Mechanisms of Karyotypic Diversification in Ancistrus (Siluriformes, Loricariidae): Inferences from Repetitive Sequence Analysis
Abstract
:1. Introduction
2. Results
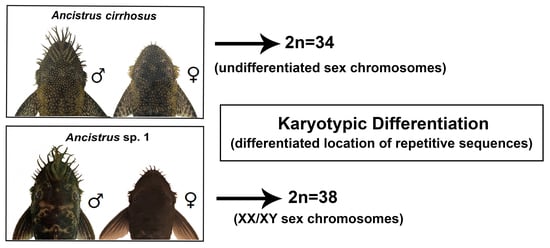
2.1. Karyotypes in Ancistrus
2.2. In Situ Localization of Repetitive Sequences in Ancistrus
2.3. U snDNA Sequences from Ancistrus
3. Discussion
4. Materials and Methods
4.1. Samples
4.2. Isolation and Amplification of Repetitive Sequences
4.3. Cloning, Sequencing, and Characterization of Repetitive Sequences
4.4. Obtaining and Analyzing Metaphases
4.5. Probes and Fluorescent In Situ Hybridization (FISH)
4.6. Images and Karyotypic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fricke, R.; Eschmeyer, W.N.; Van der Laan, R. (Eds.) Eschmeyer’s Catalog of Fishes: Genera, Species, References. 2023. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 12 July 2023).
- Borba, R.S.; Mariotto, S.; Centofante, L.; Zawadzki, C.H.; Parise-Maltempi, P.P. Molecular discrimination of Ancistrus lineages (Siluriformes: Loricariidae) using barcode DNA tool. Mitochondrial DNA Part A 2019, 30, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, E.B.; Britto, M.R.; Birindelli, J.L.O.; Sousa, L.M. A new species of Ancistrus (Siluriformes: Loricariidae) from Tapajós and Xingu basins, Brazil. Neotrop. Ichthyol. 2022, 20, e210129. [Google Scholar] [CrossRef]
- Mariotto, S.; Artoni, R.F.; Miyazawa, C.S. Occurrence of sexual chromosome, of the type ZZ/ZW, in Ancistrus cf. dubius (Loricariidae, Ancistrinae) of the Paraguay River Basin, Mato Grosso, Brazil. Caryologia 2004, 57, 327–331. [Google Scholar]
- Alves, A.L.; Oliveira, C.; Nirchio, M.; Granado, A.; Foresti, F. Karyotypic relationships among the tribes of Hypostominae (Siluriformes: Loricariidae) with description of XO sex chromosome system in a Neotropical fish species. Genetica 2006, 128, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mariotto, S.; Miyazawa, C.S. Ancistrus cf. dubius (Siluriformes, Ancistrinae), a complex of species. 1. Chromosomic characterization of four populations and occurence of sexual chromosomes of type XX/XY, in the pantanal basin of Mato Grosso, Brazil. Caryologia 2006, 59, 299–304. [Google Scholar] [CrossRef]
- De Oliveira, R.R.; Feldberg, E.; Anjos, M.B.D.; Zuanon, J. Karyotype characterization and ZZ/ZW sex chromosome heteromorphism in two species of the catfish genus Ancistrus Kner, 1854 (Siluriformes: Loricariidae) from the Amazon basin. Neotrop. Ichthyol. 2007, 5, 301–306. [Google Scholar] [CrossRef]
- De Oliveira, R.R.; Feldberg, E.; Dos Anjos, M.B.; Zuanon, J. Occurrence of multiple sexual chromosomes (XX/XY1Y2 and Z1Z1Z2Z2/Z1Z2W1W2) in catfishes of the genus Ancistrus (Siluriformes: Loricariidae) from the Amazon basin. Genetica 2008, 134, 243. [Google Scholar] [CrossRef]
- Santos da Silva, K.; Glugoski, L.; Vicari, M.R.; de Souza, A.C.P.; Noronha, R.C.R.; Pieczarka, J.C.; Nagamachi, C.Y. Chromosomal Diversification in Ancistrus Species (Siluriformes: Loricariidae) Inferred from Repetitive Sequence Analysis. Front.Genet. 2022, 13, 838462. [Google Scholar] [CrossRef]
- Nirchio, M.; Oliveira, C.; Cioffi, M.B.; de Menezes Cavalcante Sassi, F.; Valdiviezo, J.; Paim, F.G.; Soares, L.B.; Rossi, A.R. Occurrence of Sex Chromosomes in Fish of the Genus Ancistrus with a New Description of Multiple Sex Chromosomes in the Ecuadorian Endemic Ancistrus clementinae (Loricariidae). Genes 2023, 14, 306. [Google Scholar] [CrossRef]
- Bueno, V.; Konerat, J.T.; Zawadzki, C.H.; Venere, P.C.; Blanco, D.R.; Margarido, V.P. Divergent Chromosome Evolution in Hypostominae Tribes (Siluriformes: Loricariidae): Correlation of Chromosomal Data with Morphological and Molecular Phylogenies. Zebrafish 2018, 15, 492–503. [Google Scholar] [CrossRef]
- Barros, A.V.; Wolski, M.A.V.; Nogaroto, V.; Almeida, M.C.; Moreira-Filho, O.; Vicari, M.R. Fragile sites, dysfunctional telomere and chromosome fusions: What is 5S rDNA role? Gene 2017, 608, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Glugoski, L.; Deon, G.; Schott, S.; Vicari, M.R.; Nogaroto, V.; Moreira-Filho, O. Comparative cytogenetic analyses in Ancistrus species (Siluriformes: Loricariidae). Neotrop. Ichthyol. 2020, 18, e200013. [Google Scholar] [CrossRef]
- Venturelli, N.B.; Takagui, F.H.; Pompeo, L.R.S.; Rodriguez, M.S.; da Rosa, R.; Giuliano-Caetano, L. Cytogenetic markers to understand chromosome diversification and conflicting taxonomic issues in Rineloricaria (Loricariidae: Loricariinae) from Rio Grande do Sul coastal drainages. Biologia 2021, 76, 2561–2572. [Google Scholar] [CrossRef]
- Centofante, L.; Bertollo, L.A.C.; Moreira-Filho, O. Cytogenetic characterization and description of an XX/XY1Y2 sex chromosome system in catfish Harttia carvalhoi (Siluriformes, Loricariidae). Cytogenet. Genome Res. 2006, 112, 320–324. [Google Scholar] [CrossRef]
- Marajó, L.; Viana, P.F.; Ferreira, A.M.V.; Py-Daniel, L.H.R.; Cioffi, M.D.B.; Sember, A.; Feldberg, E. Chromosomal rearrangements and the first indication of an♀ X1X1X2X2/♂ X1X2Y sex chromosome system in Rineloricaria fishes (Teleostei: Siluriformes). J. Fish Biol. 2023, 102, 443–454. [Google Scholar] [CrossRef]
- Long, E.O.; Dawid, I.B. Repeated genes in eukaryotes. Annu. Rev. Biochem. 1980, 49, 727–764. [Google Scholar] [CrossRef]
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420. [Google Scholar] [CrossRef]
- Carbone, L.; Harris, R.A.; Vessere, G.M.; Mootnick, A.R.; Humphray, S.; Rogers, J.; Kim, K.S.; Wall, J.D.; Martin, D.; Jurka, J.; et al. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet. 2009, 5, e1000538. [Google Scholar] [CrossRef]
- Farré, M.; Bosch, M.; López-Giráldez, F.; Ponsa, M.; Ruiz-Herrera, A. Assessing the role of tandem repeats in shaping the genomic architecture of great apes. PLoS ONE 2011, 6, e27239. [Google Scholar] [CrossRef]
- Bailey, J.A.; Baertsch, R.; Kent, W.J.; Haussler, D.; Eichler, E.E. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004, 5, R23. [Google Scholar] [CrossRef]
- Gornung, E. Twenty years of physical mapping of major ribosomal RNA genes across the teleosts: A review of research. Cytogenet. Genome Res. 2013, 141, 90–102. [Google Scholar] [CrossRef]
- Rebordinos, L.; Cross, I.; Merlo, A. High evolutionary dynamism in 5S rDNA of fish: State of the art. Cytogenet. Genome Res. 2013, 141, 103–113. [Google Scholar] [CrossRef]
- Favarato, R.M.; da Silva, M.; de Oliveira, R.R.; Artoni, R.F.; Feldberg, E.; Matoso, D.A. Cytogenetic diversity and the evolutionary dynamics of rDNA genes and telomeric sequences in the Ancistrus genus (Loricariidae: Ancistrini). Zebrafish 2016, 13, 103–111. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Valente, G.T.; Nakajima, R.T.; Martins, C. Genomic organization and comparative chromosome mapping of the U1 snRNA gene in cichlid fish, with an emphasis in Oreochromis niloticus. Chromosome Res. 2012, 20, 279–292. [Google Scholar] [CrossRef]
- Santos da Silva, K.; de Souza, A.C.P.; Pety, A.M.; Noronha, R.C.R.; Vicari, M.R.; Pieczarka, J.C.; Nagamachi, C.Y. Comparative Cytogenetics Analysis among Peckoltia Species (Siluriformes, Loricariidae): Insights on Karyotype Evolution and Biogeography in the Amazon Region. Front. Genet. 2021, 12, 779464. [Google Scholar] [CrossRef]
- Azambuja, M.; Schemberger, M.O.; Nogaroto, V.; Moreira-Filho, O.; Martins, C.; Vicari, M.R. Major and minor U small nuclear RNAs genes characterization in a neotropical fish genome: Chromosomal remodeling and repeat units dispersion in Parodontidae. Gene 2022, 826, 146459. [Google Scholar] [CrossRef] [PubMed]
- Santos da Silva, K.; de Souza, A.C.P.; Rodrigues, L.R.R.; Pieczarka, J.C.; Nagamachi, C.Y. Chromosomal Diversification in Pseudacanthicus Species (Loricariidae, Hypostominae) Revealed by Comparative Mapping of Repetitive Sequences. Animals 2022, 12, 2612. [Google Scholar] [CrossRef]
- Schott, S.C.Q.; Glugoski, L.; Azambuja, M.; Moreira-Filho, O.; Vicari, M.R.; Nogaroto, V. Comparative cytogenetic and sequence analysis of U Small Nuclear RNA genes in three Ancistrus species (Siluriformes: Loricariidae). Zebrafish 2022, 19, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Usso, M.C.; Santos, A.R.D.; Gouveia, J.G.; Frantine-Silva, W.; Araya-Jaime, C.; Oliveira, M.L.M.D.; Foresti, F.; Giuliano-Caetano, L.; Dias, A.L. Genetic and chromosomal differentiation of Rhamdia quelen (Siluriformes, Heptapteridae) revealed by repetitive molecular markers and DNA barcoding. Zebrafish 2019, 16, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Busch, H.; Reddy, R.; Rothblum, L.; Choi, Y.C. SnRNAs, SnRNPs, and RNA processing. Annu. Rev. Biochem. 1982, 51, 617–654. [Google Scholar] [CrossRef]
- Yano, C.F.; Merlo, M.A.; Portela-Bens, S.; Cioffi, M.D.B.; Bertollo, L.A.; Santos-Júnior, C.D.; Rebordinos, L. Evolutionary dynamics of multigene families in Triportheus (Characiformes, Triportheidae): A transposon mediated mechanism? Front. Mar. Sci. 2020, 7, 6. [Google Scholar] [CrossRef]
- Yano, C.F.; Bertollo, L.A.C.; Rebordinos, L.; Merlo, M.A.; Liehr, T.; Portela-Bens, S.; Cioffi, M.D.B. Evolutionary dynamics of rDNAs and U2 small nuclear DNAs in Triportheus (Characiformes, Triportheidae): High variability and particular syntenic organization. Zebrafish 2017, 14, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Molina, W.F.; Yano, C.F.; Zhang, Y.; de Oliveira, E.A.; Lou, B.; de Bello Cioffi, M. Comparative cytogenetics in three Sciaenid species (Teleostei, Perciformes): Evidence of interspecific chromosomal diversification. Mol. Cytogenet. 2017, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Utsunomia, R.; Scacchetti, P.C.; Pansonato-Alves, J.C.; Oliveira, C.; Foresti, F. Comparative chromosome mapping of U2 snRNA and 5S rRNA genes in Gymnotus species (Gymnotiformes, Gymnotidae): Evolutionary dynamics and sex chromosome linkage in G. pantanal. Cytogenet. Genome Res. 2014, 142, 286–292. [Google Scholar] [CrossRef]
- Artoni, R.F.; Bertollo, L.A.C. Trends in the karyotype evolution of Loricariidae fish (Siluriformes). Hereditas 2001, 134, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Prizon, A.C.; Bruschi, D.P.; Gazolla, C.B.; Borin-Carvalho, L.A.; Portela-Castro, A.L.D.B. Chromosome spreading of the retrotransposable Rex-3 element and microsatellite repeats in karyotypes of the Ancistrus populations. Zebrafish 2018, 15, 504–514. [Google Scholar] [CrossRef]
- Slijepcevic, P. Telomeres and Mechanisms of Robertsonian Fusion. Chromossoma 1998, 107, 136–140. [Google Scholar] [CrossRef]
- Ziemniczak, K.; Barros, A.V.; Rosa, K.O.; Nogaroto, V.; Almeida, M.C.; Cestari, M.M.; Moreira-Filho, O.; Artoni, R.F.; Vicari, M.R. Comparative cytogenetics of Loricariidae (Actinopterygii: Siluriformes): Emphasis in Neoplecostominae and Hypoptopomatinae. Ital. J. Zool. 2012, 79, 492–501. [Google Scholar] [CrossRef]
- Cazaux, B.; Catalan, J.; Veyrunes, F.; Douzery, E.J.; Britton-Davidian, J. Are ribosomal DNA clusters rearrangement hotspots? A case study in the genus Mus (Rodentia, Muridae). BMC Evolut. Biol. 2011, 11, 124. [Google Scholar] [CrossRef]
- Huang, J.; Ma, L.; Yang, F.; Fei, S.Z.; Li, L. 45S rDNA regions are chromosome fragile sites expressed as gaps in vitro on metaphase chromosomes of root-tip meristematic cells in Lolium spp. PLoS ONE 2008, 3, e2167. [Google Scholar] [CrossRef]
- Martins, C.; Wasko, A.P. Organization and evolution of 5S ribosomal DNA in the fish genome. Focus Genome Res. 2004, 289, 318. [Google Scholar]
- Ubeda-Manzanaro, M.; Merlo, M.A.; Palazon, J.L.; Cross, I.; Sarasquete, C.; Rebordinos, L. Chromosomal mapping of the major and minor ribosomal genes, (GATA)n and U2 snRNA gene by double-colour FISH in species of the Batrachoididae family. Genetica 2010, 138, 787–794. [Google Scholar] [CrossRef]
- Gross, M.C.; Schneider, C.H.; Valente, G.T.; Martins, C.; Feldberg, E. Variability of 18S rDNA locus among Symphysodon fishes: Chromosomal rearrangements. J. Fish Biol. 2010, 76, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.; Galetti, P.M., Jr. Chromosomal localization of 5S rDNA genes in Leporinus Fish (Anostomidae, Characiformes). Chromosome Res. 1999, 7, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, C.; Duran, T.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Bernhart, S.H.; Hofacker, I.L.; Will, S.; Gruber, A.R.; Stadler, P.F. RNAalifold: Improved consensus structure prediction for RNA alignments. BMC Bioinform. 2008, 9, 474. [Google Scholar] [CrossRef]
- Bertollo, L.A.C.; Takahashi, C.S.; Moreira-Filho, O. Cytotaxonomic considerations on Hoplias lacerdae (Pisces Erythrinidae). Braz. J. Genet. 1978, 1, 103–120. [Google Scholar]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Pinkel, D.; Straume, T.; Gray, J.W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 2934–2938. [Google Scholar] [CrossRef]
- Ijdo, J.W.; Wells, R.A.; Baldini, A.; Reeders, S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucl. Acids Res. 1991, 19, 4780. [Google Scholar] [CrossRef] [PubMed]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]





| Species | Populations | 2n | FN | KF | SC | rDNA | snDNA | Ref. | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 18S | 5S | U1 | U2 | |||||||
| Ancistrus sp. 1 | Quianduba (*) | 38 | 72 | 20 m + 14 sm + 4 st | XX/XY | 2q | 4p | 14q | 2, 18p | 1, 2 |
| Maracapucú | ||||||||||
| Capim island | 4p, 5p | 2, 18p, homologue 18q | ||||||||
| Ancistrus cirrhosus | Maracapucú (*) | 34 | 68 | 20 m + 14 sm | Not found | 3p | 4p, 6p, 7p, 16 | 13q | 11 | 1, 2 |
| Capim island (*) | ||||||||||
| Sequences | BLASTn (GenBank) | ||||
|---|---|---|---|---|---|
| Gene Family | Organism | Ident (%) | E-Value | Access Number | |
| U1 (157-pb) * | U1 spliceosomal RNA | Pygocentrus nattereri | 88.61 | 9-10−44 | XR_005129869.1 |
| U2 (1112-pb) | U2 spliceosomal RNA | Megaleporinus obtusidens | 90.05 | 2-10−61 | MT_563075.1 |
| U5 spliceosomal RNA | Astyanax paranae | 92.31 | 3-10−31 | MG_963291.1 | |
| U2 spliceosomal RNA | Anguilla anguilla | 91.39 | 2-10−49 | XR_004763517.1 | |
| U2 (190-pb) * | U2 spliceosomal RNA | Tachysurus fulvidraco | 87.33 | 4-10−38 | XR_007138734.1 |
| Sequences | Rfam | |||||
|---|---|---|---|---|---|---|
| Gene Family | Start | End | Bit Score | E-Value | Access Number | |
| U1 (157-pb) * | U1 spliceosomal RNA | 1 | 196 | 107.2 | 6.2-10−29 | RF00003 |
| U2 (1112-pb) | U2 spliceosomal RNA | 1 | 191 | 179.7 | 7-10−31 | RF00004 |
| U5 spliceosomal RNA | 562 | 676 | 80.6 | 2-10−13 | RF00020 | |
| U2 spliceosomal RNA | 954 | 1112 | 142.4 | 8.1-10−40 | RF00004 | |
| U2 (190-pb) * | U2 spliceosomal RNA | 1 | 190 | 119 | 4.9-10−26 | RF00004 |
| Species | Populations | Sex | River | Locality | Voucher (#) | Coordinates | |
|---|---|---|---|---|---|---|---|
| Ancistrus sp. 1 | Quianduba (*) | 5♂ | 2♀ | A | Abaetetuba/PA | P4229 | S01°45′18.2″/W49°00′38.8″ |
| Maracapucú | 2♂ | -♀ | B | Abaetetuba/PA | P4264 | S01°45′29.2″/W48°56′57″ | |
| Capim island | 1♂ | -♀ | C | Abaetetuba/PA | P4250 | S01°34′02.8″/W48°51′49.1″ | |
| Ancistrus cirrhosus | Maracapucú (*) | 8♂ | 1♀ | B | Abaetetuba/PA | P4263 | S01°45′29.2″/W48°56′57″ |
| Capim island (*) | -♂ | 1♀ | C | Abaetetuba/PA | P4251 | S01°34′02.8″/W48°51′49.1″ | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos da Silva, K.; Glugoski, L.; Vicari, M.R.; de Souza, A.C.P.; Akama, A.; Pieczarka, J.C.; Nagamachi, C.Y. Mechanisms of Karyotypic Diversification in Ancistrus (Siluriformes, Loricariidae): Inferences from Repetitive Sequence Analysis. Int. J. Mol. Sci. 2023, 24, 14159. https://doi.org/10.3390/ijms241814159
Santos da Silva K, Glugoski L, Vicari MR, de Souza ACP, Akama A, Pieczarka JC, Nagamachi CY. Mechanisms of Karyotypic Diversification in Ancistrus (Siluriformes, Loricariidae): Inferences from Repetitive Sequence Analysis. International Journal of Molecular Sciences. 2023; 24(18):14159. https://doi.org/10.3390/ijms241814159
Chicago/Turabian StyleSantos da Silva, Kevin, Larissa Glugoski, Marcelo Ricardo Vicari, Augusto César Paes de Souza, Alberto Akama, Julio Cesar Pieczarka, and Cleusa Yoshiko Nagamachi. 2023. "Mechanisms of Karyotypic Diversification in Ancistrus (Siluriformes, Loricariidae): Inferences from Repetitive Sequence Analysis" International Journal of Molecular Sciences 24, no. 18: 14159. https://doi.org/10.3390/ijms241814159
APA StyleSantos da Silva, K., Glugoski, L., Vicari, M. R., de Souza, A. C. P., Akama, A., Pieczarka, J. C., & Nagamachi, C. Y. (2023). Mechanisms of Karyotypic Diversification in Ancistrus (Siluriformes, Loricariidae): Inferences from Repetitive Sequence Analysis. International Journal of Molecular Sciences, 24(18), 14159. https://doi.org/10.3390/ijms241814159