Validation of the Reference Genes for Expression Analysis in the Hippocampus after Transient Ischemia/Reperfusion Injury in Gerbil Brain
Abstract
:1. Introduction
2. Results
2.1. qRT-PCR Primer Efficiency and Specificity and Expression Levels of Candidate Reference Genes
2.2. Comprehensive Ranking of Candidate Reference Genes in CA1 and CA2-3,DG of Gerbil Hippocampus
2.2.1. ΔCt Method
2.2.2. BestKeeper
2.2.3. NormFinder
2.2.4. GeNorm
2.2.5. RefFinder in CA1 and CA2-3,DG
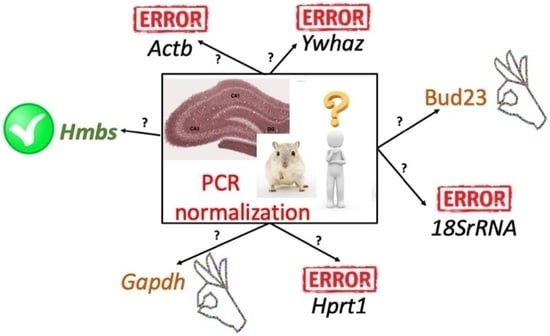
2.3. RefFinder in Combined Hippocampal Samples
3. Discussion
4. Materials and Methods
4.1. Ethical Statement and Animals
4.2. Transient Brain Ischemia in Gerbils
4.3. RNA Isolation and Synthesis of cDNA
4.4. Gene Selection and Primer Design
4.5. qRT-PCR
4.6. Statistical Data Analysis
4.7. Data Analysis of Gene Expression Stability
4.7.1. Delta Ct (ΔCt) Method
4.7.2. BestKeeper
4.7.3. NormFinder
4.7.4. GeNorm
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beresewicz-Haller, M. Hippocampal region-specific endogenous neuroprotection as an approach in the search for new neuroprotective strategies in ischemic stroke. Fiction or fact? Neurochem. Int. 2023, 162, 105455. [Google Scholar] [CrossRef] [PubMed]
- Sanganalmath, S.K.; Gopal, P.; Parker, J.R.; Downs, R.K.; Parker, J.C., Jr.; Dawn, B. Global cerebral ischemia due to circulatory arrest: Insights into cellular pathophysiology and diagnostic modalities. Mol. Cell Biochem. 2017, 426, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Zablocka, B.; Maternicka, K.; Zalewska, T.; Domanska-Janik, K. Expression of Ca2+-dependent (classical) PKC mRNA isoforms after transient cerebral ischemia in gerbil hippocampus. Brain Res. 1998, 779, 254–258. [Google Scholar] [CrossRef]
- Dos-Anjos, S.; Martinez-Villayandre, B.; Montori, S.; Regueiro-Purrinos, M.M.; Gonzalo-Orden, J.M.; Fernandez-Lopez, A. Transient global ischemia in rat brain promotes different NMDA receptor regulation depending on the brain structure studied. Neurochem. Int. 2009, 54, 180–185. [Google Scholar] [CrossRef]
- Kharlamov, E.A.; Downey, K.L.; Jukkola, P.I.; Grayson, D.R.; Kelly, K.M. Expression of GABA A receptor alpha1 subunit mRNA and protein in rat neocortex following photothrombotic infarction. Brain Res. 2008, 1210, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Yoo, D.Y.; Cho, S.B.; Jung, H.Y.; Kim, W.; Nam, S.M.; Kim, J.W.; Moon, S.M.; Yoon, Y.S.; Kim, D.W.; Choi, S.Y.; et al. Differential roles of exogenous protein disulfide isomerase A3 on proliferating cell and neuroblast numbers in the normal and ischemic gerbils. Brain Behav. 2020, 10, e01534. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Kang, C.; Hou, X.; Zhang, Q.; Meng, Q.; Jiang, J.; Hao, W. Blue Light Deprivation Produces Depression-Like Responses in Mongolian Gerbils. Front. Psychiatry 2020, 11, 233. [Google Scholar] [CrossRef]
- Kovac, S.; Angelova, P.R.; Holmstrom, K.M.; Zhang, Y.; Dinkova-Kostova, A.T.; Abramov, A.Y. Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim. Biophys. Acta 2015, 1850, 794–801. [Google Scholar] [CrossRef]
- Schwarz, A.P.; Kovalenko, A.A.; Malygina, D.A.; Postnikova, T.Y.; Zubareva, O.E.; Zaitsev, A.V. Reference Gene Validation in the Brain Regions of Young Rats after Pentylenetetrazole-Induced Seizures. Biomedicines 2020, 8, 239. [Google Scholar] [CrossRef]
- Otto, E.; Kohli, P.; Appelt, J.; Menzel, S.; Fuchs, M.; Bahn, A.; Graef, F.; Duda, G.N.; Tsitsilonis, S.; Keller, J.; et al. Validation of reference genes for expression analysis in a murine trauma model combining traumatic brain injury and femoral fracture. Sci. Rep. 2020, 10, 15057. [Google Scholar] [CrossRef]
- Kohsler, M.; Leitsch, D.; Muller, N.; Walochnik, J. Validation of reference genes for the normalization of RT-qPCR gene expression in Acanthamoeba spp. Sci. Rep. 2020, 10, 10362. [Google Scholar] [CrossRef] [PubMed]
- Feria-Romero, I.A.; Bribiesca-Cruz, I.; Coyoy-Salgado, A.; Segura-Uribe, J.J.; Bautista-Poblet, G.; Granados-Cervantes, A.; Guerra-Araiza, C. Validation of housekeeping genes as an internal control for gene expression studies in the brain of ovariectomized rats treated with tibolone. Gene 2021, 769, 145255. [Google Scholar] [CrossRef] [PubMed]
- Sieber, M.W.; Guenther, M.; Kohl, M.; Witte, O.W.; Claus, R.A.; Frahm, C. Inter-age variability of bona fide unvaried transcripts Normalization of quantitative PCR data in ischemic stroke. Neurobiol. Aging 2010, 31, 654–664. [Google Scholar] [CrossRef]
- Pomierny, B.; Krzyzanowska, W.; Jurczyk, J.; Strach, B.; Skorkowska, A.; Leonovich, I.; Budziszewska, B.; Pera, J. Identification of optimal reference genes for gene expression studies in a focal cerebral ischaemia model-Spatiotemporal effects. J. Cell Mol. Med. 2022, 26, 3060–3067. [Google Scholar] [CrossRef]
- Pluta, R.; Kocki, J.; Ułamek-Kozioł, M.; Petniak, A.; Gil-Kulik, P.; Januszewski, S.; Bogucki, J.; Jabłoński, M.; Brzozowska, J.; Furmaga-Jabłońska, W.; et al. Discrepancy in Expression of β-Secretase and Amyloid-β Protein Precursor in Alzheimer-Related Genes in the Rat Medial Temporal Lobe Cortex Following Transient Global Brain Ischemia. JAD 2016, 51, 1023–1031. [Google Scholar] [CrossRef]
- Beresewicz, M.; Majewska, M.; Makarewicz, D.; Vayro, S.; Zablocka, B.; Gorecki, D.C. Changes in the expression of insulin-like growth factor 1 variants in the postnatal brain development and in neonatal hypoxia-ischaemia. Int. J. Dev. Neurosci. 2010, 28, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Turovsky, E.A.; Mal’tseva, V.N.; Sarimov, R.M.; Simakin, A.V.; Gudkov, S.V.; Plotnikov, E.Y. Features of the cytoprotective effect of selenium nanoparticles on primary cortical neurons and astrocytes during oxygen-glucose deprivation and reoxygenation. Sci. Rep. 2022, 12, 1710. [Google Scholar] [CrossRef]
- Nishida, Y.; Sugahara-Kobayashi, M.; Takahashi, Y.; Nagata, T.; Ishikawa, K.; Asai, S. Screening for control genes in mouse hippocampus after transient forebrain ischemia using high-density oligonucleotide array. J. Pharmacol. Sci. 2006, 101, 52–57. [Google Scholar] [CrossRef]
- Kobayashi, M.S.; Takahashi, Y.; Nagata, T.; Nishida, Y.; Murata, A.; Ishikawa, K.; Asai, S. Screening for control genes in rat global cerebral ischemia using high-density oligonucleotide array. J. Neurosci. Res. 2004, 76, 512–518. [Google Scholar] [CrossRef]
- Kang, Y.; Wu, Z.; Cai, D.; Lu, B. Evaluation of reference genes for gene expression studies in mouse and N2a cell ischemic stroke models using quantitative real-time PCR. BMC Neurosci. 2018, 19, 3. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.F.; Zhang, P.B.; Xiao, X.L.; Zhang, J.S.; Zhao, J.J.; Kang, Q.Y.; Chen, X.L.; Qiu, F.; Liu, Y. The quantification of ADAMTS expression in an animal model of cerebral ischemia using real-time PCR. Acta Anaesthesiol. Scand. 2007, 51, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Kirino, T. Delayed neuronal death in the gerbil hippocampus following ischemia. Brain Res. 1982, 239, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Krupska, O.; Kowalczyk, T.; Beresewicz-Haller, M.; Samczuk, P.; Pietrowska, K.; Zablocki, K.; Kretowski, A.; Ciborowski, M.; Zablocka, B. Hippocampal Sector-Specific Metabolic Profiles Reflect Endogenous Strategy for Ischemia-Reperfusion Insult Resistance. Mol. Neurobiol. 2020, 58, 1621–1633. [Google Scholar] [CrossRef]
- Krupska, O.; Sarnowska, A.; Fedorczyk, B.; Gewartowska, M.; Misicka, A.; Zablocka, B.; Beresewicz, M. Ischemia/Reperfusion-Induced Translocation of PKCbetaII to Mitochondria as an Important Mediator of a Protective Signaling Mechanism in an Ischemia-Resistant Region of the Hippocampus. Neurochem. Res. 2017, 42, 2392–2403. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Li, Z.; Sardana, R.; Bujnicki, J.M.; Marcotte, E.M.; Johnson, A.W. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell Biol. 2008, 28, 3151–3161. [Google Scholar] [CrossRef] [PubMed]
- Black, J.J.; Johnson, A.W. Genetics animates structure: Leveraging genetic interactions to study the dynamics of ribosome biogenesis. Curr. Genet. 2021, 67, 729–738. [Google Scholar] [CrossRef]
- Baxter, M.; Voronkov, M.; Poolman, T.; Galli, G.; Pinali, C.; Goosey, L.; Knight, A.; Krakowiak, K.; Maidstone, R.; Iqbal, M.; et al. Cardiac mitochondrial function depends on BUD23 mediated ribosome programming. eLife 2020, 9, e50705. [Google Scholar] [CrossRef]
- Chi, Y.; Liang, Z.; Guo, Y.; Chen, D.; Lu, L.; Lin, J.; Qiu, S.; Wang, X.; Qiu, E.; Lin, F.; et al. WBSCR22 confers cell survival and predicts poor prognosis in glioma. Brain Res. Bull. 2020, 161, 1–12. [Google Scholar] [CrossRef]
- Jangani, M.; Poolman, T.M.; Matthews, L.; Yang, N.; Farrow, S.N.; Berry, A.; Hanley, N.; Williamson, A.J.; Whetton, A.D.; Donn, R.; et al. The methyltransferase WBSCR22/Merm1 enhances glucocorticoid receptor function and is regulated in lung inflammation and cancer. J. Biol. Chem. 2014, 289, 8931–8946. [Google Scholar] [CrossRef]
- Guo, Y.Y.; Chi, Q.S.; Zhang, X.Y.; Liu, W.; Hao, S.Y.; Wang, D.H. Brown adipose tissue plays thermoregulatory role within the thermoneutral zone in Mongolian gerbils (Meriones unguiculatus). J. Therm. Biol. 2019, 81, 137–145. [Google Scholar] [CrossRef]
- Ishijima, N.; Suzuki, M.; Ashida, H.; Ichikawa, Y.; Kanegae, Y.; Saito, I.; Borén, T.; Haas, R.; Sasakawa, C.; Mimuro, H. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J. Biol. Chem. 2011, 286, 25256–25264. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Chapman, J.R.; Waldenström, J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef] [PubMed]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Gozzelino, R. The Pathophysiology of Heme in the Brain. Curr. Alzheimer Res. 2016, 13, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.G.; Raven, E.L.; Chernova, T. The regulatory role of heme in neurons. Metallomics 2011, 3, 955–962. [Google Scholar] [CrossRef]
- Simon, N.G.; Herkes, G.K. The neurologic manifestations of the acute porphyrias. J. Clin. Neurosci. 2011, 18, 1147–1153. [Google Scholar] [CrossRef]
- Larsen, R.; Gouveia, Z.; Soares, M.P.; Gozzelino, R. Heme cytotoxicity and the pathogenesis of immune-mediated inflammatory diseases. Front. Pharmacol. 2012, 3, 77. [Google Scholar] [CrossRef]
- Fiorito, V.; Allocco, A.L.; Petrillo, S.; Gazzano, E.; Torretta, S.; Marchi, S.; Destefanis, F.; Pacelli, C.; Audrito, V.; Provero, P.; et al. The heme synthesis-export system regulates the tricarboxylic acid cycle flux and oxidative phosphorylation. Cell Rep. 2021, 35, 109252. [Google Scholar] [CrossRef]
- Medlock, A.E.; Dailey, H.A. New Avenues of Heme Synthesis Regulation. Int. J. Mol. Sci. 2022, 23, 7467. [Google Scholar] [CrossRef]
- Sun, F.; Cheng, Y.; Chen, C. Regulation of heme biosynthesis and transport in metazoa. Sci. China Life Sci. 2015, 58, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Lewczuk, A.; Zablocka, B.; Beresewicz-Haller, M. Is Nrf2 Behind Endogenous Neuroprotection of the Hippocampal CA2-4,DG Region? Mol. Neurobiol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Kannan, M.; Steinert, J.R.; Forsythe, I.D.; Smith, A.G.; Chernova, T. Mevastatin accelerates loss of synaptic proteins and neurite degeneration in aging cortical neurons in a heme-independent manner. Neurobiol. Aging 2010, 31, 1543–1553. [Google Scholar] [CrossRef] [PubMed]
- Iadecola, C.; Zhang, F.; Xu, S.; Casey, R.; Ross, M.E. Inducible nitric oxide synthase gene expression in brain following cerebral ischemia. J. Cereb. Blood Flow Metab. 1995, 15, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Bémeur, C.; Ste-Marie, L.; Desjardins, P.; Hazell, A.S.; Vachon, L.; Butterworth, R.; Montgomery, J. Decreased beta-actin mRNA expression in hyperglycemic focal cerebral ischemia in the rat. Neurosci. Lett. 2004, 357, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Vesentini, N.; Barsanti, C.; Martino, A.; Kusmic, C.; Ripoli, A.; Rossi, A.; L’Abbate, A. Selection of reference genes in different myocardial regions of an in vivo ischemia/reperfusion rat model for normalization of antioxidant gene expression. BMC Res. Notes 2012, 5, 124. [Google Scholar] [CrossRef]
- Stassen, Q.E.; Riemers, F.M.; Reijmerink, H.; Leegwater, P.A.; Penning, L.C. Reference genes for reverse transcription quantitative PCR in canine brain tissue. BMC Res. Notes 2015, 8, 761. [Google Scholar] [CrossRef]
- Gubern, C.; Hurtado, O.; Rodríguez, R.; Morales, J.R.; Romera, V.G.; Moro, M.A.; Lizasoain, I.; Serena, J.; Mallolas, J. Validation of housekeeping genes for quantitative real-time PCR in in-vivo and in-vitro models of cerebral ischaemia. BMC Mol. Biol. 2009, 10, 57. [Google Scholar] [CrossRef]
- Julian, G.S.; de Oliveira, R.W.; Perry, J.C.; Tufik, S.; Chagas, J.R. Validation of housekeeping genes in the brains of rats submitted to chronic intermittent hypoxia, a sleep apnea model. PLoS ONE 2014, 9, e109902. [Google Scholar] [CrossRef]
- Zorbas, C.; Nicolas, E.; Wacheul, L.; Huvelle, E.; Heurgué-Hamard, V.; Lafontaine, D.L. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell 2015, 26, 2080–2095. [Google Scholar] [CrossRef]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control selection for RNA quantitation. BioTechniques 2000, 29, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, R.; Fujita, K.; Takahashi, S.; Yoneda, H.; Nagao, K.; Masuda, W.; Naito, M.; Tsuruo, T.; Miyatake, K.; Inui, H.; et al. Hypoxia up-regulates glyceraldehyde-3-phosphate dehydrogenase in mouse brain capillary endothelial cells: Involvement of Na+/Ca2+ exchanger. Biochim. Biophys. Acta 2003, 1593, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Simons, J.W. Direct comparison of GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem. Biophys. Res. Commun. 1999, 259, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Graven, K.K.; Troxler, R.F.; Kornfeld, H.; Panchenko, M.V.; Farber, H.W. Regulation of endothelial cell glyceraldehyde-3-phosphate dehydrogenase expression by hypoxia. J. Biol. Chem. 1994, 269, 24446–24453. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.C.; Medhurst, A.D.; Bond, B.C.; Campbell, C.A.; Davis, R.P.; Philpott, K.L. The use of quantitative RT-PCR to measure mRNA expression in a rat model of focal ischemia—Caspase-3 as a case study. Brain Res. Mol. Brain Res. 2000, 75, 143–149. [Google Scholar] [CrossRef]
- Tanaka, R.; Mochizuki, H.; Suzuki, A.; Katsube, N.; Ishitani, R.; Mizuno, Y.; Urabe, T. Induction of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in rat brain after focal ischemia/reperfusion. J. Cereb. Blood Flow Metab. 2002, 22, 280–288. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, B.H.; Wu, S.; Wu, C.H.; Suo, J.L.; Gui, Y.; Zhou, C.M.; Li, Y.C. Different changes in pre- and postsynaptic components in the hippocampal CA1 subfield after transient global cerebral ischemia. Brain Struct. Funct. 2022, 227, 345–360. [Google Scholar] [CrossRef]
- Xie, Y.X.; Herget, T.; Hallmayer, J.; Starzinski-Powitz, A.; Hossmann, K.A. Determination of RNA content in postischemic gerbil brain by in situ hybridization. Metab. Brain Dis. 1989, 4, 239–251. [Google Scholar] [CrossRef]
- Kumar, K.; Wu, X.L. Expression of beta-actin and alpha-tubulin mRNA in gerbil brain following transient ischemia and reperfusion up to 1 month. Mol. Brain Res. 1995, 30, 149–157. [Google Scholar] [CrossRef]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [Green Version]





| Gene | Accession Number | Primer Sequence (5′–3′) | Amplicon Size (bp) | Primer Efficiency | R2 | Reference |
|---|---|---|---|---|---|---|
| Gapdh | XM_021636934 | F: AGTATGACTCTACCCACGGC | 150 | 103.94% | 0.999 | [30] |
| R: ACTCCACAACATACTCGGCA | ||||||
| Actb | XM_021652973 | F: CTCTGGCTCCTAGCACCATG | 129 | 96.39% | 0.999 | |
| R: ACTCCTGCTTGCTGATCCAC | ||||||
| 18S rRNA | AJ877917 | F: GGCTACCACATCCAAGGAAGG | 101 | 99.67% | 0.999 | [31] |
| R: AGGGCCTCGAAAGAGTCCTG | ||||||
| Hprt1 | XM_021660111 | F: TTACGGCTTTCCTGGAGGTG | 136 | 90.10% | 0.999 | |
| R: TAGCCTGGTTCATCATCGCC | ||||||
| Hmbs | XM_021659401 | F: GAAGAGTGGCCCAGCTACAG | 108 | 103.57% | 0.999 | |
| R: CACTGAACTCCTGCTGCTCA | ||||||
| Ywhaz | XM_021657529 | F: CACTCACTCCGGACACAGAA | 197 | 107.05% | 0.997 | |
| R: CCTACGGGCTCCTACAACAT | ||||||
| Bud23 | XM_021664516 | F: GTGGCTCTGCAATGCAAACA | 132 | 104.37% | 0.998 | |
| R: TGCTCCGAGTTCTCAGGGTA |
| Region of Hippocampus | Gene Name | RefFinder | Delta Ct | BestKeeper | NormFinder | geNorm | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SV | R | SV | R | SV | R | SV | R | SV | R | ||
| Combined | Gapdh | 2.632 | 3 | 0.598 | 2 | 0.575 | 4 | 0.319 | 2 | 0.455 | 3 |
| Actb | 5.733 | 6 | 0.741 | 6 | 0.593 | 5 | 0.585 | 6 | 0.57 | 6 | |
| 18S rRNA | 7.00 | 7 | 1.018 | 7 | 0.962 | 7 | 0.936 | 7 | 0.698 | 7 | |
| Hprt1 | 3.976 | 4 | 0.67 | 5 | 0.525 | 2 | 0.447 | 5 | 0.529 | 5 | |
| Hmbs | 1.00 | 1 | 0.59 | 1 | 0.452 | 1 | 0.26 | 1 | 0.422 | 1 | |
| Ywhaz | 4.427 | 5 | 0.656 | 4 | 0.647 | 6 | 0.432 | 4 | 0.507 | 4 | |
| Bud23 | 2.28 | 2 | 0.612 | 3 | 0.534 | 3 | 0.341 | 3 | 0.422 | 1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewczuk, A.; Boratyńska-Jasińska, A.; Zabłocka, B. Validation of the Reference Genes for Expression Analysis in the Hippocampus after Transient Ischemia/Reperfusion Injury in Gerbil Brain. Int. J. Mol. Sci. 2023, 24, 2756. https://doi.org/10.3390/ijms24032756
Lewczuk A, Boratyńska-Jasińska A, Zabłocka B. Validation of the Reference Genes for Expression Analysis in the Hippocampus after Transient Ischemia/Reperfusion Injury in Gerbil Brain. International Journal of Molecular Sciences. 2023; 24(3):2756. https://doi.org/10.3390/ijms24032756
Chicago/Turabian StyleLewczuk, Anita, Anna Boratyńska-Jasińska, and Barbara Zabłocka. 2023. "Validation of the Reference Genes for Expression Analysis in the Hippocampus after Transient Ischemia/Reperfusion Injury in Gerbil Brain" International Journal of Molecular Sciences 24, no. 3: 2756. https://doi.org/10.3390/ijms24032756
APA StyleLewczuk, A., Boratyńska-Jasińska, A., & Zabłocka, B. (2023). Validation of the Reference Genes for Expression Analysis in the Hippocampus after Transient Ischemia/Reperfusion Injury in Gerbil Brain. International Journal of Molecular Sciences, 24(3), 2756. https://doi.org/10.3390/ijms24032756