Effects of Long-Term Physical Activity and BCAA Availability on the Subcellular Associations between Intramyocellular Lipids, Perilipins and PGC-1α
Abstract
:1. Introduction
2. Results
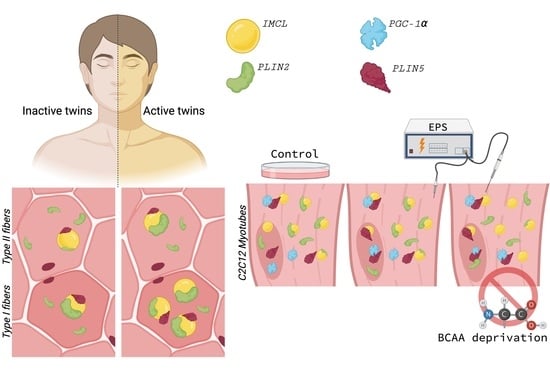
2.1. Active Co-Twins Have Increased IMCL in Type I Fibers
2.2. Inactive Twins Show Decreased IMCL-PLIN2 Association
2.3. PLIN5 Abounds in C2C12 Myotube Nuclei, PLIN2 Detected
2.4. PLIN2 Dissociates from IMCL upon BCAA Deprivation in Myotubes
2.5. PLIN5 Moves to Myotube Nuclei upon Stimulation, Further Associating with IMCL and PGC-1
3. Discussion
3.1. Overview
3.2. Active Twins Resemble Athlete Phenotype
3.3. Myotubes Resembling Type II Fibers
3.4. BCAA Necessary for PLIN2 Coating of IMCL
3.5. Setting Transcription in Cytosol?
3.6. Nuclear Affairs
3.7. Limitations and Strengths
4. Materials and Methods
4.1. Human Twin Pairs
4.2. Myotube Experiments
4.3. Protein Extraction and Western Blotting
4.4. Gene-Expression Arrays
4.5. Histology
4.6. Image Acquisition
4.7. Image Analyses
4.8. Data Cleaning and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACAA2 | Acetyl-Coenzyme A acyltransferase 2 |
| ACAT1 | Acetyl-CoA acetyltransferase, mitochondrial |
| ACAT2 | Acetyl-CoA acetyltransferase, cytosolic |
| ATGL | Adipose triglyceride lipase |
| BCAA | Branched-chain amino acids |
| EPS | Electrical pulse stimulation |
| ERR | Estrogen-related receptor alpha |
| HMGCS2 | 3-hydroxy-3-methylglutaryl-CoA synthase 2, mitochondrial |
| HSL | Hormone-sensitive lipase |
| ICA | Intensity correlation analysis |
| IMCL | Intramyocellular lipids |
| LDs | Lipid droplets |
| LTPA | Leisure time physical activity |
| mTORC1 | Mammalian target of rapamycin complex 1 |
| MUFAs | Monounsaturated fatty acids |
| MyHC | Myosin heavy chain |
| PGC-1 | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha |
| PLIN2 | Perilipin 2 |
| PLIN3 | Perilipin 3 |
| PLIN5 | Perilipin 5 |
| PLINs | Perilipin protein family |
| PPAR- | Peroxisome proliferator-activated receptor alpha |
| SIRT1 | Sirtuin 1 |
| SIRT3 | Sirtuin 3 |
| TAG | Triacylglycerol |
References
- Pedersen, B.K. Muscle as a Secretory Organ. In Comprehensive Physiology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; pp. 1337–1362. [Google Scholar] [CrossRef]
- Goodpaster, B.H.; He, J.; Watkins, S.; Kelley, D.E. Skeletal Muscle Lipid Content and Insulin Resistance: Evidence for a Paradox in Endurance-Trained Athletes. J. Clin. Endocrinol. Metab. 2001, 86, 5755–5761. [Google Scholar] [CrossRef]
- Moro, C.; Bajpeyi, S.; Smith, S.R. Determinants of intramyocellular triglyceride turnover: Implications for insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 2008, 294, E203–E213. [Google Scholar] [CrossRef]
- van Loon, L.J.C.; Koopman, R.; Manders, R.; van der Weegen, W.; van Kranenburg, G.P.; Keizer, H.A. Intramyocellular lipid content in type 2 diabetes patients compared with overweight sedentary men and highly trained endurance athletes. Am. J. Physiol. Endocrinol. Metab. 2004, 287, E558–E565. [Google Scholar] [CrossRef]
- Sztalryd, C.; Brasaemle, D.L. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1221–1232. [Google Scholar] [CrossRef]
- MacPherson, R.E.; Peters, S.J. Piecing together the puzzle of perilipin proteins and skeletal muscle lipolysis. Appl. Physiol. Nutr. Metab. 2015, 40, 641–651. [Google Scholar] [CrossRef]
- Prats, C.; Donsmark, M.; Qvortrup, K.; Londos, C.; Sztalryd, C.; Holm, C.; Galbo, H.; Ploug, T. Decrease in intramuscular lipid droplets and translocation of HSL in response to muscle contraction and epinephrine. J. Lipid Res. 2006, 47, 2392–2399. [Google Scholar] [CrossRef] [Green Version]
- Whytock, K.; Shepherd, S.; Wagenmakers, A.; Strauss, J. Hormone sensitive lipase preferentially redistributes to perilipin-5 lipid droplets in human skeletal muscle during moderate-intensity exercise. J. Physiol. 2018, 596, 2077–2090. [Google Scholar] [CrossRef] [Green Version]
- Unraveling the roles of PLIN5: Linking cell biology to physiology. Trends Endocrinol. Metab. 2015, 26, 144–152. [CrossRef]
- Gemmink, A.; Bosma, M.; Kuijpers, H.J.; Hoeks, J.; Schaart, G.; van Zandvoort, M.A.; Schrauwen, P.; Hesselink, M.K. Decoration of intramyocellular lipid droplets with PLIN5 modulates fasting-induced insulin resistance and lipotoxicity in humans. Diabetologia 2016, 59, 1040–1048. [Google Scholar] [CrossRef] [Green Version]
- Gemmink, A.; Daemen, S.; Brouwers, B.; Hoeks, J.; Schaart, G.; Knoops, K.; Schrauwen, P.; Hesselink, M.K. Decoration of myocellular lipid droplets with perilipins as a marker for in vivo lipid droplet dynamics: A super-resolution microscopy study in trained athletes and insulin resistant individuals. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158852. [Google Scholar] [CrossRef]
- Gemmink, A.; Daemen, S.; Brouwers, B.; Huntjens, P.R.; Schaart, G.; Moonen-Kornips, E.; Jörgensen, J.; Hoeks, J.; Schrauwen, P.; Hesselink, M.K.C. Dissociation of intramyocellular lipid storage and insulin resistance in trained athletes and type 2 diabetes patients; involvement of perilipin 5? J. Physiol. 2018, 596, 857–868. [Google Scholar] [CrossRef] [Green Version]
- Shepherd, S.O.; Cocks, M.; Tipton, K.D.; Ranasinghe, A.M.; Barker, T.A.; Burniston, J.G.; Wagenmakers, A.J.M.; Shaw, C.S. Sprint interval and traditional endurance training increase net intramuscular triglyceride breakdown and expression of perilipin 2 and 5. J. Physiol. 2013, 591, 657–675. [Google Scholar] [CrossRef]
- Shaw, C.S.; Sherlock, M.; Stewart, P.M.; Wagenmakers, A.J. Adipophilin distribution and colocalisation with lipid droplets in skeletal muscle. Histochem. Cell Biol. 2009, 131, 575–581. [Google Scholar] [CrossRef]
- Koh, H.C.E.; Ørtenblad, N.; Winding, K.M.; Hellsten, Y.; Mortensen, S.P.; Nielsen, J. High-intensity interval, but not endurance, training induces muscle fiber type-specific subsarcolemmal lipid droplet size reduction in type 2 diabetic patients. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E872–E884. [Google Scholar] [CrossRef]
- Costes, S.V.; Daelemans, D.; Cho, E.H.; Dobbin, Z.; Pavlakis, G.; Lockett, S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004, 86, 3993–4003. [Google Scholar] [CrossRef] [Green Version]
- Gallardo-Montejano, V.I.; Saxena, G.; Kusminski, C.M.; Yang, C.; McAfee, J.L.; Hahner, L.; Hoch, K.; Dubinsky, W.; Narkar, V.A.; Bickel, P.E. Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1α/SIRT1-dependent transcriptional regulation of mitochondrial function. Nat. Commun. 2016, 7, 12723. [Google Scholar] [CrossRef] [Green Version]
- Najt, C.P.; Khan, S.A.; Heden, T.D.; Witthuhn, B.A.; Perez, M.; Heier, J.L.; Mead, L.E.; Franklin, M.P.; Karanja, K.K.; Graham, M.J.; et al. Lipid Droplet-Derived Monounsaturated Fatty Acids Traffic via PLIN5 to Allosterically Activate SIRT1. Mol. Cell 2020, 77, 810–824. [Google Scholar] [CrossRef]
- Hatazawa, Y.; Tadaishi, M.; Nagaike, Y.; Morita, A.; Ogawa, Y.; Ezaki, O.; Takai-Igarashi, T.; Kitaura, Y.; Shimomura, Y.; Kamei, Y.; et al. PGC-1α-Mediated Branched-Chain Amino Acid Metabolism in the Skeletal Muscle. PLoS ONE 2014, 9, e91006. [Google Scholar] [CrossRef] [Green Version]
- Sjögren, R.J.; Rizo-Roca, D.; Chibalin, A.V.; Chorell, E.; Furrer, R.; Katayama, S.; Harada, J.; Karlsson, H.K.; Handschin, C.; Moritz, T.; et al. Branched-chain amino acid metabolism is regulated by ERRα in primary human myotubes and is further impaired by glucose loading in type 2 diabetes. Diabetologia 2021, 64, 2077–2091. [Google Scholar] [CrossRef]
- Lerin, C.; Goldfine, A.B.; Boes, T.; Liu, M.; Kasif, S.; Dreyfuss, J.M.; De Sousa-Coelho, A.L.; Daher, G.; Manoli, I.; Sysol, J.R.; et al. Defects in muscle branched-chain amino acid oxidation contribute to impaired lipid metabolism. Mol. Metab. 2016, 5, 926–936. [Google Scholar] [CrossRef]
- Kainulainen, H.; Hulmi, J.J.; Kujala, U.M. Potential role of branched-chain amino acid catabolism in regulating fat oxidation. Exerc. Sport Sci. Rev. 2013, 41, 194–200. [Google Scholar] [CrossRef]
- Nye, C.K.; Hanson, R.W.; Kalhan, S.C. Glyceroneogenesis is the dominant pathway for triglyceride glycerol synthesis in vivo in the rat. J. Biol. Chem. 2008, 283, 27565–27574. [Google Scholar] [CrossRef] [Green Version]
- Leskinen, T.; Rinnankoski-Tuikka, R.; Rintala, M.; Seppänen-Laakso, T.; Pöllänen, E.; Alen, M.; Sipilä, S.; Kaprio, J.; Kovanen, V.; Rahkila, P.; et al. Differences in Muscle and Adipose Tissue Gene Expression and Cardio-Metabolic Risk Factors in the Members of Physical Activity Discordant Twin Pairs. PLoS ONE 2010, 5, e12609. [Google Scholar] [CrossRef]
- Lautaoja, J.H.; O’Connell, T.M.; Mäntyselkä, S.; Peräkylä, J.; Kainulainen, H.; Pekkala, S.; Permi, P.; Hulmi, J.J. Higher glucose availability augments the metabolic responses of the C2C12 myotubes to exercise-like electrical pulse stimulation. Am. J. Physiol. Endocrinol. Metab. 2021, 321, E229–E245. [Google Scholar] [CrossRef]
- Nikolić, N.; Bakke, S.S.; Kase, E.T.; Rudberg, I.; Halle, I.F.; Rustan, A.C.; Thoresen, G.H.; Aas, V. Correction: Electrical Pulse Stimulation of Cultured Human Skeletal Muscle Cells as an In Vitro Model of Exercise. PLoS ONE 2013, 8, e33203. [Google Scholar] [CrossRef]
- Kankaanpää, A.; Tolvanen, A.; Bollepalli, S.; Leskinen, T.; Kujala, U.M.; Kaprio, J.; Ollikainen, M.; Sillanpää, E. Leisure-time and occupational physical activity associates differently with epigenetic aging. Med. Sci. Sport. Exerc. 2021, 53, 487. [Google Scholar] [CrossRef]
- Ko, K.; Woo, J.; Bae, J.Y.; Roh, H.T.; Lee, Y.H.; Shin, K.O. Exercise training improves intramuscular triglyceride lipolysis sensitivity in high-fat diet induced obese mice. Lipids Health Dis. 2018, 17, 81. [Google Scholar] [CrossRef] [Green Version]
- Ramos, S.V.; MacPherson, R.E.K.; Turnbull, P.C.; Bott, K.N.; LeBlanc, P.; Ward, W.E.; Peters, S.J. Higher PLIN5 but not PLIN3 content in isolated skeletal muscle mitochondria following acute in vivo contraction in rat hindlimb. Physiol. Rep. 2014, 2, e12154. [Google Scholar] [CrossRef]
- Rinnankoski-Tuikka, R.; Hulmi, J.J.; Torvinen, S.; Silvennoinen, M.; Lehti, M.; Kivelä, R.; Reunanen, H.; Kujala, U.M.; Kainulainen, H. Lipid droplet-associated proteins in high-fat fed mice with the effects of voluntary running and diet change. Metabolism 2014, 63, 1031–1040. [Google Scholar] [CrossRef] [Green Version]
- Bosma, M.; Hesselink, M.K.; Sparks, L.M.; Timmers, S.; Ferraz, M.J.; Mattijssen, F.; van Beurden, D.; Schaart, G.; de Baets, M.H.; Verheyen, F.K.; et al. Perilipin 2 improves insulin sensitivity in skeletal muscle despite elevated intramuscular lipid levels. Diabetes 2012, 61, 2679–2690. [Google Scholar] [CrossRef] [Green Version]
- Bonen, A.; Luiken, J.; Liu, S.; Dyck, D.; Kiens, B.; Kristiansen, S.; Turcotte, L.; Van Der Vusse, G.; Glatz, J. Palmitate transport and fatty acid transporters in red and white muscles. Am. J. Physiol. Endocrinol. Metab. 1998, 275, E471–E478. [Google Scholar] [CrossRef]
- Fachada, V.; Rahkila, P.; Fachada, N.; Turpeinen, T.; Kujala, U.M.; Kainulainen, H. Enlarged PLIN5-uncoated lipid droplets in inner regions of skeletal muscle type II fibers associate with type 2 diabetes. Acta Histochem. 2022, 124, 151869. [Google Scholar] [CrossRef]
- Yang, B.; Yu, Q.; Chang, B.; Guo, Q.; Xu, S.; Yi, X.; Cao, S. MOTS-c interacts synergistically with exercise intervention to regulate PGC-1α expression, attenuate insulin resistance and enhance glucose metabolism in mice via AMPK signaling pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166126. [Google Scholar] [CrossRef]
- Philp, A.; Belew, M.Y.; Evans, A.; Pham, D.; Sivia, I.; Chen, A.; Schenk, S.; Baar, K. The PGC-1α-related coactivator promotes mitochondrial and myogenic adaptations in C2C12 myotubes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, R864–R872. [Google Scholar] [CrossRef]
- Melouane, A.; Yoshioka, M.; Kanzaki, M.; St-Amand, J. Sparc, an EPS-induced gene, modulates the extracellular matrix and mitochondrial function via ILK/AMPK pathways in C2C12 cells. Life Sci. 2019, 229, 277–287. [Google Scholar] [CrossRef]
- Lee, I.H.; Lee, Y.J.; Seo, H.; Kim, Y.S.; Nam, J.O.; Jeon, B.D.; Kwon, T.D. Study of muscle contraction induced by electrical pulse stimulation and nitric oxide in C2C12 myotube cells. J. Exerc. Nutr. Biochem. 2018, 22, 22. [Google Scholar] [CrossRef]
- Finck, B.N.; Kelly, D.P. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J. Clin. Investig. 2006, 116, 615–622. [Google Scholar] [CrossRef] [Green Version]
- Karvinen, S.; Fachada, V.; Sahinaho, U.M.; Pekkala, S.; Lautaoja, J.H.; Mäntyselkä, S.; Permi, P.; Hulmi, J.J.; Silvennoinen, M.; Kainulainen, H. Branched-Chain Amino Acid Deprivation Decreases Lipid Oxidation and Lipogenesis in C2C12 Myotubes. Metabolites 2022, 12, 328. [Google Scholar] [CrossRef]
- Son, Y.H.; Lee, S.M.; Lee, S.H.; Yoon, J.H.; Kang, J.S.; Yang, Y.R.; Kwon, K.S. Comparative molecular analysis of endurance exercise in vivo with electrically stimulated in vitro myotube contraction. J. Appl. Physiol. 2019, 127, 1742–1753. [Google Scholar] [CrossRef]
- Philp, A.; Perez-Schindler, J.; Green, C.; Hamilton, D.L.; Baar, K. Pyruvate suppresses PGC1α expression and substrate utilization despite increased respiratory chain content in C2C12 myotubes. Am. J. Physiol. Cell Physiol. 2010, 299, C240–C250. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.M.; Parr, T.; Brameld, J.M. Myosin heavy chain mRNA isoforms are expressed in two distinct cohorts during C2C12 myogenesis. J. Muscle Res. Cell Motil. 2012, 32, 383–390. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, A.L.; Senthivinayagam, S.; Moon, K.C.; Gupta, S.; Lwande, J.S.; Murphy, C.C.; Storey, S.M.; Atshaves, B.P. Direct interaction of Plin2 with lipids on the surface of lipid droplets: A live cell FRET analysis. Am. J. Physiol. Cell Physiol. 2012, 303, C728–C742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Zhang, F.; Sun, D.; Wang, X.; Zhang, X.; Zhang, J.; Yan, F.; Huang, C.; Xie, H.; Lin, C.; et al. Branched-chain amino acids exacerbate obesity-related hepatic glucose and lipid metabolic disorders via attenuating Akt2 signaling. Diabetes 2020, 69, 1164–1177. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jeon, J.M.; Kwon, O.K.; Choe, M.S.; Yeo, H.C.; Peng, X.; Cheng, Z.; Lee, M.Y.; Lee, S. Comparative Proteomic Analysis Reveals the Upregulation of Ketogenesis in Cardiomyocytes Differentiated from Induced Pluripotent Stem Cells. Proteomics 2019, 19, 1800284. [Google Scholar] [CrossRef] [PubMed]
- Halama, A.; Horsch, M.; Kastenmüller, G.; Möller, G.; Kumar, P.; Prehn, C.; Laumen, H.; Hauner, H.; Hrabĕ de Angelis, M.; Beckers, J.; et al. Metabolic switch during adipogenesis: From branched chain amino acid catabolism to lipid synthesis. Arch. Biochem. Biophys. 2016, 589, 93–107. [Google Scholar] [CrossRef] [Green Version]
- Itabe, H.; Yamaguchi, T.; Nimura, S.; Sasabe, N. Perilipins: A diversity of intracellular lipid droplet proteins. Lipids Health Dis. 2017, 16, 83. [Google Scholar] [CrossRef] [Green Version]
- Rossi, M.; Colecchia, D.; Iavarone, C.; Strambi, A.; Piccioni, F.; Verrotti di Pianella, A.; Chiariello, M. Extracellular Signal-regulated Kinase 8 (ERK8) Controls Estrogen-related Receptor α (ERRα) Cellular Localization and Inhibits Its Transcriptional Activity. J. Biol. Chem. 2011, 286, 8507–8522. [Google Scholar] [CrossRef] [Green Version]
- Nasrin, N.; Kaushik, V.K.; Fortier, E.; Wall, D.; Pearson, K.J.; De Cabo, R.; Bordone, L. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PloS ONE 2009, 4, e8414. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Schwaid, A.G.; Wang, X.; Wang, X.; Chen, S.; Chu, Q.; Saghatelian, A.; Wan, Y. Ligand activation of ERRα by cholesterol mediates statin and bisphosphonate effects. Cell Metab. 2016, 23, 479–491. [Google Scholar] [CrossRef] [Green Version]
- Neishabouri, S.H.; Hutson, S.; Davoodi, J. Chronic activation of mTOR complex 1 by branched chain amino acids and organ hypertrophy. Amino Acids 2015, 47, 1167–1182. [Google Scholar] [CrossRef]
- Cunningham, J.T.; Rodgers, J.T.; Arlow, D.H.; Vazquez, F.; Mootha, V.K.; Puigserver, P. mTOR controls mitochondrial oxidative function through a YY1–PGC-1α transcriptional complex. Nature 2007, 450, 736–740. [Google Scholar] [CrossRef]
- Ohsaki, Y.; Kawai, T.; Yoshikawa, Y.; Cheng, J.; Jokitalo, E.; Fujimoto, T. PML isoform II plays a critical role in nuclear lipid droplet formation. J. Cell Biol. 2016, 212, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Gemmink, A.; Daemen, S.; Kuijpers, H.J.; Schaart, G.; Duimel, H.; López-Iglesias, C.; van Zandvoort, M.A.; Knoops, K.; Hesselink, M.K. Super-resolution microscopy localizes perilipin 5 at lipid droplet-mitochondria interaction sites and at lipid droplets juxtaposing to perilipin 2. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Leskinen, T.; Waller, K.; Mutikainen, S.; Aaltonen, S.; Ronkainen, P.H.A.; Alén, M.; Sipilä, S.; Kovanen, V.; Perhonen, M.; Pietiläinen, K.H.; et al. Effects of 32-Year Leisure Time Physical Activity Discordance in Twin Pairs on Health (TWINACTIVE Study): Aims, Design and Results for Physical Fitness. Twin Res. Hum. Genet. 2009, 12, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.Y.; Qiu, Y.T.; Sun, H.; Yan, J.w.; Zhang, L. A lysosome-targeting dual-functional fluorescent probe for imaging intracellular viscosity and beta-amyloid. Chem. Commun. 2019, 55, 2688–2691. [Google Scholar] [CrossRef]
- Spandl, J.; White, D.J.; Peychl, J.; Thiele, C. Live Cell Multicolor Imaging of Lipid Droplets with a New Dye, LD540. Traffic 2009, 10, 1579–1584. [Google Scholar] [CrossRef] [PubMed]
- Arganda-Carreras, I.; Kaynig, V.; Rueden, C.; Eliceiri, K.W.; Schindelin, J.; Cardona, A.; Sebastian Seung, H. Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 2017, 33, 2424–2426. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Pandas Development Team. pandas-dev/pandas: Pandas, 2020. Available online: https://doi.org/10.5281/zenodo.3509134 (accessed on 1 August 2022).
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [Green Version]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and statistical modeling with python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010. [Google Scholar]
- Waskom, M. The Seaborn Development Team. Mwaskom/Seaborn, 2020. Available online: https://doi.org/10.5281/zenodo.592845 (accessed on 1 August 2022).
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]







| Inactive | Active | |
|---|---|---|
| Number of Participants | 4 | 4 |
| LTPA (MET-hours · day−1) ** | 2.9 +/− 1.4 | 13.8 +/− 1.0 |
| Age (years) | 58.0 +/− 2.9 | 58.0 +/− 2.9 |
| VO2 max (mL · min−1 · kg−1) | 30.2 +/− 1.4 | 32.8 +/− 1.8 |
| Body weight (kg) | 71.5 +/− 3.4 | 69.8 +/− 5.1 |
| BMI (kg · m−2) | 25.0 +/− 0.6 | 24.6 +/− 1.1 |
| Body fat (%) | 24.1 +/− 2.9 | 20.2 +/− 3.3 |
| Triglycerides (mmol · L−1) | 0.9 +/− 0.2 | 1.0 +/− 0.3 |
| HOMA index | 1.9 +/− 0.3 | 1.5 +/− 0.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fachada, V.; Silvennoinen, M.; Sahinaho, U.-M.; Rahkila, P.; Kivelä, R.; Hulmi, J.J.; Kujala, U.; Kainulainen, H. Effects of Long-Term Physical Activity and BCAA Availability on the Subcellular Associations between Intramyocellular Lipids, Perilipins and PGC-1α. Int. J. Mol. Sci. 2023, 24, 4282. https://doi.org/10.3390/ijms24054282
Fachada V, Silvennoinen M, Sahinaho U-M, Rahkila P, Kivelä R, Hulmi JJ, Kujala U, Kainulainen H. Effects of Long-Term Physical Activity and BCAA Availability on the Subcellular Associations between Intramyocellular Lipids, Perilipins and PGC-1α. International Journal of Molecular Sciences. 2023; 24(5):4282. https://doi.org/10.3390/ijms24054282
Chicago/Turabian StyleFachada, Vasco, Mika Silvennoinen, Ulla-Maria Sahinaho, Paavo Rahkila, Riikka Kivelä, Juha J. Hulmi, Urho Kujala, and Heikki Kainulainen. 2023. "Effects of Long-Term Physical Activity and BCAA Availability on the Subcellular Associations between Intramyocellular Lipids, Perilipins and PGC-1α" International Journal of Molecular Sciences 24, no. 5: 4282. https://doi.org/10.3390/ijms24054282
APA StyleFachada, V., Silvennoinen, M., Sahinaho, U. -M., Rahkila, P., Kivelä, R., Hulmi, J. J., Kujala, U., & Kainulainen, H. (2023). Effects of Long-Term Physical Activity and BCAA Availability on the Subcellular Associations between Intramyocellular Lipids, Perilipins and PGC-1α. International Journal of Molecular Sciences, 24(5), 4282. https://doi.org/10.3390/ijms24054282