respiratory Tract. Annu. Rev. Physiol. 2016, 78, 481–504. [Google Scholar] [CrossRef] [Green Version]
- Wensel, C.R.; Pluznick, J.L.; Salzberg, S.L.; Sears, C.L. Next-generation sequencing: Insights to advance clinical investigations of the microbiome. J. Clin. Investig. 2022, 132, 154944. [Google Scholar] [CrossRef]
- Wypych, T.P.; Wickramasinghe, L.C.; Marsland, B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019, 20, 1279–1290. [Google Scholar] [CrossRef]
- Kogut, M.H.; Lee, A.; Santin, E. Microbiome and pathogen interaction with the immune system. Poult. Sci. 2020, 99, 1906–1913. [Google Scholar] [CrossRef]
- Natalini, J.G.; Singh, S.; Segal, L.N. The dynamic lung microbiome in health and disease. Nat. Rev. Microbiol. 2022, 21, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Nuñez, M.; Millares, L.; Pomares, X.; Ferrari, R.; Pérez-Brocal, V.; Gallego, M.; Espasa, M.; Moya, A.; Monsó, E. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J. Clin. Microbiol. 2014, 52, 4217–4223. [Google Scholar] [CrossRef] [Green Version]
- Leng, Q.; Holden, V.K.; Deepak, J.; Todd, N.W.; Jiang, F. Microbiota biomarkers for lung cancer. Diagnostics 2021, 11, 407. [Google Scholar] [CrossRef]
- Gunaydin, G.; Gedik, M.E.; Ayan, S. Photodynamic Therapy for the Treatment and Diagnosis of Cancer—A Review of the Current Clinical Status. Front. Chem. 2021, 9, 686303. [Google Scholar] [CrossRef] [PubMed]
- Perrone, F.; Belluomini, L.; Mazzotta, M.; Bianconi, M.; Di Noia, V.; Meacci, F.; Montrone, M.; Pignataro, D.; Prelaj, A.; Rinaldi, S.; et al. Exploring the role of respiratory microbiome in lung cancer: A systematic review. Crit. Rev. Oncol. Hematol. 2021, 164, 103404. [Google Scholar] [CrossRef] [PubMed]
- Russo, C.; Colaianni, V.; Ielo, G.; Valle, M.S.; Spicuzza, L.; Malaguarnera, L. Impact of Lung Microbiota on COPD. Biomedicines 2022, 10, 1337. [Google Scholar] [CrossRef] [PubMed]
- Safiri, S.; Carson-Chahhoud, K.; Noori, M.; Nejadghaderi, S.A.; Sullman, M.J.M.; Ahmadian Heris, J.; Ansarin, K.; Mansournia, M.A.; Collins, G.S.; Kolahi, A.A.; et al. Burden of chronic obstructive pulmonary disease and its attributable risk factors in 204 countries and territories, 1990–2019: Results from the Global Burden of Disease Study 2019. BMJ 2022, 378, e069679. [Google Scholar] [CrossRef] [PubMed]
- Adeloye, D.; Song, P.; Zhu, Y.; Campbell, H.; Sheikh, A.; Rudan, I. Global, regional, and national prevalence of, and risk factors for, chronic obstructive pulmonary disease (COPD) in 2019: A systematic review and modelling analysis. Lancet Respir. Med. 2022, 10, 447–458. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Hou, W.; Hu, S.; Li, C.; Ma, H.; Wang, Q.; Meng, G.; Guo, T.; Zhang, J. Cigarette Smoke Induced Lung Barrier Dysfunction, EMT, and Tissue Remodeling: A Possible Link between COPD and Lung Cancer. Biomed Res. Int. 2019, 2019, 2025636. [Google Scholar] [CrossRef]
- Parris, B.A.; O’Farrell, H.E.; Fong, K.M.; Yang, I.A. Chronic obstructive pulmonary disease (COPD) and lung cancer: Common pathways for pathogenesis. J. Thorac. Dis. 2019, 11, 2155–2172. [Google Scholar] [CrossRef]
- Zheng, Y.; Huang, Y.; Zheng, X.; Peng, J.; Chen, Y.; Yu, K.; Yang, Y.; Wang, X.; Yang, X.; Qian, J.; et al. Deaths from COPD in patients with cancer: A population-based study. Aging 2021, 13, 12641–12659. [Google Scholar] [CrossRef]
- Taucher, E.; Mykoliuk, I.; Lindenmann, J.; Smolle-Juettner, F.M. Implications of the Immune Landscape in COPD and Lung Cancer: Smoking Versus Other Causes. Front. Immunol. 2022, 13, 846605. [Google Scholar] [CrossRef]
- Wang, G.; Ma, A.; Zhang, L.; Guo, J.; Liu, Q.; Petersen, F.; Wang, Z.; Yu, X. Acute exacerbations of chronic obstructive pulmonary disease in a cohort of Chinese never smokers goes along with decreased risks of recurrent acute exacerbation, emphysema and comorbidity of lung cancer as well as decreased levels of circulating eosinophils and basophils. Front. Med. 2022, 9, 907893. [Google Scholar]
- de Alencar, V.T.L.; Figueiredo, A.B.; Corassa, M.; Gollob, K.J.; Cordeiro de Lima, V.C. Lung cancer in never smokers: Tumor immunology and challenges for immunotherapy. Front. Immunol. 2022, 13, 984349. [Google Scholar] [CrossRef]
- Zhao, G.; Li, X.; Lei, S.; Zhao, H.; Zhang, H.; Li, J. Prevalence of lung cancer in chronic obstructive pulmonary disease: A systematic review and meta-analysis. Front. Oncol. 2022, 12, 947981. [Google Scholar] [CrossRef]
- Park, H.Y.; Kang, D.; Shin, S.H.; Yoo, K.H.; Rhee, C.K.; Suh, G.Y.; Kim, H.; Shim, Y.M.; Guallar, E.; Cho, J.; et al. Chronic obstructive pulmonary disease and lung cancer incidence in never smokers: A cohort study. Thorax 2020, 75, 506–509. [Google Scholar] [CrossRef] [Green Version]
- Belkaid, Y.; Hand, T. Role of the Microbiota in Immunity and inflammation Yasmine. Early Hum. Dev. 2014, 157, 121–141. [Google Scholar]
- Ahn, S.V.; Lee, E.; Park, B.; Jung, J.H.; Park, J.E.; Sheen, S.S.; Park, K.J.; Hwang, S.C.; Park, J.B.; Park, H.S.; et al. Cancer development in patients with COPD: A retrospective analysis of the National Health Insurance Service-National Sample Cohort in Korea. BMC Pulm. Med. 2020, 20, 170. [Google Scholar] [CrossRef] [PubMed]
- Czarnecka-Chrebelska, K.H.; Mukherjee, D.; Maryanchik, S.V.; Rudzinska-Radecka, M. Biological and Genetic Mechanisms of COPD, Its Diagnosis, Treatment, and Relationship with Lung Cancer. Biomedicines 2023, 11, 448. [Google Scholar] [CrossRef] [PubMed]
- Forder, A.; Zhuang, R.; Souza, V.G.P.; Brockley, L.J.; Pewarchuk, M.E.; Telkar, N.; Stewart, G.L.; Benard, K.; Marshall, E.A.; Reis, P.P.; et al. Mechanisms Contributing to the Comorbidity of COPD and Lung Cancer. Int. J. Mol. Sci. 2023, 24, 2859. [Google Scholar] [CrossRef]
- Caramori, G.; Ruggeri, P.; Mumby, S.; Ieni, A.; Lo Bello, F.; Chaminka, V.; Donovan, C.; Andò, F.; Nucera, F.; Coppolino, I.; et al. Molecular links between COPD and lung cancer: New targets for drug discovery? Expert Opin. Ther. Targets 2019, 23, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Durham, A.L.; Adcock, I.M. The relationship between COPD and lung cancer. Lung Cancer 2015, 90, 121–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paudel, K.R.; Dharwal, V.; Patel, V.K.; Galvao, I.; Wadhwa, R.; Malyla, V.; Shen, S.S.; Budden, K.F.; Hansbro, N.G.; Vaughan, A.; et al. Role of Lung Microbiome in Innate Immune Response Associated With Chronic Lung Diseases. Front. Med. 2020, 7, 554. [Google Scholar] [CrossRef]
- Bou Zerdan, M.; Kassab, J.; Meouchy, P.; Haroun, E.; Nehme, R.; Bou Zerdan, M.; Fahed, G.; Petrosino, M.; Dutta, D.; Graziano, S. The Lung Microbiota and Lung Cancer: A Growing Relationship. Cancers 2022, 14, 4813. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, Y.; Li, S.; Peng, Z.; Liu, X.; Chen, J.; Zheng, X. Role of lung and gut microbiota on lung cancer pathogenesis. J. Cancer Res. Clin. Oncol. 2021, 147, 2177–2186. [Google Scholar] [CrossRef]
- Ito, N.; Tsujimoto, H.; Ueno, H.; Xie, Q.; Shinomiya, N. Helicobacter pylori-Mediated Immunity and Signaling Transduction in Gastric Cancer. J. Clin. Med. 2020, 9, 3699. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Sadler, P.J. Metals in Medicine: Therapeutic Agents. In Wiley Encyclopedia of Chemical Biology; Wiley: Hoboken, NJ, USA, 2009; pp. 1–47. [Google Scholar]
- Lemire, J.A.; Harrison, J.J.; Turner, R.J. Antimicrobial activity of metals: Mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 2013, 11, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.; Kavanagh, K.A. Evaluation of metal-based antimicrobial compounds for the treatment of bacterial pathogens. J. Med. Microbiol. 2021, 70, 001363. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.J. Metal-based antimicrobial strategies. Microb. Biotechnol. 2017, 10, 1062–1065. [Google Scholar] [CrossRef] [PubMed]
- Frei, A. Metal complexes, an untapped source of antibiotic potential? Antibiotics 2020, 9, 90. [Google Scholar] [CrossRef] [Green Version]
- Abate, C.; Carnamucio, F.; Giuffrè, O.; Foti, C. Metal-Based Compounds in Antiviral Therapy. Biomolecules 2022, 12, 933. [Google Scholar] [CrossRef]
- Claudel, M.; Schwarte, J.V.; Fromm, K.M. New Antimicrobial Strategies Based on Metal Complexes. Chemistry 2020, 2, 849–899. [Google Scholar] [CrossRef]
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. Third row transition metals for the treatment of cancer. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2015, 373, 20140185. [Google Scholar] [CrossRef] [Green Version]
- Anthony, E.J.; Bolitho, E.M.; Bridgewater, H.E.; Carter, O.W.L.; Donnelly, J.M.; Imberti, C.; Lant, E.C.; Lermyte, F.; Needham, R.J.; Palau, M.; et al. Metallodrugs are unique: Opportunities and challenges of discovery and development. Chem. Sci. 2020, 11, 12888–12917. [Google Scholar] [CrossRef]
- Lucaciu, R.L.; Hangan, A.C.; Sevastre, B.; Oprean, L.S. Metallo-Drugs in Cancer Therapy: Past, Present and Future. Molecules 2022, 27, 6485. [Google Scholar] [CrossRef]
- Diaz-Ochoa, V.E.; Jellbauer, S.; Klaus, S.; Raffatellu, M. Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis. Front. Cell. Infect. Microbiol. 2014, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Fong, W.; Li, Q.; Yu, J. Gut microbiota modulation: A novel strategy for prevention and treatment of colorectal cancer. Oncogene 2020, 39, 4925–4943. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Spiga, L.; Winter, S. Transition metals and host-microbe interactions in the inflamed intestine. BioMetals 2019, 32, 369–384. [Google Scholar] [CrossRef] [PubMed]
- She, P.; Zhou, L.; Li, S.; Liu, Y.; Xu, L.; Chen, L.; Luo, Z.; Wu, Y. Synergistic microbicidal effect of auranofin and antibiotics against planktonic and biofilm-encased S. aureus and E. faecalis. Front. Microbiol. 2019, 10, 2453. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Wang, J.; Gao, Y.; Ren, X.; Rottenberg, M.E.; Lu, J.; Holmgren, A. Synergistic antibacterial activity of silver with antibiotics correlating with the upregulation of the ROS production. Sci. Rep. 2018, 8, 11131. [Google Scholar] [CrossRef] [Green Version]
- Abutaleb, N.S.; Seleem, M.N. Auranofin, at clinically achievable dose, protects mice and prevents recurrence from Clostridioides difficile infection. Sci. Rep. 2020, 10, 7701. [Google Scholar] [CrossRef]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between drugs and the gut microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef]
- Conti, G.; D’Amico, F.; Fabbrini, M.; Brigidi, P.; Barone, M.; Turroni, S. Pharmacomicrobiomics in Anticancer Therapies: Why the Gut Microbiota Should Be Pointed Out. Genes 2023, 14, 55. [Google Scholar] [CrossRef]
- Sommariva, M.; Le Noci, V.; Bianchi, F.; Camelliti, S.; Balsari, A.; Tagliabue, E.; Sfondrini, L. The lung microbiota: Role in maintaining pulmonary immune homeostasis and its implications in cancer development and therapy. Cell. Mol. Life Sci. 2020, 77, 2739–2749. [Google Scholar] [CrossRef] [Green Version]
- Moffatt, M.F.; Cookson, W.O. The Lung Microbiome in Health and Respiratory Diseases. Clin. Pulm. Med. 2017, 17, 525–529. [Google Scholar] [CrossRef]
- Pattaroni, C.; Watzenboeck, M.L.; Schneidegger, S.; Kieser, S.; Wong, N.C.; Bernasconi, E.; Pernot, J.; Mercier, L.; Knapp, S.; Nicod, L.P.; et al. Early-Life Formation of the Microbial and Immunological Environment of the Human Airways. Cell Host Microbe 2018, 24, 857–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 2015, 6, 00037. [Google Scholar] [CrossRef] [Green Version]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal Immunol. 2017, 10, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, 8578. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Xing, Y.; Song, X.; Qian, Y. The impact of lung microbiota dysbiosis on inflammation. Immunology 2020, 159, 156–166. [Google Scholar] [CrossRef] [Green Version]
- Whiteside, S.A.; McGinniss, J.E.; Collman, R.G. The lung microbiome: Progress and promise. J. Clin. Investig. 2021, 131, e150473. [Google Scholar] [CrossRef] [PubMed]
- Yagi, K.; Huffnagle, G.B.; Lukacs, N.W.; Asai, N. The lung microbiome during health and disease. Int. J. Mol. Sci. 2021, 22, 10872. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Gao, J.; Wang, Z. The human lung microbiome—A hidden link between microbes and human health and diseases. iMeta 2022, 1, 33. [Google Scholar] [CrossRef]
- Sze, M.A.; Hogg, J.C.; Sin, D.D. Bacterial microbiome of lungs in COPD. Int. J. COPD 2014, 9, 229–238. [Google Scholar]
- Ramsheh, M.Y.; Haldar, K.; Esteve-Codina, A.; Purser, L.F.; Richardson, M.; Müller-Quernheim, J.; Greulich, T.; Nowinski, A.; Barta, I.; Stendardo, M.; et al. Lung microbiome composition and bronchial epithelial gene expression in patients with COPD versus healthy individuals: A bacterial 16S rRNA gene sequencing and host transcriptomic analysis. Lancet Microbe 2021, 2, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Whelan, F.J.; Heirali, A.A.; Rossi, L.; Rabin, H.R.; Parkins, M.D.; Surette, M.G. Longitudinal sampling of the lung microbiota in individuals with cystic fibrosis. PLoS ONE 2017, 12, 0172811. [Google Scholar] [CrossRef] [Green Version]
- Cox, M.J.; Turek, E.M.; Hennessy, C.; Mirza, G.K.; James, P.L.; Coleman, M.; Jones, A.; Wilson, R.; Bilton, D.; Cookson, W.O.C.; et al. Longitudinal assessment of sputum microbiome by sequencing of the 16S rRNA gene in non-cystic fibrosis bronchiectasis patients. PLoS ONE 2017, 12, 0170622. [Google Scholar] [CrossRef] [Green Version]
- Hong, B.; Paulson, J.N.; Stine, O.C.; Weinstock, G.M.; Cervantes, J.L. Meta-analysis of the lung microbiota in pulmonary tuberculosis. Tuberculosis 2018, 109, 102–108. [Google Scholar] [CrossRef]
- Tsay, J.C.J.; Wu, B.G.; Badri, M.H.; Clemente, J.C.; Shen, N.; Meyn, P.; Li, Y.; Yie, T.A.; Lhakhang, T.; Olsen, E.; et al. Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am. J. Respir. Crit. Care Med. 2018, 198, 1188–1198. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.A.; Knight, R. Species divergence and the measurement of microbial diversity. FEMS Microbiol. Rev. 2008, 32, 557–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kriss, M.; Hazleton, K.Z.; Nusbacherd, N.M.; Martine, C.G.; Lozuponed, C.A. Low Diversity Gut Microbiota Dysbiosis: Drivers, Functional Implications and Recovery. Curr Opin Microbiol 2018, 44, 34–40. [Google Scholar] [CrossRef]
- Segal, L.N.; Clemente, J.C.; Tsay, J.C.J.; Koralov, S.B.; Keller, B.C.; Wu, B.G.; Li, Y.; Shen, N.; Ghedin, E.; Morris, A.; et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 2016, 1, 16031. [Google Scholar] [CrossRef] [Green Version]
- Gustafson, A.M.; Soldi, R.; Anderlind, C.; Scholand, M.B.; Qian, J.; Zhang, X.; Cooper, K.; Walker, D.; Mcwilliams, A.; Gang, L.; et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci. Transl. Med. 2010, 2, 2625. [Google Scholar] [CrossRef] [Green Version]
- Jungnickel, C.; Schmidt, L.H.; Bittigkoffer, L.; Wolf, L.; Wolf, A.; Ritzmann, F.; Kamyschnikow, A.; Herr, C.; Menger, M.D.; Spieker, T.; et al. IL-17C mediates the recruitment of tumor-associated neutrophils and lung tumor growth. Oncogene 2017, 36, 4182–4190. [Google Scholar] [CrossRef] [PubMed]
- Ritzmann, F.; Lunding, L.P.; Bals, R.; Wegmann, M.; Beisswenger, C. IL-17 Cytokines and Chronic Lung Diseases. Cells 2022, 11, 2132. [Google Scholar] [CrossRef]
- Segal, L.N.; Clemente, J.C.; Li, Y.; Ruan, C.; Cao, J.; Danckers, M.; Morris, A.; Tapyrik, S.; Wu, B.G.; Diaz, P.; et al. Anaerobic Bacterial Fermentation Products Increase Tuberculosis Risk in Antiretroviral-Drug-Treated HIV Patients. Cell Host Microbe 2017, 21, 530–537. [Google Scholar] [CrossRef] [Green Version]
- Nazir, S.A.; Erbland, M.L. Chronic Obstructive Pulmonary Disease An Update on Diagnosis and Management Issues in Older Adults. Drugs Aging Vol. 2009, 26, 813–831. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, B.D.; Morrow, J.D.; Wang, X.W.; Liu, Y.Y.; DeMeo, D.L.; Hersh, C.P.; Celli, B.R.; Bueno, R.; Criner, G.J.; Silverman, E.K.; et al. Identifying chronic obstructive pulmonary disease from integrative omics and clustering in lung tissue. BMC Pulm. Med. 2023, 23, 115. [Google Scholar] [CrossRef]
- Kim, V.; Criner, G.J. Chronic bronchitis and chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2013, 187, 228–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, V.; Criner, G.J. The Chronic Bronchitis Phenotype in COPD: Features and Implications Victor. Curr. Opin. Pulm. Med. 2016, 21, 133–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erb-Downward, J.R.; Thompson, D.L.; Han, M.K.; Freeman, C.M.; McCloskey, L.; Schmidt, L.A.; Young, V.B.; Toews, G.B.; Curtis, J.L.; Sundaram, B.; et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE 2011, 6, 16384. [Google Scholar] [CrossRef] [Green Version]
- Pragman, A.A.; Kim, H.B.; Reilly, C.S.; Wendt, C.; Isaacson, R.E. The Lung Microbiome in Moderate and Severe Chronic Obstructive Pulmonary Disease. PLoS ONE 2012, 7, 47305. [Google Scholar] [CrossRef] [Green Version]
- Han, M.L.K.; Huang, Y.J.; LiPuma, J.J.; Boushey, H.A.; Boucher, R.C.; Cookson, W.O.; Curtis, J.L.; Erb-Downward, J.; Lynch, S.V.; Sethi, S.; et al. Significance of the microbiome in obstructive lung disease. Thorax 2012, 67, 456–463. [Google Scholar] [CrossRef] [Green Version]
- Larsen, J.M.; Musavian, H.S.; Butt, T.M.; Ingvorsen, C.; Thysen, A.H.; Brix, S. Chronic obstructive pulmonary disease and asthma-associated Proteobacteria, but not commensal Prevotella spp., promote Toll-like receptor 2-independent lung inflammation and pathology. Immunology 2014, 144, 333–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaeckle, N.T.; Pragman, A.A.; Pendleton, K.M.; Baldomero, A.K.; Criner, G.J. The oral-lung axis: The impact of oral health on lung health. Respir. Care 2020, 65, 1211–1220. [Google Scholar] [CrossRef]
- Garcia-Nuñez, M.; Marti, S.; Puig, C.; Perez-Brocal, V.; Millares, L.; Santos, S.; Ardanuy, C.; Moya, A.; Linãres, J.; Monsó, E. Bronchial microbiome, PA biofilm-forming capacity and exacerbation in severe COPD patients colonized by P. aeruginosa. Future Microbiol. 2017, 12, 379–392. [Google Scholar] [CrossRef]
- Wang, Z.; Bafadhel, M.; Haldar, K.; Spivak, A.; Mayhew, D.; Miller, B.E.; Tal-Singer, R.; Johnston, S.L.; Ramsheh, M.Y.; Barer, M.R.; et al. Lung microbiome dynamics in COPD exacerbations. Eur. Respir. J. 2016, 47, 1082–1092. [Google Scholar] [CrossRef] [Green Version]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Beck, J.M.; Huffnagle, G.B.; Curtis, J.L. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann. Am. Thorac. Soc. 2015, 12, 821–830. [Google Scholar] [CrossRef] [Green Version]
- Xue, Q.; Xie, Y.; He, Y.; Yu, Y.; Fang, G.; Yu, W.; Wu, J.; Li, J.; Zhao, L.; Deng, X.; et al. Lung microbiome and cytokine profiles in different disease states of COPD: A cohort study. Sci. Rep. 2023, 13, 5715. [Google Scholar] [CrossRef]
- Dicker, A.J.; Huang, J.T.J.; Lonergan, M.; Keir, H.R.; Fong, C.J.; Tan, B.; Cassidy, A.J.; Finch, S.; Mullerova, H.; Miller, B.E.; et al. The sputum microbiome, airway inflammation, and mortality in chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2021, 147, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Singh, R.; Miller, B.E.; Tal-Singer, R.; Van Horn, S.; Tomsho, L.; MacKay, A.; Allinson, J.P.; Webb, A.J.; Brookes, A.J.; et al. Sputum microbiome temporal variability and dysbiosis in chronic obstructive pulmonary disease exacerbations: An analysis of the COPDMAP study. Thorax 2018, 73, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Zheng, D.; Lin, Y.; Liu, Z.; Liang, Z.; Su, J.; Chen, R.; Zhou, H.; Wang, Z. Association of sputum microbiome with clinical outcome of initial antibiotic treatment in hospitalized patients with acute exacerbations of COPD. Pharmacol. Res. 2020, 160, 105095. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, Y.; Yan, Z.; Liu, H.; Chen, B.; Liang, Z.; Wang, F.; Miller, B.E.; Tal-Singer, R.; Yi, X.; et al. Multi-omic meta-analysis identifies functional signatures of airway microbiome in chronic obstructive pulmonary disease. ISME J. 2020, 14, 2748–2765. [Google Scholar] [CrossRef]
- MacLeod, M.; Papi, A.; Contoli, M.; Beghé, B.; Celli, B.R.; Wedzicha, J.A.; Fabbri, L.M. Chronic obstructive pulmonary disease exacerbation fundamentals: Diagnosis, treatment, prevention and disease impact. Respirology 2021, 26, 532–551. [Google Scholar] [CrossRef] [PubMed]
- Fishbein, S.R.S.; Mahmud, B.; Dantas, G. Antibiotic perturbations to the gut microbiome. Nat. Rev. Microbiol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.; Guarner, F.; Bustos Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as Major Disruptors of Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Bajinka, O.; Jarju, P.O.; Tan, Y.; Taal, A.M.; Ozdemir, G. The varying effects of antibiotics on gut microbiota. AMB Express 2021, 11, 116. [Google Scholar] [CrossRef] [PubMed]
- Anthony, W.E.; Wang, B.; Sukhum, K.V.; D’Souza, A.W.; Hink, T.; Cass, C.; Seiler, S.; Reske, K.A.; Coon, C.; Dubberke, E.R.; et al. Acute and persistent effects of commonly used antibiotics on the gut microbiome and resistome in healthy adults. Cell Rep. 2022, 39, 110649. [Google Scholar] [CrossRef]
- Dickson, R.; Martinez, F.; Huffnagle, G. The Role of the Microbiome in Exacerbations of Chronic Lung Diseases. Hosp. Peer Rev. 2014, 384, 691–702. [Google Scholar] [CrossRef] [Green Version]
- Assefa, M. Multi-drug resistant gram-negative bacterial pneumonia: Etiology, risk factors, and drug resistance patterns. Pneumonia 2022, 14, 4. [Google Scholar] [CrossRef]
- Merker, M.; Tueffers, L.; Vallier, M.; Groth, E.E.; Sonnenkalb, L.; Unterweger, D.; Baines, J.F.; Niemann, S.; Schulenburg, H. Evolutionary Approaches to Combat Antibiotic Resistance: Opportunities and Challenges for Precision Medicine. Front. Immunol. 2020, 11, 1938. [Google Scholar] [CrossRef]
- Toraldo, D.M.; Conte, L. Influence of the Lung Microbiota Dysbiosis in Chronic Obstructive Pulmonary Disease Exacerbations: The Controversial Use of Corticosteroid and Antibiotic Treatments and the Role of Eosinophils as a Disease Marker. J. Clin. Med. Res. 2019, 11, 667–675. [Google Scholar] [CrossRef] [Green Version]
- Ren, L.; Zhang, R.; Rao, J.; Xiao, Y.; Zhang, Z.; Yang, B.; Cao, D.; Zhong, H.; Ning, P.; Shang, Y.; et al. Transcriptionally Active Lung Microbiome and Its Association with Bacterial Biomass and Host Inflammatory Status. mSystems 2018, 3, 00199-18. [Google Scholar] [CrossRef] [Green Version]
- Von, S.T.; Seng, H.L.; Lee, H.B.; Ng, S.W.; Kitamura, Y.; Chikira, M.; Ng, C.H. DNA molecular recognition and cellular selectivity of anticancer metal(II) complexes of ethylenediaminediacetate and phenanthroline: Multiple targets. J. Biol. Inorg. Chem. 2012, 17, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Pragman, A.A.; Knutson, K.A.; Gould, T.J.; Isaacson, R.E.; Reilly, C.S.; Wendt, C.H. Chronic obstructive pulmonary disease upper airway microbiota alpha diversity is associated with exacerbation phenotype: A case-control observational study. Respir. Res. 2019, 20, 114. [Google Scholar] [CrossRef] [Green Version]
- Sze, M.A.; Dimitriu, P.A.; Hayashi, S.; Elliott, W.M.; McDonough, J.E.; Gosselink, J.V.; Cooper, J.; Sin, D.D.; Mohn, W.W.; Hogge, J.C. The lung tissue microbiome in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2012, 185, 1073–1080. [Google Scholar] [CrossRef] [Green Version]
- Zhu, K.; Zhou, S.; Xu, A.; Sun, L.; Li, M.; Jiang, H.; Zhang, B.; Zeng, D.; Fei, G.; Wang, R. Microbiota Imbalance Contributes to COPD Deterioration by Enhancing IL-17a Production via miR-122 and miR-30a. Mol. Ther. Nucleic Acids 2020, 22, 520–529. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Anusewicz, D.; Orzechowska, M.; Bednarek, A.K. Lung squamous cell carcinoma and lung adenocarcinoma differential gene expression regulation through pathways of Notch, Hedgehog, Wnt, and ErbB signalling. Sci. Rep. 2020, 10, 21128. [Google Scholar] [CrossRef]
- Testa, U.; Castelli, G.; Pelosi, E. Lung cancers: Molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers 2018, 10, 248. [Google Scholar] [CrossRef]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.-K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [PubMed]
- Eapen, M.S.; Hansbro, P.M.; Larsson-Callerfelt, A.K.; Jolly, M.K.; Myers, S.; Sharma, P.; Jones, B.; Rahman, M.A.; Markos, J.; Chia, C.; et al. Chronic Obstructive Pulmonary Disease and Lung Cancer: Underlying Pathophysiology and New Therapeutic Modalities. Drugs 2018, 78, 1717–1740. [Google Scholar] [CrossRef]
- Gomes, S.; Cavadas, B.; Ferreira, J.C.; Marques, P.I.; Monteiro, C.; Sucena, M.; Sousa, C.; Vaz Rodrigues, L.; Teixeira, G.; Pinto, P.; et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci. Rep. 2019, 9, 12838. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Yang, M.; Liu, J.; Gao, R.; Hu, J.; Li, J.; Zhang, L.; Shi, Y.; Guo, H.; Cheng, J.; et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 2015, 5, 3111–3122. [Google Scholar]
- Lee, S.H.; Sung, J.Y.; Yong, D.; Chun, J.; Kim, S.Y.S.K.; Song, J.H.; Chung, K.S.; Kim, E.Y.; Jung, J.Y.; Kang, Y.A.; et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 2016, 102, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Apopa, P.L.; Alley, L.; Penney, R.B.; Arnaoutakis, K.; Steliga, M.A.; Jeffus, S.; Bircan, E.; Gopalan, B.; Jin, J.; Patumcharoenpol, P.; et al. PARP1 is up-regulated in non-small cell lung cancer tissues in the presence of the Cyanobacterial toxin microcystin. Front. Microbiol. 2018, 9, 1757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, I.; Araya, P.; Roj, A. Helicobacter pylori infection and lung cancer: New insights and future challenges. Chin. J. Lung Cancer 2018, 21, 658–662. [Google Scholar]
- Xu, N.; Wang, L.; Li, C.; Ding, C.; Li, C.; Fan, W.; Cheng, C.; Gu, B. Microbiota dysbiosis in lung cancer: Evidence of association and potential mechanisms. Transl. Lung Cancer Res. 2020, 9, 1554–1568. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.N.; Ma, Q.; Ge, Y.; Yi, C.X.; Wei, L.Q.; Tan, J.C.; Chu, Q.; Li, J.Q.; Zhang, P.; Wang, H. Microbiome dysbiosis in lung cancer: From composition to therapy. npj Precis. Oncol. 2020, 4, 33. [Google Scholar] [CrossRef]
- Karakasidis, E.; Kotsiou, O.S.; Gourgoulianis, K.I. Lung and Gut Microbiome in COPD. J. Pers. Med. 2023, 13, 804. [Google Scholar] [CrossRef]
- He, J.Q.; Chen, Q.; Wu, S.J.; Wang, D.Q.; Zhang, S.Y.; Zhang, S.Z.; Chen, R.L.; Wang, J.F.; Wang, Z.; Yu, C.H. Potential Implications of the Lung Microbiota in Patients with Chronic Obstruction Pulmonary Disease and Non-Small Cell Lung Cancer. Front. Cell. Infect. Microbiol. 2022, 12, 937864. [Google Scholar] [CrossRef]
- Yu, G.; Gail, M.H.; Consonni, D.; Carugno, M.; Humphrys, M.; Pesatori, A.C.; Caporaso, N.E.; Goedert, J.J.; Ravel, J.; Landi, M.T. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016, 17, 163. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Su, X.; Yuan, M.; Zhang, S.; Jing, H.; Deng, Q.; Qiu, W.; Dong, H.; Cai, S. The Characterization of Lung Microbiome in Sputum of Lung Cancer Patients with Different Clinicopathology. Am. J. Cancer Res. 2019, 9, 2047–2063. [Google Scholar]
- Najafi, S.; Abedini, F.; Azimzadeh Jamalkandi, S.; Shariati, P.; Ahmadi, A.; Gholami Fesharaki, M. The composition of lung microbiome in lung cancer: A systematic review and meta-analysis. BMC Microbiol. 2021, 21, 315. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.V.; Jakobsson, H.E.; Holmén-Larsson, J.; Schütte, A.; Ermund, A.; Rodríguez-Piñeiro, A.M.; Arike, L.; Wising, C.; Svensson, F.; Bäckhed, F.; et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 2015, 18, 582–592. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, H.; Cheng, L.; Wang, Y.; Zhang, Y.K.; Zhao, M.F.; Liang, G.D.; Zhang, M.C.; Li, Y.G.; Zhao, J.B.; Gao, Y.N.; et al. Dysbiosis of the gut microbiome in lung cancer. Front. Cell. Infect. Microbiol. 2019, 9, 112. [Google Scholar] [CrossRef]
- Wang, Z.; Bai, C.; Hu, T.; Luo, C.; Yu, H.; Ma, X.; Liu, T.; Gu, X. Emerging trends and hotspot in gut–lung axis research from 2011 to 2021: A bibliometrics analysis. Biomed. Eng. Online 2022, 21, 27. [Google Scholar] [CrossRef] [PubMed]
- Enaud, R.; Prevel, R.; Ciarlo, E.; Beaufils, F.; Wieërs, G.; Guery, B.; Delhaes, L. The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front. Cell. Infect. Microbiol. 2020, 10, 9. [Google Scholar] [CrossRef] [Green Version]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S.; et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell 2019, 176, 998–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; García Rodríguez, L.A.; Hernández-Díaz, S. Antibiotic use and the risk of lung cancer. Cancer Epidemiol. Biomarkers Prev. 2008, 17, 1308–1315. [Google Scholar] [CrossRef] [Green Version]
- Francescone, R.; Hou, V.; Grivennikov, S.I. Microbiome, inflammation, and cancer. Cancer J. 2014, 20, 181–189. [Google Scholar] [CrossRef] [Green Version]
- Sadrekarimi, H.; Gardanova, Z.R.; Bakhshesh, M.; Ebrahimzadeh, F.; Yaseri, A.F.; Thangavelu, L.; Hasanpoor, Z.; Zadeh, F.A.; Kahrizi, M.S. Emerging role of human microbiome in cancer development and response to therapy: Special focus on intestinal microflora. J. Transl. Med. 2022, 20, 301. [Google Scholar] [CrossRef]
- Hou, X.; Zheng, Z.; Wei, J.; Zhao, L. Effects of gut microbiota on immune responses and immunotherapy in colorectal cancer. Front. Immunol. 2022, 13, 1030745. [Google Scholar] [CrossRef] [PubMed]
- Sheflin, A.M.; Whitney, A.K.; Weir, T.L. Cancer-Promoting Effects of Microbial Dysbiosis. Curr. Oncol. Rep. 2014, 16, 406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Sharma, P.; Sarma, D.K.; Kumawat, M.; Tiwari, R.; Verma, V.; Nagpal, R.; Kumar, M. Implication of Obesity and Gut Microbiome Dysbiosis in the Etiology of Colorectal Cancer. Cancers 2023, 15, 1913. [Google Scholar] [CrossRef] [PubMed]
- Mills, K.H.G. IL-17 and IL-17-producing cells in protection versus pathology. Nat. Rev. Immunol. 2023, 23, 38–54. [Google Scholar] [CrossRef]
- Rivas-Domínguez, A.; Pastor, N.; Martínez-López, L.; Colón-Pérez, J.; Bermúdez, B.; Orta, M.L. The role of dna damage response in dysbiosis-induced colorectal cancer. Cells 2021, 10, 1934. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, J.; Qi, J.; Jin, Y.; Tong, L. Activation of NADPH/ROS pathway contributes to angiogenesis through JNK signaling in brain endothelial cells. Microvasc. Res. 2020, 131, 104012. [Google Scholar] [CrossRef]
- Ochoa Perez, C.E.; Mirabolfathinejad, S.G.; Venado, A.R.; Evans, S.E.; Gagea, M.; Evans, C.M.; Dickey, B.F.; Moghaddam, S.J. Interleukin 6, but not T helper 2 cytokines, promotes lung carcinogenesis. Cancer Prev. Res. 2011, 4, 51–64. [Google Scholar] [CrossRef] [Green Version]
- Caetano, M.S.; Zhang, H.; Cumpian, A.M.; Gong, L.; Unver, N.; Ostrin, E.J.; Daliri, S.; Chang, S.H.; Ochoa, C.E.; Hanash, S.; et al. IL-6 blockade reprograms the lung tumor microenvironment to limit the development and progression of K-ras mutant lung cancer. Cancer Res. 2016, 76, 3189–3199. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.H.; Mirabolfathinejad, S.G.; Katta, H.; Cumpian, A.M.; Gong, L.; Caetano, M.S.; Moghaddam, S.J.; Dong, C. T helper 17 cells play a critical pathogenic role in lung cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 5664–5669. [Google Scholar] [CrossRef]
- Weeks, J.R.; Staples, K.J.; Spalluto, C.M.; Watson, A.; Wilkinson, T.M.A. The Role of Non-Typeable Haemophilus influenzae Biofilms in Chronic Obstructive Pulmonary Disease. Front. Cell. Infect. Microbiol. 2021, 11, 720742. [Google Scholar] [CrossRef]
- Sriram, K.B.; Cox, A.J.; Sivakumaran, P.; Singh, M.; Watts, A.M.; West, N.P.; Cripps, A.W. Non-typeable Haemophilus influenzae detection in the lower airways of patients with lung cancer and chronic obstructive pulmonary disease. Multidiscip. Respir. Med. 2018, 13, 11. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.C.; Jalalvand, F.; Thegerström, J.; Riesbeck, K. The interplay between immune response and bacterial infection in COPD: Focus Upon non-typeable Haemophilus influenzae. Front. Immunol. 2018, 9, 2530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobili, S.; Mini, E.; Landini, I.; Gabbiani, C.; Casini, A.; Messori, L. Gold compounds as anticancer agents: Chemistry, cellular pharmacology, and preclinical studies. Med. Res. Rev. 2009, 30, 550–580. [Google Scholar] [CrossRef] [PubMed]
- Dilda, P.J.; Hogg, P.J. Arsenical-based cancer drugs. Cancer Treat. Rev. 2007, 33, 542–564. [Google Scholar] [CrossRef]
- Klasen, H.J. A historical review of the use of silver in the treatment of burns. II. Renewed interest for silver. Burns 2000, 26, 131–138. [Google Scholar] [CrossRef]
- Ehrlich, P. About Salvarsan. Abhandlungen über Salvarsan 1912, 2, 547–563. [Google Scholar]
- Ehrlich, P.; Bertheim, A. On the hydrochloric acid 3,3′-diamino-4,4′-dioxyarsenobenzene and its closest relatives. Rep. Ger. Chem. Soc. 1912, 45, 756–766. [Google Scholar]
- Rosenberg, B.; Van Camp, L.; Krigas, T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature 1965, 205, 698–699. [Google Scholar] [CrossRef]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [Green Version]
- Aldossary, S.A. Review on pharmacology of cisplatin: Clinical use, toxicity and mechanism of resistance of cisplatin. Biomed. Pharmacol. J. 2019, 12, 7–15. [Google Scholar] [CrossRef]
- Makovník, M.; Rejleková, K.; Uhrin, I.; Mego, M.; Chovanec, M. Intricacies of Radiographic Assessment in Testicular Germ Cell Tumors. Front. Oncol. 2021, 10, 587523. [Google Scholar] [CrossRef]
- Nieder, C.; Pawinski, A.; Andratschke, N.H. Combined radio- and chemotherapy for non-small cell lung cancer: Systematic review of landmark studies based on acquired citations. Front. Oncol. 2013, 3, 176. [Google Scholar] [CrossRef] [Green Version]
- Tchounwou, P.B.; Dasari, S.; Noubissi, F.K.; Ray, P.; Kumar, S. Advances in our understanding of the molecular mechanisms of action of cisplatin in cancer therapy. J. Exp. Pharmacol. 2021, 13, 303–328. [Google Scholar] [CrossRef]
- Cocetta, V.; Ragazzi, E.; Montopoli, M. Mitochondrial involvement in cisplatin resistance. Int. J. Mol. Sci. 2019, 20, 3384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kryczka, J.; Kryczka, J.; Czarnecka-Chrebelska, K.H.; Brzeziańska-Lasota, E. Molecular mechanisms of chemoresistance induced by cisplatin in NSCLC cancer therapy. Int. J. Mol. Sci. 2021, 22, 8885. [Google Scholar] [CrossRef]
- Dasari, S.; Njiki, S.; Mbemi, A.; Yedjou, C.G.; Tchounwou, P.B. Pharmacological Effects of Cisplatin Combination with Natural Products in Cancer Chemotherapy. Int. J. Mol. Sci. 2022, 23, 1532. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; La Manna, S.; Cipollone, I.; Can, L.; Cozzolino, F. From the Discovery of Targets to Delivery Systems: How to Decipher and Improve the Metallodrugs’ Actions at a Molecular Level. Pharm. Rev. 2023, 15, 1997. [Google Scholar] [CrossRef]
- Gou, Y.; Liu, L.; Liang, H. The developments of metal-based agents against lung cancer Yi. Front. Pharmacol. 2022, 13, 5106–5131. [Google Scholar] [CrossRef]
- Riccardi, C.; Piccolo, M. Metal-Based Complexes in Cancer. Int. J. Mol. Sci. 2023, 24, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shambhwani, D.; Pandey, S.; Singh, J.; Lalhlenmawia, H.; Kumarasamy, M.; Singh, S.K.; Chellappan, D.K.; Gupta, G.; Prasher, P.; et al. Advances in Lung Cancer Treatment Using Nanomedicines. ACS Omega 2022, 8, 10–41. [Google Scholar] [CrossRef]
- Guo, Q.; Liu, L.; Chen, Z.; Fan, Y.; Zhou, Y.; Yuan, Z.; Zhang, W. Current treatments for non-small cell lung cancer. Front. Oncol. 2022, 12, 945102. [Google Scholar] [CrossRef]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef]
- Zhang, C.; Leighl, N.B.; Wu, Y.L.; Zhong, W.Z. Emerging therapies for non-small cell lung cancer. J. Hematol. Oncol. 2019, 12, 45. [Google Scholar] [CrossRef] [Green Version]
- Anand, U.; Dey, A.; Chandel, A.K.S.; Sanyal, R.; Mishra, A.; Pandey, D.K.; De Falco, V.; Upadhyay, A.; Kandimalla, R.; Chaudhary, A.; et al. Cancer chemotherapy and beyond: Current status, drug candidates, associated risks and progress in targeted therapeutics. Genes Dis. 2023, 10, 1367–1401. [Google Scholar] [CrossRef]
- Alessio, E.; Messori, L. NAMI-A and KP1019/1339, two iconic ruthenium anticancer drug candidates face-to-face: A case story in medicinal inorganic chemistry. Molecules 2019, 24, 1995. [Google Scholar] [CrossRef] [Green Version]
- Leijen, S.; Burgers, S.A.; Baas, P.; Pluim, D.; Tibben, M.; Van Werkhoven, E.; Alessio, E.; Sava, G.; Beijnen, J.H.; Schellens, J.H.M. Phase I/II study with ruthenium compound NAMI-A and gemcitabine in patients with non-small cell lung cancer after first line therapy. Investig. New Drugs 2015, 33, 201–214. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, C.Y.; Nam, T.G. Ruthenium complexes as anticancer agents: A brief history and perspectives. Drug Des. Devel. Ther. 2020, 14, 5375–5392. [Google Scholar] [CrossRef] [PubMed]
- Abdalbari, F.H.; Telleria, C.M. The gold complex auranofin: New perspectives for cancer therapy. Discov. Oncol. 2021, 12, 42. [Google Scholar] [CrossRef]
- Gordon, E.M.; Angel, N.L.; Omelchenko, N.; Chua-Alcala, V.S.; Moradkhani, A.; Quon, D.; Wong, S. A Phase I/II Investigation of Safety and Efficacy of Nivolumab and nab-Sirolimus in Patients with a Variety of Tumors with Genetic Mutations in the mTOR Pathway. Anticancer Res. 2023, 43, 1993–2002. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, C.; Martoriati, A.; Pelinski, L.; Cailliau, K. Copper complexes as anticancer agents targeting topoisomerases i and ii. Cancers 2020, 12, 2863. [Google Scholar] [CrossRef]
- Liu, Y.L.; Bager, C.L.; Willumsen, N.; Ramchandani, D.; Kornhauser, N.; Ling, L.; Cobham, M.; Andreopoulou, E.; Cigler, T.; Moore, A.; et al. Tetrathiomolybdate (TM)-associated copper depletion influences collagen remodeling and immune response in the pre-metastatic niche of breast cancer. npj Breast Cancer 2021, 7, 108. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Li, L.; Hou, S.; Yuan, Z.; Li, C.; Zhang, W.; Zheng, L.; Li, X. The Role of Iron in Cancer Progression. Front. Oncol. 2021, 11, 778492. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, S.P.; Gadre, S.; Kamisetti, R.T.; Patra, M. Challenges and opportunities in the development of metal-based anticancer theranostic agents. Biosci. Rep. 2022, 42, BSR20212160. [Google Scholar] [CrossRef]
- Kroschinsky, F.; Stölzel, F.; von Bonin, S.; Beutel, G.; Kochanek, M.; Kiehl, M.; Schellongowski, P. New drugs, new toxicities: Severe side effects of modern targeted and immunotherapy of cancer and their management. Crit. Care 2017, 21, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gou, Y.; Huang, G.J.; Li, J.; Yang, F.; Liang, H. Versatile delivery systems for non-platinum metal-based anticancer therapeutic agents. Coord. Chem. Rev. 2021, 441, 213975. [Google Scholar] [CrossRef]
- Balali-Mood, M.; Naseri, K.; Tahergorabi, Z.; Khazdair, M.R.; Sadeghi, M. Toxic Mechanisms of Five Heavy Metals: Mercury, Lead, Chromium, Cadmium, and Arsenic. Front. Pharmacol. 2021, 12, 643972. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Chen, X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019, 2, 141–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boros, E.; Dyson, P.J.; Gasser, G. Classification of Metal-Based Drugs according to Their Mechanisms of Action. Chem 2020, 6, 41–60. [Google Scholar] [CrossRef]
- Butler, M.S.; Gigante, V.; Sati, H.; Paulin, S.; Al-Sulaiman, L.; Rex, J.H.; Fernandes, P.; Arias, C.A.; Paul, M.; Thwaites, G.E.; et al. Analysis of the Clinical Pipeline of Treatments for Drug-Resistant Bacterial Infections: Despite Progress, More Action Is Needed. Antimicrob. Agents Chemother. 2022, 66, 0199121. [Google Scholar] [CrossRef]
- Reig, S.; Le Gouellec, A.; Bleves, S. What Is New in the Anti–Pseudomonas aeruginosa Clinical Development Pipeline Since the 2017 WHO Alert? Front. Cell. Infect. Microbiol. 2022, 12, 909731. [Google Scholar] [CrossRef]
- Wang, C.; Yang, D.; Wang, Y.; Ni, W. Cefiderocol for the Treatment of Multidrug-Resistant Gram-Negative Bacteria: A Systematic Review of Currently Available Evidence. Front. Pharmacol. 2022, 13, 896971. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Golden, A.R.; Zelenitsky, S.; Wiebe, K.; Lawrence, C.K.; Adam, H.J.; Idowu, T.; Domalaon, R.; Schweizer, F.; Zhanel, M.A.; et al. Cefiderocol: A Siderophore Cephalosporin with Activity Against Carbapenem-Resistant and Multidrug-Resistant Gram-Negative Bacilli. Drugs 2019, 79, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Theuretzbacher, U.; Piddock, L.J.V. Non-traditional Antibacterial Therapeutic Options and Challenges. Cell Host Microbe 2019, 26, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Theuretzbacher, U. Dual-mechanism antibiotics. Nat. Microbiol. 2020, 5, 984–985. [Google Scholar] [CrossRef]
- Rex, J.H.; Fernandez Lynch, H.; Cohen, I.G.; Darrow, J.J.; Outterson, K. Designing development programs for non-traditional antibacterial agents. Nat. Commun. 2019, 10, 3416. [Google Scholar] [CrossRef] [Green Version]
- Langendonk, R.F.; Neill, D.R.; Fothergill, J.L. The Building Blocks of Antimicrobial Resistance in Pseudomonas aeruginosa: Implications for Current Resistance-Breaking Therapies. Front. Cell. Infect. Microbiol. 2021, 11, 665759. [Google Scholar] [CrossRef]
- Nasiri Sovari, S.; Zobi, F. Recent Studies on the Antimicrobial Activity of Transition Metal Complexes of Groups 6–12. Chemistry 2020, 2, 418–452. [Google Scholar] [CrossRef]
- Frei, A.; Zuegg, J.; Elliott, A.G.; Baker, M.; Braese, S.; Brown, C.; Chen, F.; Dowson, C.G.; Dujardin, G.; Jung, N.; et al. Metal complexes as a promising source for new antibiotics. Chem. Sci. 2020, 11, 2627–2639. [Google Scholar] [CrossRef] [Green Version]
- Viganor, L.; Howe, O.; McCarron, P.; McCann, M.; Devereux, M. The Antibacterial Activity of Metal Complexes Containing 1,10- phenanthroline: Potential as Alternative Therapeutics in the Era of Antibiotic Resistance. Curr. Top. Med. Chem. 2016, 17, 1280–1302. [Google Scholar] [CrossRef]
- Tempera, P.J.; Michael, M.; Tageldin, O.; Hasak, S. Gastric Cancer Due to Chronic H. pylori Infection: What We Know and Where We Are Going. Diseases 2022, 10, 57. [Google Scholar] [CrossRef]
- Peek, R.M.; Blaser, M.J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2002, 2, 28–37. [Google Scholar] [CrossRef]
- Suerbaum, S.; Michetti, P. INCE the first culture of. N. Engl. J. Med. 2002, 347, 1175–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robin Warren, J.; Marshall, B. Unidentified Curved Bacilli on Gastric Epithelium in Active Chronic Gastritis. Lancet 1983, 321, 1273–1275. [Google Scholar] [CrossRef]
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liou, J.M.; Malfertheiner, P.; Lee, Y.C.; Sheu, B.S.; Sugano, K.; Cheng, H.C.; Yeoh, K.G.; Hsu, P.I.; Goh, K.L.; Mahachai, V.; et al. Screening and eradication of Helicobacter pylori for gastric cancer prevention: The Taipei global consensus. Gut 2020, 69, 2093–2112. [Google Scholar] [CrossRef] [PubMed]
- Piscione, M.; Mazzone, M.; Di Marcantonio, M.C.; Muraro, R.; Mincione, G. Eradication of Helicobacter pylori and Gastric Cancer: A Controversial Relationship. Front. Microbiol. 2021, 12, 630852. [Google Scholar] [CrossRef]
- Boyanova, L.; Hadzhiyski, P.; Gergova, R.; Markovska, R. Evolution of Helicobacter pylori Resistance to Antibiotics: A Topic of Increasing Concern. Antibiotics 2023, 12, 332. [Google Scholar] [CrossRef]
- Wang, H.; Yan, A.; Liu, Z.; Yang, X.; Xu, Z.; Wang, Y.; Wang, R.; Koohi-Moghadam, M.; Hu, L.; Xia, W.; et al. Deciphering molecular mechanism of silver by integrated omic approaches enables enhancing its antimicrobial efficacy in E. coli. PLoS Biol. 2019, 17, 3000292. [Google Scholar] [CrossRef] [Green Version]
- Frei, A.; Verderosa, A.D.; Elliott, A.G.; Zuegg, J.; Blaskovich, M.A.T. Metals to combat antimicrobial resistance. Nat. Rev. Chem. 2023, 7, 202–224. [Google Scholar] [CrossRef]
- Maier, R.J.; Benoit, S.L. Role of nickel in microbial pathogenesis. Inorganics 2019, 7, 80. [Google Scholar] [CrossRef] [Green Version]
- Keogan, D.M.; Griffith, D.M. Current and potential applications of bismuth-based drugs. Molecules 2014, 19, 15258–15297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shetu, S.A.; Sanchez-Palestino, L.M.; Rivera, G.; Bandyopadhyay, D. Medicinal bismuth: Bismuth-organic frameworks as pharmaceutically privileged compounds. Tetrahedron 2022, 129, 133117. [Google Scholar] [CrossRef]
- Abdelkhalek, A.; Abutaleb, N.S.; Mohammad, H.; Mohamed, N.; Lafayette, W.; Disease, I.; Lafayette, W. Antibacterial and antivirulence activities of auranofin against Clostridium difficile. Int. J. Antimicrob. Agents 2020, 53, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Thangamani, S.; Mohammad, H.; Abushahba, M.F.N.; Sobreira, T.J.P.; Hedrick, V.E.; Paul, L.N.; Seleem, M.N. Antibacterial activity and mechanism of action of auranofin against multi-drug resistant bacterial pathogens. Sci. Rep. 2016, 6, 22571. [Google Scholar] [CrossRef] [Green Version]
- AbdelKhalek, A.; Abutaleb, N.S.; Elmagarmid, K.A.; Seleem, M.N. Repurposing auranofin as an intestinal decolonizing agent for vancomycin-resistant enterococci. Sci. Rep. 2018, 8, 8353. [Google Scholar] [CrossRef] [Green Version]
- Abutaleb, N.S.; Seleem, M.N. Antivirulence activity of auranofin against vancomycin-resistant enterococci: In vitro and in vivo studies. Int. J. Antimicrob. Agents 2020, 55, 105828. [Google Scholar] [CrossRef]
- Kim, N.-H.; Lee, M.-Y.; Park, S.-J.; Choi, J.-S.; Oh, M.-K.; Kim, I.-S. Auranofin blocks interleukin-6 signalling by inhibiting phosphorylation of JAK1 and STAT3 Nam-Hoon. Immunology 2007, 122, 607–614. [Google Scholar] [CrossRef]
- Han, Y.; Chen, P.; Zhang, Y.; Lu, W.; Ding, W.; Luo, Y.; Wen, S.; Xu, R.; Liu, P.; Huang, P. Synergy between auranofin and celecoxib against colon cancer in vitro and in vivo through a novel redox-mediated mechanism. Cancers 2019, 11, 931. [Google Scholar] [CrossRef] [Green Version]
- Fiskus, W.; Saba, N.; Shen, M.; Ghias, M.; Liu, J.; Gupta, S.D.; Chauhan, L.; Rao, R.; Gunewardena, S.; Schorno, K.; et al. Auranofin induces lethal oxidative and endoplasmic reticulum stress and exerts potent preclinical activity against chronic lymphocytic leukemia. Cancer Res. 2014, 74, 2520–2532. [Google Scholar] [CrossRef]
- Sharlow, E.R.; Leimgruber, S.; Murray, S.; Lira, A.; Sciotti, R.J.; Hickman, M.; Hudson, T.; Leed, S.; Caridha, D.; Barrios, A.M.; et al. Auranofin is an apoptosis-simulating agent with in vitro and in vivo anti-leishmanial activity. ACS Chem. Biol. 2014, 9, 663–672. [Google Scholar] [CrossRef]
- Yang, L.; Wang, H.; Yang, X.; Wu, Q.; An, P.; Jin, X.; Liu, W.; Huang, X.; Li, Y.; Yan, S.; et al. Auranofin mitigates systemic iron overload and induces ferroptosis via distinct mechanisms. Signal Transduct. Target. Ther. 2020, 5, 138. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Perrin, M.W.; Sedgwick, A.C.; Lynch, V.M.; Sessler, J.L.; Arambula, J.F. Covalent and non-covalent albumin binding of Au(i) bis-NHCsviapost-synthetic amide modification. Chem. Sci. 2021, 12, 7547–7553. [Google Scholar] [CrossRef] [PubMed]
- Martín-Encinas, E.; Conejo-Rodríguez, V.; Miguel, J.A.; Martínez-Ilarduya, J.M.; Rubiales, G.; Knudsen, B.R.; Palacios, F.; Alonso, C. Novel phosphine sulphide gold(i) complexes: Topoisomerase I inhibitors and antiproliferative agents. Dalton Trans. 2020, 49, 7852–7861. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, H.; Cao, M.; Wang, L.; Wu, S.; Fang, B. Auranofin enhances ibrutinib’s anticancer activity in EGFR-mutant lung adenocarcinoma. Mol. Cancer Ther. 2018, 17, 2156–2163. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-J.; Kim, I.-S. The role of p38 MAPK activation in auranofin-induced apoptosis of human promyelocytic leukaemia HL-60 cells. Br. J. Pharmacol. 2005, 146, 506–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Li, X.; Huang, H.; Zhao, C.; Liao, S.; Yang, C.; Liu, S.; Song, W.; Lu, X.; Lan, X.; et al. Clinically used antirheumatic agent auranofin is a proteasomal deubiquitinase inhibitor and inhibits tumor growth. Oncotarget 2014, 5, 5453–5471. [Google Scholar] [CrossRef] [Green Version]
- Husain, S.; Nandi, A.; Simnani, F.Z.; Saha, U.; Ghosh, A.; Sinha, A.; Sahay, A.; Samal, S.K.; Panda, P.K.; Verma, S.K. Emerging Trends in Advanced Translational Applications of Silver Nanoparticles: A Progressing Dawn of Nanotechnology. J. Funct. Biomater. 2023, 14, 47. [Google Scholar] [CrossRef]
- O’Shaughnessy, M.; Hurley, J.; Dillon, S.C.; Herra, C.; McCarron, P.; McCann, M.; Devereux, M.; Howe, O. Antibacterial activity of metal–phenanthroline complexes against multidrug-resistant Irish clinical isolates: A whole genome sequencing approach. J. Biol. Inorg. Chem. 2023, 28, 153–171. [Google Scholar] [CrossRef]
- Wang, H.; Wang, M.; Xu, X.; Gao, P.; Xu, Z.; Zhang, Q.; Li, H.; Yan, A.; Kao, R.Y.T.; Sun, H. Multi-target mode of action of silver against Staphylococcus aureus endows it with capability to combat antibiotic resistance. Nat. Commun. 2021, 12, 3331. [Google Scholar] [CrossRef]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial Activity and Mechanism of Action of the Silver Ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McQuillan, J.S.; Groenaga Infante, H.; Stokes, E.; Shaw, A.M. Silver nanoparticle enhanced silver ion stress response in Escherichia coli K12. Nanotoxicology 2012, 6, 857–866. [Google Scholar] [CrossRef]
- Arakawa, H.; Neault, J.F.; Tajmir-Riahi, H.A. Silver(I) complexes with DNA and RNA studied by fourier transform infrared spectroscopy and capillary electrophoresis. Biophys. J. 2001, 81, 1580–1587. [Google Scholar] [CrossRef] [Green Version]
- Gordon, O.; Slenters, T.V.; Brunetto, P.S.; Villaruz, A.E.; Sturdevant, D.E.; Otto, M.; Landmann, R.; Fromm, K.M. Silver coordination polymers for prevention of implant infection: Thiol interaction, impact on respiratory chain enzymes, and hydroxyl radical induction. Antimicrob. Agents Chemother. 2010, 54, 4208–4218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, M.; Imlay, J. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol. Microbiol. 2011, 4, 1136–1150. [Google Scholar] [CrossRef] [Green Version]
- Barras, F.; Aussel, L.; Ezraty, B. Silver and antibiotic, new facts to an old story. Antibiotics 2018, 7, 79. [Google Scholar] [CrossRef] [Green Version]
- Saulou-Bérion, C.; Gonzalez, I.; Enjalbert, B.; Audinot, J.N.; Fourquaux, I.; Jamme, F.; Cocaign-Bousquet, M.; Mercier-Bonin, M.; Girbal, L. Escherichia coli under ionic silver stress: An integrative approach to explore transcriptional, physiological and biochemical responses. PLoS ONE 2015, 10, 0145748. [Google Scholar] [CrossRef] [Green Version]
- Surwade, P.; Ghildyal, C.; Weikel, C.; Luxton, T.; Peloquin, D.; Fan, X.; Shah, V. Augmented antibacterial activity of ampicillin with silver nanoparticles against methicillin-resistant Staphylococcus aureus (MRSA). J. Antibiot. 2019, 72, 50–53. [Google Scholar] [CrossRef]
- Morones-Ramirez, J.; Winkler, J.A.; Spina, C.S.; Collins, J.J.; Morones-Ramirez, J.R.; Winkler, J.A.; Spina, C.S.; Collins, J.J.; RubenMorones-Ramirez, J.; Winkler, J.A.; et al. Silver Enhances Antibiotic Activity Against Gram-negative Bacteria. Sci. Transl. Med. 2013, 5, 190ra81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panáček, A.; Smékalová, M.; Večeřová, R.; Bogdanová, K.; Röderová, M.; Kolář, M.; Kilianová, M.; Hradilová, Š.; Froning, J.P.; Havrdová, M.; et al. Silver nanoparticles strongly enhance and restore bactericidal activity of inactive antibiotics against multiresistant Enterobacteriaceae. Colloids Surf. B Biointerfaces 2016, 142, 392–399. [Google Scholar] [CrossRef]
- Habash, M.B.; Goodyear, M.C.; Park, A.J.; Surette, M.D.; Vis, E.C.; Harris, R.J.; Khursigara, C.M.; Khursigara, M. Potentiation of Tobramycin by Silver Nanoparticles against Pseudomonas aeruginosa Biofilm. Antimicrob. Agents Chemother. 2017, 61, e00415-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Shaughnessy, M.; McCarron, P.; Viganor, L.; McCann, M.; Devereux, M.; Howe, O. The antibacterial and anti-biofilm activity of metal complexes incorporating 3,6,9-trioxaundecanedioate and 1,10-phenanthroline ligands in clinical isolates of Pseudomonas aeruginosa from irish cystic fibrosis patients. Antibiotics 2020, 9, 674. [Google Scholar] [CrossRef]
- Williams, K.; Milner, J.; Boudreau, M.D.; Gokulan, K.; Cerniglia, C.E.; Khare, S. Effects of subchronic exposure of silver nanoparticles on intestinal microbiota and gut-associated immune responses in the ileum of Sprague-Dawley rats. Nanotoxicology 2015, 9, 279–289. [Google Scholar] [CrossRef]
- Hadrup, N.; Gao, X.; Lam, H.R.; Loeschner, K.; Vogel, U.; Bergström, A.; Frandsen, H.L.; Mortensen, A.; Wilcks, A.; Larsen, E.H. Subacute oral toxicity investigation of nanoparticulate and ionic silver in rats. Arch. Toxicol. 2012, 86, 543–551. [Google Scholar] [CrossRef]
- Javurek, A.B.; Suresh, D.; Spollen, W.G.; Hart, M.L.; Hansen, A.; Ellersieck, M.R.; Bivens, N.J.; Givan, S.A. Gut Dysbiosis and Neurobehavioral Alterations in Rats Exposed to Silver Nanoparticles. Sci. Rep. 2017, 7, 2822. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Z.; Ghoshdastidar, S.; Rekha, K.R.; Suresh, D. Developmental exposure to silver nanoparticles leads to long term gut dysbiosis and neurobehavioral alterations. Sci. Rep. 2021, 11, 6558. [Google Scholar] [CrossRef] [PubMed]
- Kankala, S.; Thota, N.; Björkling, F.; Taylor, M.K.; Vadde, R.; Balusu, R. Silver carbene complexes: An emerging class of anticancer agents. Drug Dev. Res. 2019, 80, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Akkoç, M.; Khan, S.; Yüce, H.; Türkmen, N.B.; Yaşar, Ş.; Yaşar, S.; Özdemir, İ. Molecular docking and in vitro anticancer studies of silver(I)-N-heterocyclic carbene complexes. Heliyon 2022, 8, 10133. [Google Scholar] [CrossRef]
- Sammes, P.G.; Yahioglu, G. 1,10-Phenanthroline: A versatile ligand. Chem. Soc. Rev. 1994, 23, 327–334. [Google Scholar] [CrossRef]
- Bencini, A.; Lippolis, V. 1,10-Phenanthroline: A versatile building block for the construction of ligands for various purposes. Coord. Chem. Rev. 2010, 254, 2096–2180. [Google Scholar] [CrossRef]
- Turian, G. Tuberculostatic action of o-phenanthroline. Schweiz. Z. Pathol. Bakteriol. 1951, 14, 338–344. [Google Scholar]
- Kilah, N.L.; Meggers, E. Sixty years young: The diverse biological activities of metal polypyridyl complexes pioneered by Francis P. Dwyer. Aust. J. Chem. 2012, 65, 1325–1332. [Google Scholar] [CrossRef]
- McCann, M.; Santos, A.L.S.; Da Silva, B.A.; Romanos, M.T.V.; Pyrrho, A.S.; Devereux, M.; Kavanagh, K.; Fichtner, I.; Kellett, A. In vitro and in vivo studies into the biological activities of 1,10-phenanthroline, 1,10-phenanthroline-5,6-dione and its copper(ii) and silver(i) complexes. Toxicol. Res. 2012, 1, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Santos, A.L.; Sodre, C.L.; Valle, R.S.; Silva, B.A.; Abi-Chacra, E.A.; Silva, L.V.; Souza-Goncalves, A.L.; Sangenito, L.S.; Goncalves, D.S.; Souza, L.O.; et al. Antimicrobial Action of Chelating Agents: Repercussions on the Microorganism Development, Virulence and Pathogenesis. Curr. Med. Chem. 2012, 19, 2715–2737. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, M.H.B.; Da Costa, A.F.E.; Santos, G.D.S.; Dos Santos, A.L.S.; Nagao, P.E. Effect of chelating agents on the growth, surface polypeptide synthesis and interaction of Streptococcus agalactiae with human epithelial cells. Mol. Med. Rep. 2009, 2, 81–84. [Google Scholar] [PubMed] [Green Version]
- Husseini, R.; Stretton, R.J. Studies on the antibacterial activity of phanquone: Chelating properties in relation to mode of action against Escherichia coli and Staphylococcus aureus. Microbios 1980, 29, 109–125. [Google Scholar]
- Zoroddu, M.A.; Zanetti, S.; Pogni, R.; Basosi, R. An electron spin resonance study and antimicrobial activity of copper(II)-phenanthroline complexes. J. Inorg. Biochem. 1996, 63, 291–300. [Google Scholar] [CrossRef]
- Kellett, A.; Howe, O.; O’Connor, M.; McCann, M.; Creaven, B.S.; McClean, S.; Foltyn-Arfa Kia, A.; Casey, A.; Devereux, M. Radical-induced DNA damage by cytotoxic square-planar copper(II) complexes incorporating o-phthalate and 1,10-phenanthroline or 2,2′-dipyridyl. Free Radic. Biol. Med. 2012, 53, 564–576. [Google Scholar] [CrossRef]
- Rochford, G.; Molphy, Z.; Browne, N.; Surlis, C.; Devereux, M.; McCann, M.; Kellett, A.; Howe, O.; Kavanagh, K. In-vivo evaluation of the response of Galleria mellonella larvae to novel copper(II) phenanthroline-phenazine complexes. J. Inorg. Biochem. 2018, 186, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Rochford, G.; Molphy, Z.; Kavanagh, K.; McCann, M.; Devereux, M.; Kellett, A.; Howe, O. Cu(ii) phenanthroline-phenazine complexes dysregulate mitochondrial function and stimulate apoptosis. Metallomics 2020, 12, 65–78. [Google Scholar] [CrossRef]
- Thornton, L.; Dixit, V.; Assad, L.O.N.; Ribeiro, T.P.; Queiroz, D.D.; Kellett, A.; Casey, A.; Colleran, J.; Pereira, M.D.; Rochford, G.; et al. Water-soluble and photo-stable silver(I) dicarboxylate complexes containing 1,10-phenanthroline ligands: Antimicrobial and anticancer chemotherapeutic potential, DNA interactions and antioxidant activity. J. Inorg. Biochem. 2016, 159, 120–132. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Fang, R.; Wei, W.; Wang, Y.; Jin, J.; Yang, F.; Chen, J. Organometallic gold(I) and gold(III) complexes for lung cancer treatment. Front. Pharmacol. 2022, 13, 979951. [Google Scholar] [CrossRef]
- Iglesias, S.; Alvarez, N.; Torre, M.H.; Kremer, E.; Ellena, J.; Ribeiro, R.R.; Barroso, R.P.; Costa-Filho, A.J.; Kramer, G.M.; Facchin, G. Synthesis, structural characterization and cytotoxic activity of ternary copper(II)-dipeptide-phenanthroline complexes. A step towards the development of new copper compounds for the treatment of cancer. J. Inorg. Biochem. 2014, 139, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Pivetta, T.; Isaia, F.; Verani, G.; Cannas, C.; Serra, L.; Castellano, C.; Demartin, F.; Pilla, F.; Manca, M.; Pani, A. Mixed-1,10-phenanthroline-Cu(II) complexes: Synthesis, cytotoxic activity versus hematological and solid tumor cells and complex formation equilibria with glutathione. J. Inorg. Biochem. 2012, 114, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Masuri, S.; Cadoni, E.; Cabiddu, M.G.; Isaia, F.; Demuru, M.G.; Morán, L.; Morán, L.; Bucek, D.; Vanhara, P.; Vanhara, P.; et al. The first copper(ii) complex with 1,10-phenanthroline and salubrinal with interesting biochemical properties. Metallomics 2020, 12, 891–901. [Google Scholar] [CrossRef] [PubMed]
- Olsen, P.M.; Ruiz, C.; Lussier, D.; Le, B.K.; Angel, N.; Smith, M.; Hwang, C.; Khatib, R.; Jenkins, J.; Adams, K.; et al. Synthesis, characterization, and antitumor activity of unusual pseudo five coordinate gold(III) complexes: Distinct cytotoxic mechanism or expensive ligand delivery systems? J. Inorg. Biochem. 2014, 141, 121–131. [Google Scholar] [CrossRef]
- Wenzel, M.N.; Mósca, A.F.; Graziani, V.; Aikman, B.; Thomas, S.R.; De Almeida, A.; Platts, J.A.; Re, N.; Coletti, C.; Marrone, A.; et al. Insights into the Mechanisms of Aquaporin-3 Inhibition by Gold(III) Complexes: The Importance of Non-Coordinative Adduct Formation. Inorg. Chem. 2019, 58, 2140–2148. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-j.; Liu, J.-p.; Hao, Z.-f.; He, J.; Sun, M.; Hu, S.; Yu, L.; Chao, H. Synthesis, characterization and biological evaluation of ruthenium(II) complexes [Ru(dtzp)(dppz)Cl]+ and [Ru(dtzp)(dppz)CH3CN]2+ for photodynamic therapy. Dye. Pigment. 2017, 136, 416–426. [Google Scholar] [CrossRef]
- Deo, K.M.; Pages, B.J.; Ang, D.L.; Gordon, C.P.; Aldrich-Wright, J.R. Transition Metal Intercalators as Anticancer Agents-Recent Advances. Int. J. Mol. Sci. 2016, 17, 1818. [Google Scholar] [CrossRef] [Green Version]
- Sidambaram, P.; Colleran, J. Evaluating the anticancer properties and real-time electrochemical extracellular bio-speciation of bis (1,10-phenanthroline) silver (I) acetate monohydrate in the presence of A549 lung cancer cells. Biosens. Bioelectron. 2021, 175, 112876. [Google Scholar] [CrossRef]
- Gandra, R.M.; Carron, P.M.; Fernandes, M.F.; Ramos, L.S.; Mello, T.P.; Aor, A.C.; Branquinha, M.H.; McCann, M.; Devereux, M.; Santos, A.L.S. Antifungal potential of copper(II), Manganese(II) and silver(I) 1,10-phenanthroline chelates against multidrug-resistant fungal species forming the Candida haemulonii Complex: Impact on the planktonic and biofilm lifestyles. Front. Microbiol. 2017, 8, 1257. [Google Scholar] [CrossRef]
- Gandra, R.M.; McCarron, P.; Viganor, L.; Fernandes, M.F.; Kavanagh, K.; McCann, M.; Branquinha, M.H.; Santos, A.L.S.; Howe, O.; Devereux, M. In vivo Activity of Copper(II), Manganese(II), and Silver(I) 1,10-Phenanthroline Chelates Against Candida haemulonii Using the Galleria mellonella Model. Front. Microbiol. 2020, 11, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granato, M.Q.; Gonçalves, D.d.S.; Seabra, S.H.; McCann, M.; Devereux, M.; dos Santos, A.L.S.; Kneipp, L.F. 1,10-phenanthroline-5,6-dione-based compounds are effective in disturbing crucial physiological events of Phialophora verrucosa. Front. Microbiol. 2017, 8, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granato, M.Q.; Mello, T.P.; Nascimento, R.S.; Pereira, M.D.; Rosa, T.L.S.A.; Pessolani, M.C.V.; McCann, M.; Devereux, M.; Branquinha, M.H.; Santos, A.L.S.; et al. Silver(I) and Copper(II) Complexes of 1,10-Phenanthroline-5,6-Dione Against Phialophora verrucosa: A Focus on the Interaction With Human Macrophages and Galleria mellonella Larvae. Front. Microbiol. 2021, 12, 641258. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.K.C.; Elias, C.G.R.; Oliveira, S.S.C.; Santos-Mallet, J.R.; McCann, M.; Devereux, M.; Branquinha, M.H.; Dutra, P.M.L.; Santos, A.L.S. Anti-Leishmania braziliensis activity of 1,10-phenanthroline-5,6-dione and its Cu(II) and Ag(I) complexes. Parasitol. Res. 2021, 120, 3273–3285. [Google Scholar] [CrossRef] [PubMed]
- Vargas Rigo, G.; Petro-Silveira, B.; Devereux, M.; McCann, M.; Souza Dos Santos, A.L.; Tasca, T. Anti-Trichomonas vaginalis activity of 1,10-phenanthroline-5,6-dione-based metallodrugs and synergistic effect with metronidazole. Parasitology 2019, 146, 1179–1183. [Google Scholar] [CrossRef]
- Rigo, G.V.; Cardoso, F.G.; Pereira, M.M.; Devereux, M. Peptidases Are Potential Targets of and Potent New Drug against Trichomonas vaginalis. Pathogens 2023, 12, 745. [Google Scholar] [CrossRef]
- Silva-oliveira, R.; Sangenito, L.S.; Reddy, A.; Velasco-torrijos, T. Tropical Medicine and Infectious Disease In Vitro Effects of Aminopyridyl Ligands Complexed to Copper (II) on the Physiology and Interaction Process of In Vitro Effects of Aminopyridyl Ligands Complexed to Copper (II) on the Physiology and Interaction. Trop. Med. Infect. Dis. 2023, 8, 288. [Google Scholar] [CrossRef]
- Papadia, P.; Margiotta, N.; Bergamo, A.; Sava, G.; Natile, G. Platinum(II) complexes with antitumoral/antiviral aromatic heterocycles: Effect of glutathione upon in vitro cell growth inhibition. J. Med. Chem. 2005, 48, 3364–3371. [Google Scholar] [CrossRef]
- Shulman, A.; White, D.O. Virostatic activity of 1,10-phenanthroline transition metal chelates: A structure-activity analysis. Chem. Biol. Interact. 1973, 6, 407–413. [Google Scholar] [CrossRef]
- Mazumder, A.; Gupta, M.; Perrin, D.M.; Sigman, D.S.; Rabinovitz, M.; Pommier, Y. Inhibition of Human Immunodeficiency Virus Type 1 Integrase by a Hydrophobic Cation: The Phenanthroline-Cuprous Complex. AIDS Res. Hum. Retroviruses 1995, 11, 115–125. [Google Scholar] [CrossRef]
- Chang, E.L.; Simmers, C.; Knight, D.A. Cobalt complexes as antiviral and antibacterial agents. Pharmaceuticals 2010, 3, 1711–1728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viganor, L.; Galdino, A.C.M.; Nunes, A.P.F.; Santos, K.R.N.; Branquinha, M.H.; Devereux, M.; Kellett, A.; McCann, M.; Santos, A.L.S. Anti-Pseudomonas aeruginosa activity of 1,10-phenanthroline-based drugs against both planktonic- and biofilm-growing cells. J. Antimicrob. Chemother. 2016, 71, 128–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarron, P.; McCann, M.; Devereux, M.; Kavanagh, K.; Skerry, C.; Karakousis, P.C.; Aor, A.C.; Mello, T.P.; Santos, A.L.S.; Campos, D.L.; et al. Unprecedented in vitro antitubercular activitiy of manganese(II) complexes containing 1,10-phenanthroline and dicarboxylate ligands: Increased activity, superior selectivity, and lower toxicity in comparison to their copper(II) analogs. Front. Microbiol. 2018, 9, 1432. [Google Scholar] [CrossRef]
- Ahmed, M.; Rooney, D.; McCann, M.; Devereux, M.; Twamley, B.; Galdino, A.C.M.; Sangenito, L.S.; Souza, L.O.P.; Lourenço, M.C.; Gomes, K.; et al. Synthesis and antimicrobial activity of a phenanthroline-isoniazid hybrid ligand and its Ag+ and Mn2+ complexes. BioMetals 2019, 32, 671–682. [Google Scholar] [CrossRef]
- Ventura, R.F.; Galdino, A.C.M.; Viganor, L.; Schuenck, R.P.; Devereux, M.; McCann, M.; Santos, A.L.S.; Nunes, A.P.F. Antimicrobial action of 1,10-phenanthroline-based compounds on carbapenemase-producing Acinetobacter baumannii clinical strains: Efficacy against planktonic- and biofilm-growing cells. Braz. J. Microbiol. 2020, 51, 1703–1710. [Google Scholar] [CrossRef] [PubMed]
- Vianez Peregrino, I.; Ferreira Ventura, R.; Borghi, M.; Pinto Schuenck, R.; Devereux, M.; McCann, M.; Souza dos Santos, A.L.; FerreiraNunes, A.P. Antibacterial activity and carbapenem re-sensitizing ability of 1,10-phenanthroline-5,6-dione and its metal complexes against KPC-producing Klebsiella pneumoniae clinical strains. Lett. Appl. Microbiol. 2021, 73, 139–148. [Google Scholar] [CrossRef]
- Dwyer, F.P.; Reid, I.K.; Shulman, A.; Laycock, G.M.; Dixson, S. The biological actions of 1,10-phenanthroline and 2,2′-bipyridine hydrochlorides, quaternary salts and metal chelates and related compounds. Aust. J. Exp. Biol. Med. 1969, 47, 203–218. [Google Scholar] [CrossRef]
- McCann, M.; Kellett, A.; Kavanagh, K.; Devereux, M.; Santos, A.L.S. Deciphering the Antimicrobial Activity of Phenanthroline Chelators. Curr. Med. Chem. 2012, 19, 2703–2714. [Google Scholar] [CrossRef]
- Raman, N.; Dhaveethu Raja, J.; Sakthivel, A. Synthesis, spectral characterization of Schiff base transition metal complexes: DNA cleavage and antimicrobial activity studies. J. Chem. Sci. 2007, 119, 303–310. [Google Scholar] [CrossRef]
- Butler, H.M.; Hurse, A.; Thursky, E.; Shulman, A. Bactericidal action of selected phenanthroline chelates and related compounds. Aust. J. Exp. Biol. Med. Sci. 1969, 47, 541–552. [Google Scholar] [CrossRef]
- Wang, S.; König, G.; Roth, H.J.; Fouché, M.; Rodde, S.; Riniker, S. Effect of Flexibility, Lipophilicity, and the Location of Polar Residues on the Passive Membrane Permeability of a Series of Cyclic Decapeptides. J. Med. Chem. 2021, 64, 12761–12773. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, F.P.; Gyarfas, E.C.; Rogers, W.P.; Koch, J.H. Biological activity of complex ions. Nature 1952, 170, 190–191. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Scottwell, S.O.; Waugh, E.; McAdam, C.J.; Hanton, L.R.; Brooks, H.J.L.; Crowley, J.D. Antimicrobial Properties of Tris(homoleptic) Ruthenium(II) 2-Pyridyl-1,2,3-triazole “click” Complexes against Pathogenic Bacteria, Including Methicillin-Resistant Staphylococcus aureus (MRSA). Inorg. Chem. 2016, 55, 9767–9777. [Google Scholar] [CrossRef]
- Yang, X.; Sun, B.; Zhang, L.; Li, N.; Han, J.; Zhang, J.; Sun, X.; He, Q. Chemical Interference with Iron Transport Systems to Suppress Bacterial Growth of Streptococcus pneumoniae. PLoS ONE 2014, 9, 105953. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Zhang, L.; Liu, J.; Li, N.; Yu, G.; Cao, K.; Han, H.; Zeng, G.; Pan, Y.; Sun, X.; et al. Proteomic analysis on the antibacterial activity of a Ru(II) complex against Streptococcus pneumoniae. J. Proteom. 2015, 115, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.K.; Sani, M.A.; Downton, M.T.; Separovic, F.; Keene, F.R.; Collins, J.G. Membrane Insertion of a Dinuclear Polypyridylruthenium(II) Complex Revealed by Solid-State NMR and Molecular Dynamics Simulation: Implications for Selective Antibacterial Activity. J. Am. Chem. Soc. 2016, 138, 15267–15277. [Google Scholar] [CrossRef]
- Li, F.; Harry, E.J.; Bottomley, A.L.; Edstein, M.D.; Birrell, G.W.; Woodward, C.E.; Keene, F.R.; Collins, J.G. Dinuclear ruthenium(ii) antimicrobial agents that selectively target polysomes in vivo. Chem. Sci. 2014, 5, 685–693. [Google Scholar] [CrossRef] [Green Version]
- Gorle, A.K.; Feterl, M.; Warner, J.M.; Wallace, L.; Keene, F.R.; Collins, J.G. Tri- and tetra-nuclear polypyridyl ruthenium(ii) complexes as antimicrobial agents. Dalton Trans. 2014, 43, 16713–16725. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Collins, J.G.; Keene, F.R. Ruthenium complexes as antimicrobial agents. Chem. Soc. Rev. 2015, 44, 2529–2542. [Google Scholar] [CrossRef] [Green Version]
- Gorle, A.K.; Li, X.; Primrose, S.; Li, F.; Feterl, M.; Kinobe, R.T.; Heimann, K.; Warner, J.M.; Richard Keene, F.; Grant Collins, J. Oligonuclear polypyridylruthenium(II) complexes: Selectivity between bacteria and eukaryotic cells. J. Antimicrob. Chemother. 2016, 71, 1547–1555. [Google Scholar] [CrossRef]
- Sigman, D.S.; Graham, D.R.; D’Aurora, V.; Stern, A.M. Inhibition polymerase of. J. Biol. Chem. 1979, 254, 12269–12272. [Google Scholar] [CrossRef]
- Bolhuis, A.; Aldrich-Wright, J.R. DNA as a target for antimicrobials. Bioorg. Chem. 2014, 55, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Psomas, G.; Kessissoglou, D.P. Quinolones and non-steroidal anti-inflammatory drugs interacting with copper(ii), nickel(ii), cobalt(ii) and zinc(ii): Structural features, biological evaluation and perspectives. Dalton Trans. 2013, 42, 6252–6276. [Google Scholar] [CrossRef]
- Sousa, I.; Claro, V.; Pereira, J.L.; Amaral, A.L.; Cunha-Silva, L.; De Castro, B.; Feio, M.J.; Pereira, E.; Gameiro, P. Synthesis, characterization and antibacterial studies of a copper(II) levofloxacin ternary complex. J. Inorg. Biochem. 2012, 110, 64–71. [Google Scholar] [CrossRef]
- Fernandes, P.; Sousa, I.; Cunha-Silva, L.; Ferreira, M.; De Castro, B.; Pereira, E.F.; Feio, M.J.; Gameiro, P. Synthesis, characterization and antibacterial studies of a copper(II) lomefloxacin ternary complex. J. Inorg. Biochem. 2014, 131, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Gameiro, P.; Rodrigues, C.; Baptista, T.; Sousa, I.; de Castro, B. Solution studies on binary and ternary complexes of copper(II) with some fluoroquinolones and 1,10-phenanthroline: Antimicrobial activity of ternary metalloantibiotics. Int. J. Pharm. 2007, 334, 129–136. [Google Scholar] [CrossRef]
- Ude, Z.; Kavanagh, K.; Twamley, B.; Pour, M.; Gathergood, N.; Kellett, A.; Marmion, C.J. A new class of prophylactic metallo-antibiotic possessing potent anti-cancer and anti-microbial properties. Dalton Trans. 2019, 48, 8578–8593. [Google Scholar] [CrossRef] [PubMed]
- Ude, Z.; Flothkötter, N.; Sheehan, G.; Brennan, M.; Kavanagh, K.; Marmion, C.J. Multi-targeted metallo-ciprofloxacin derivatives rationally designed and developed to overcome antimicrobial resistance. Int. J. Antimicrob. Agents 2021, 58, 106449. [Google Scholar] [CrossRef] [PubMed]
- Smoleński, P.; Jaros, S.W.; Pettinari, C.; Lupidi, G.; Quassinti, L.; Bramucci, M.; Vitali, L.A.; Petrelli, D.; Kochel, A.; Kirillov, A.M. New water-soluble polypyridine silver(i) derivatives of 1,3,5-triaza-7-phosphaadamantane (PTA) with significant antimicrobial and antiproliferative activities. Dalton Trans. 2013, 42, 6572–6581. [Google Scholar] [CrossRef]
- Chetana, P.R.; Srinatha, B.S.; Somashekar, M.N.; Policegoudra, R.S. Synthesis, spectroscopic characterisation, thermal analysis, DNA interaction and antibacterial activity of copper(I) complexes with N, N′- disubstituted thiourea. J. Mol. Struct. 2016, 45, 352–365. [Google Scholar] [CrossRef] [Green Version]
- Raman, N.; Raja, S.J. DNA cleavage, structural elucidation and anti-microbial studies of three novel mixed ligand Schiff base complexes of copper(II). J. Serbian Chem. Soc. 2007, 72, 983–992. [Google Scholar] [CrossRef]
- Tabassum, S.; Asim, A.; Arjmand, F.; Afzal, M.; Bagchi, V. Synthesis and characterization of copper(II) and zinc(II)-based potential chemotherapeutic compounds: Their biological evaluation viz. DNA binding profile, cleavage and antimicrobial activity. Eur. J. Med. Chem. 2012, 58, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Ng, N.S.; Leverett, P.; Hibbs, D.E.; Yang, Q.; Bulanadi, J.C.; Jie Wu, M.; Aldrich-Wright, J.R. The antimicrobial properties of some copper(ii) and platinum(ii) 1,10-phenanthroline complexes. Dalton Trans. 2013, 42, 3196–3209. [Google Scholar] [CrossRef] [PubMed]
- Shivakumar, L.; Shivaprasad, K.; Revanasiddappa, H.D. SODs, DNA binding and cleavage studies of new Mn(III) complexes with 2-((3-(benzyloxy)pyridin-2-ylimino)methyl)phenol. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 107, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Dimitrakopoulou, A.; Dendrinou-samara, C.; Pantazaki, A.A.; Raptopoulou, C.; Terzis, A.; Samaras, E.; Kessissoglou, D.P. Interaction of Fe (III) with herbicide-carboxylato ligands. Di-, tri- and tetra-nuclear compounds: Structure, antimicrobial study and DNA interaction. Inorganica Chim. Acta 2007, 360, 546–556. [Google Scholar] [CrossRef]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Fenker, D.E.; Mcdaniel, C.T.; Panmanee, W.; Panos, R.J.; Sorscher, E.J.; Sabusap, C.; Clancy, J.P.; Hassett, D.J. A Comparison between Two Pathophysiologically Different yet Microbiologically Similar Lung Diseases: Cystic Fibrosis and Chronic Obstructive Pulmonary Disease. Int. J. Respir. Pulm. Med. 2018, 5, 098. [Google Scholar]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [Green Version]
- Kolpen, M.; Kragh, K.N.; Enciso, J.B.; Faurholt-Jepsen, D.; Lindegaard, B.; Egelund, G.B.; Jensen, A.V.; Ravn, P.; Mathiesen, I.H.M.; Gheorge, A.G.; et al. Bacterial biofilms predominate in both acute and chronic human lung infections. Eur. Respir. J. 2022, 60, 4. [Google Scholar]
- Domingue, J.C.; Drewes, J.L.; Merlo, C.A.; Housseau, F.; Sears, C.L. Host Responses to Mucosal Biofilms in the Lung and Gut. Mucosal Immunol. 2020, 13, 413–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Kumar, D.; Dahiya, K.; Hawthorne, S.; Jha, S.K.; Jha, N.K.; Nand, P.; Girgis, S.; Raj, S.; Srivastava, R.; et al. Advances in pulmonary drug delivery targeting microbial biofilms in respiratory diseases. Nanomedicine 2021, 16, 1905–1923. [Google Scholar] [CrossRef] [PubMed]
- Beeton, M.L.; Aldrich-Wright, J.R.; Bolhuis, A. The antimicrobial and antibiofilm activities of copper(II) complexes. J. Inorg. Biochem. 2014, 140, 167–172. [Google Scholar] [CrossRef]
- O’Shaughnessy, M.; Piatek, M.; McCarron, P.; McCann, M.; Devereux, M.; Kavanagh, K.; Howe, O. In Vivo Activity of Metal Complexes Containing 1,10-Phenanthroline and 3,6,9-Trioxaundecanedioate Ligands against Pseudomonas aeruginosa Infection in Galleria mellonella Larvae. Biomedicines 2022, 10, 222. [Google Scholar] [CrossRef]
- Zhang, H.; Niu, Y.; Tan, J.; Liu, W.; Sun, M.; Yang, E.; Wang, Q.; Li, R.; Wang, Y.; Liu, W. Global Screening of Genomic and Transcriptomic Factors Associated with Phenotype Differences between Multidrug-Resistant and -Susceptible Candida haemulonii Strains. mSystems 2019, 4, 00459-19. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Y.H.; Wang, W.Q.; Chen, Q.J.; Liu, B.; Zhang, F.Y.; Jin, X.Y.; Hang, J.Q.; Li, H.Y.; Bao, Z.Y.; Jie, Z.J.; et al. Candida in Lower Respiratory Tract Increases the Frequency of Acute Exacerbation of Chronic Obstructive Pulmonary Disease: A Retrospective Case-Control Study. Front. Cell. Infect. Microbiol. 2020, 10, 538005. [Google Scholar] [CrossRef]
- Falkievich, D.B.; Martínez Medina, J.J.; Alegre, W.S.; López Tévez, L.L.; Franca, C.A.; Ferrer, E.G.; Williams, P.A.M. Computational studies, antimicrobial activity, inhibition of biofilm production and safety profile of the cadmium complex of 1,10-phenanthroline and cyanoguanidine. Appl. Organomet. Chem. 2022, 36, 6695. [Google Scholar] [CrossRef]
- Tay, C.X.; Quah, S.Y.; Lui, J.N.; Yu, V.S.H.; Tan, K.S. Matrix metalloproteinase inhibitor as an antimicrobial agent to eradicate Enterococcus faecalis biofilm. J. Endod. 2015, 41, 858–863. [Google Scholar] [CrossRef]
- McCann, M.; Coyle, B.; McKay, S.; McCormack, P.; Kavanagh, K.; Devereux, M.; McKee, V.; Kinsella, P.; O’Connor, R.; Clynes, M. Synthesis and X-ray crystal structure of [Ag(phendio)2]ClO4 (phendio = 1,10-phenanthroline-5,6-dione) and its effects on fungal and mammalian cells. BioMetals 2004, 17, 635–645. [Google Scholar] [CrossRef] [Green Version]
- Galdino, A.C.M.; Viganor, L.; De Castro, A.A.; Da Cunha, E.F.F.; Mello, T.P.; Mattos, L.M.; Pereira, M.D.; Hunt, M.C.; O’Shaughnessy, M.; Howe, O.; et al. Disarming Pseudomonas aeruginosa virulence by the inhibitory action of 1,10-phenanthroline-5,6-dione-based compounds: Elastase B (lasB) as a chemotherapeutic target. Front. Microbiol. 2019, 10, 1701. [Google Scholar] [CrossRef] [Green Version]
- Galdino, A.C.M.; Viganor, L.; Pereira, M.M.; Devereux, M.; McCann, M.; Branquinha, M.H.; Molphy, Z.; O’Carroll, S.; Bain, C.; Menounou, G.; et al. Copper(II) and silver(I)-1,10-phenanthroline-5,6-dione complexes interact with double-stranded DNA: Further evidence of their apparent multi-modal activity towards Pseudomonas aeruginosa. J. Biol. Inorg. Chem. 2022, 27, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, I.; Schuster, S.; Segal, L. Perspectives in lung microbiome research. Curr Opin Microbiol. 2020, 56, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, T.; Ye, C.; Zhong, J.; Huang, J.D.; Ke, Y.; Sun, H. The Lung Microbiome: A New Frontier for Lung and Brain Disease. Int. J. Mol. Sci. 2023, 24, 2170. [Google Scholar] [CrossRef] [PubMed]
- Broderick, D.; March, R.; Waite, D.; Pillarisetti, N.; Chang, A.B.; Taylor, M.W. Realising respiratory microbiomic meta-analyses: Time for a standardised framework. Microbiome 2023, 11, 57. [Google Scholar] [CrossRef]
- Pandey, A.; Boros, E. Coordination Complexes to Combat Bacterial Infections: Recent Developments, Current Directions and Future Opportunities. Chem. Eur. J. 2021, 27, 7340–7350. [Google Scholar] [CrossRef]
- Butler, M.S.; Henderson, I.R.; Capon, R.J.; Blaskovich, M.A.T. Antibiotics in the clinical pipeline as of December 2022. J Antibiot 2023, 76, 431–473. [Google Scholar] [CrossRef]
- Huynh, M.; Crane, M.J.; Jamieson, A.M. The lung, the niche, and the microbe: Exploring the lung microbiome in cancer and immunity. Front. Immunol. 2023, 13, 1094110. [Google Scholar] [CrossRef]
- Martins, D.; Mendes, F.; Schmitt, F. Microbiome: A Supportive or a Leading Actor in Lung Cancer? Pathobiology 2021, 88, 198–207. [Google Scholar] [CrossRef]



| Metal | Drug | Therapeutic Application | Year Approved |
|---|---|---|---|
| Platinum (Pt) | Cisplatin | Cancer: lung, testicular, ovarian, bladder | 1978 |
| Carboplatin | Cancer: lung, ovarian, head and neck | 1989 | |
| Oxaliplatin | Cancer: colorectal | 1999 | |
| Gold (Au) | Auranofin | Rheumatoid arthritis | 1985 |
| Sodium aurothiomalate | Rheumatoid arthritis | 1985 | |
| Iron (Fe) | Deferoxamine | Iron chelation (iron overload) | 1968 |
| Deferiprone | Iron chelation (iron overload) | 1999 | |
| Deferasirox | Iron chelation (iron overload) | 2005 | |
| Ferric carboxymaltose | Iron deficiency anaemia | 2007 | |
| Iron isomaltoside 1000 | Iron deficiency anaemia | 2009 | |
| Ferumoxytol | Iron deficiency anaemia in chronic kidney disease | 2009 | |
| Ferric citrate | Hyperphosphatemia in chronic kidney disease | 2014 | |
| Sucroferric oxyhydroxide | Hyperphosphatemia in chronic kidney disease | 2013 | |
| Bismuth (Bi) | Bismuth subsalicylate | Gastrointestinal issues (ulcers, diarrhoea, acid reflux) | 1939 |
| Bismuth subcitrate potassium | Helicobacter pylori infection, peptic ulcers | 1998 | |
| Lithium (Li) | Lithium carbonate | Bipolar disorder | 1970 |
| Gallium (Ga) | Gallium nitrate | Hypercalcemia of malignancy | 2001 |
| Lanthanum (La) | Lanthanum carbonate | Renal failure | 2004 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Shaughnessy, M.; Sheils, O.; Baird, A.-M. The Lung Microbiome in COPD and Lung Cancer: Exploring the Potential of Metal-Based Drugs. Int. J. Mol. Sci. 2023, 24, 12296. https://doi.org/10.3390/ijms241512296
O’Shaughnessy M, Sheils O, Baird A-M. The Lung Microbiome in COPD and Lung Cancer: Exploring the Potential of Metal-Based Drugs. International Journal of Molecular Sciences. 2023; 24(15):12296. https://doi.org/10.3390/ijms241512296
Chicago/Turabian StyleO’Shaughnessy, Megan, Orla Sheils, and Anne-Marie Baird. 2023. "The Lung Microbiome in COPD and Lung Cancer: Exploring the Potential of Metal-Based Drugs" International Journal of Molecular Sciences 24, no. 15: 12296. https://doi.org/10.3390/ijms241512296
APA StyleO’Shaughnessy, M., Sheils, O., & Baird, A. -M. (2023). The Lung Microbiome in COPD and Lung Cancer: Exploring the Potential of Metal-Based Drugs. International Journal of Molecular Sciences, 24(15), 12296. https://doi.org/10.3390/ijms241512296