A Comparative Study of Binding Interactions between Proteins and Flavonoids in Angelica Keiskei: Stability, α-Glucosidase Inhibition and Interaction Mechanisms
Abstract
:1. Introduction
2. Results and Discussion
2.1. Analysis of the Flavonoid Composition of A. keiskei Extract
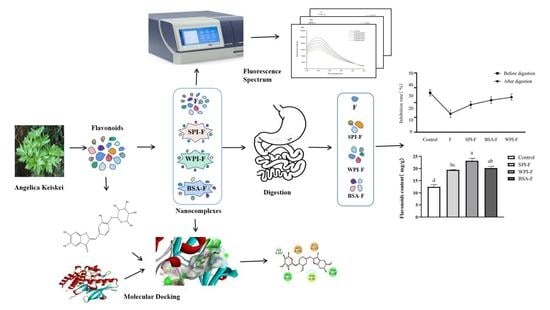
2.2. The Influence of the Combination of Flavonoids and Proteins on Flavonoid Bioavailability at Different Digestive Stages
2.3. Inhibitory Effect against α-Glucosidase
2.4. Molecular Docking Analysis
2.5. Fluorescence Quenching Analysis
3. Materials and Methods
3.1. Materials
3.2. Sample Preparation
3.2.1. Extraction of Flavonoids
3.2.2. Flavonoid Content Assays
3.3. Characterization of A. keiskei Flavonoids by UPLC-Q/TOF-MS
3.4. Preparation of Complexes with A. keiskei Flavonoid Extract
3.5. In Vitro Gastrointestinal Digestion
3.6. In Vitro α-glucosidase Inhibitory Assay
3.7. Molecular Docking
3.8. Characterization of Complexes Containing A. keiskei Flavonoid Extract and Different Proteins
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, D.W.; Curtis-Long, M.J.; Yuk, H.J.; Wang, Y.; Song, Y.H.; Jeong, S.H.; Park, K.H. Quantitative analysis of phenolic metabolites from different parts of Angelica keiskei by HPLC-ESI MS/MS and their xanthine oxidase inhibition. Food Chem. 2014, 153, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Kil, Y.S.; Pham, S.T.; Seo, E.K.; Jafari, M. Angelica keiskei, an emerging medicinal herb with various bioactive constituents and biological activities. Arch. Pharm. Res. 2017, 40, 655–675. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, G.B. In vitro bioavailability and cellular bioactivity studies of flavonoids and flavonoid-rich plant extracts: Questions, considerations and future perspectives. Proc. Nutr. Soc. 2017, 76, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.J.; Yu, Y.G.; Yin, S.W.; Tang, C.H.; Yang, X.Q. Cellular Uptake and Intracellular Antioxidant Activity of Zein/Chitosan Nanoparticles Incorporated with Quercetin. J. Agric. Food Chem. 2018, 66, 12783–12793. [Google Scholar] [CrossRef] [PubMed]
- Karadag, A.; Ozcelik, B.; Huang, Q. Quercetin nanosuspensions produced by high-pressure homogenization. J. Agric. Food Chem. 2014, 62, 1852–1859. [Google Scholar] [CrossRef] [PubMed]
- Kimpel, F.; Schmitt, J.J. Review: Milk Proteins as Nanocarrier Systems for Hydrophobic Nutraceuticals. J. Food Sci. 2015, 80, R2361–R2366. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, D.; Liu, C.; Zhu, J.; Fan, M.; Sun, X.; Wang, T.; Xu, Y.; Cao, Y. Fabrication of stable zein nanoparticles coated with soluble soybean polysaccharide for encapsulation of quercetin. Food Hydrocoll. 2019, 87, 342–351. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, W.; Li, Z.; Liu, B.; Hu, Y.; Chen, S.; de Vries, R.; Yuan, Y.; Erazo Quintero, L.E.; Hou, G.; et al. The stability and bioavailability of curcumin loaded α-lactalbumin nanocarriers formulated in functional dairy drink. Food Hydrocoll. 2022, 131, 107807. [Google Scholar] [CrossRef]
- Singh, T.K.; Øiseth, S.K.; Lundin, L.; Day, L. Influence of heat and shear induced protein aggregation on the in vitro digestion rate of whey proteins. Food Funct. 2014, 5, 2686–2698. [Google Scholar] [CrossRef]
- Sengupta, P.; Sardar, P.S.; Roy, P.; Dasgupta, S.; Bose, A. Investigation on the interaction of Rutin with serum albumins: Insights from spectroscopic and molecular docking techniques. J. Photochem. Photobiol. B 2018, 183, 101–110. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Z.; Xu, G.; Yin, B.; Yao, P. BSA-dextran emulsion for protection and oral delivery of curcumin. Food Hydrocoll. 2016, 61, 11–19. [Google Scholar] [CrossRef]
- Cao, J.; Li, F.; Li, Y.; Chen, H.; Liao, X.; Zhang, Y. Hydrophobic interaction driving the binding of soybean protein isolate and chlorophyll: Improvements to the thermal stability of chlorophyll. Food Hydrocoll. 2021, 113, 106465. [Google Scholar] [CrossRef]
- Zhang, N.; He, H.; Zhang, M.; Lv, X.; Li, W.; Wang, R.; Chang, J. Investigation of the interactions between three flavonoids and human serum albumin by isothermal titration calorimetry, spectroscopy, and molecular docking. New J. Chem. 2022, 46, 12814–12824. [Google Scholar] [CrossRef]
- Zhan, F.; Ding, S.; Xie, W.; Zhu, X.; Hu, J.; Gao, J.; Li, B.; Chen, Y. Towards understanding the interaction of β-lactoglobulin with capsaicin: Multi-spectroscopic, thermodynamic, molecular docking and molecular dynamics simulation approaches. Food Hydrocoll. 2020, 105, 105767. [Google Scholar] [CrossRef]
- Liu, C.; Lv, N.; Ren, G.; Wu, R.; Wang, B.; Cao, Z.; Xie, H. Explore the interaction mechanism between zein and EGCG using multi-spectroscopy and molecular dynamics simulation methods. Food Hydrocoll. 2021, 120, 106906. [Google Scholar] [CrossRef]
- Li, Y.; He, D.; Li, B.; Lund, M.N.; Xing, Y.; Wang, Y.; Li, F.; Cao, X.; Liu, Y.; Chen, X.; et al. Engineering polyphenols with biological functions via polyphenol-protein interactions as additives for functional foods. Trends Food Sci. Technol. 2021, 110, 470–482. [Google Scholar] [CrossRef]
- Li, J.; Tian, R.; Liang, G.; Shi, R.; Hu, J.; Jiang, Z. Interaction mechanism of flavonoids with whey protein isolate: A spectrofluorometric and theoretical investigation. Food Chem. 2021, 355, 129617. [Google Scholar] [CrossRef]
- Han, L.; Lu, K.; Zhou, S.; Zhang, S.; Xie, F.; Qi, B.; Li, Y. Development of an oil-in-water emulsion stabilized by a black bean protein-based nanocomplex for co-delivery of quercetin and perilla oil. LWT 2021, 138, 110644. [Google Scholar] [CrossRef]
- Zhang, S.; Li, X.; Zheng, L.; Zheng, X.; Yang, Y.; Xiao, D.; Ai, B.; Sheng, Z. Encapsulation of phenolics in β-lactoglobulin: Stability, antioxidant activity, and inhibition of advanced glycation end products. LWT 2022, 162, 113437. [Google Scholar] [CrossRef]
- Akihisa, T.; Tokuda, H.; Ukiya, M.; Iizuka, M.; Schneider, S.; Ogasawara, K.; Mukainaka, T.; Iwatsuki, K.; Suzuki, T.; Nishino, H. Chalcones, coumarins, and flavanones from the exudate of Angelica keiskei and their chemopreventive effects. Cancer Lett. 2003, 201, 133–137. [Google Scholar] [CrossRef]
- Sun, C.; Dai, L.; Gao, Y. Binary Complex Based on Zein and Propylene Glycol Alginate for Delivery of Quercetagetin. Biomacromolecules 2016, 17, 3973–3985. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, B.; Wang, B.; Li, C.; Fu, X.; Huang, Q. In-vitro inhibitory effects of flavonoids in Rosa roxburghii and R. sterilis fruits on α-glucosidase: Effect of stomach digestion on flavonoids alone and in combination with acarbose. J. Funct. Foods 2019, 54, 13–21. [Google Scholar] [CrossRef]
- Jeon, Y.O.; Lee, J.S.; Lee, H.G. Improving solubility, stability, and cellular uptake of resveratrol by nanoencapsulation with chitosan and gamma-poly (glutamic acid). Colloids Surf. B Biointerfaces 2016, 147, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Hamed, Y.S.; Abdin, M.; Rayan, A.M.; Saleem Akhtar, H.M.; Zeng, X. Synergistic inhibition of isolated flavonoids from Moringa oleifera leaf on α-glucosidase activity. LWT 2021, 141, 111081. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X. Binding, stability, and antioxidant activity of quercetin with soy protein isolate particles. Food Chem. 2015, 188, 24–29. [Google Scholar] [CrossRef]
- Wei, J.; Xu, D.; Yang, J.; Zhang, X.; Mu, T.; Wang, Q. Analysis of the interaction mechanism of Anthocyanins (Aronia melanocarpa Elliot) with β-casein. Food Hydrocoll. 2018, 84, 276–281. [Google Scholar] [CrossRef]
- Ren, C.; Xiong, W.; Li, J.; Li, B. Comparison of binding interactions of cyanidin-3-O-glucoside to β-conglycinin and glycinin using multi-spectroscopic and thermodynamic methods. Food Hydrocoll. 2019, 92, 155–162. [Google Scholar] [CrossRef]
- Li, Y.; Chen, C.; Zhang, C.; Duan, J.; Yao, H.; Wei, Q. Probing the binding interaction of AKR with human serum albumin by multiple fluorescence spectroscopy and molecular modeling. J. Biomol. Struct. Dyn. 2017, 35, 1189–1199. [Google Scholar] [CrossRef]
- Paul, B.K.; Ghosh, N.; Mukherjee, S. Binding interaction of a prospective chemotherapeutic antibacterial drug with beta-lactoglobulin: Results and challenges. Langmuir 2014, 30, 5921–5929. [Google Scholar] [CrossRef]
- Lin, Y.; Lu, M.F.; Liao, H.B.; Li, Y.X.; Han, W.; Yuan, K. Content determination of the flavonoids in the different parts and different species of Abelmoschus esculentus L. by reversed phase-high performance liquid chromatograph and colorimetric method. Pharmacogn. Mag. 2014, 10, 278–284. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Zha, X.Q.; Shen, W.D.; Li, Q.M.; Pan, L.H.; Luo, J.P. The hydrogel of whey protein isolate coated by lotus root amylopectin enhance the stability and bioavailability of quercetin. Carbohydr. Polym. 2020, 236, 116009. [Google Scholar] [CrossRef] [PubMed]
- Le, H.D.; Loveday, S.M.; Singh, H.; Sarkar, A. Gastrointestinal digestion of Pickering emulsions stabilised by hydrophobically modified cellulose nanocrystals: Release of short-chain fatty acids. Food Chem. 2020, 320, 126650. [Google Scholar] [CrossRef] [PubMed]
- Mulet-Cabero, A.I.; Egger, L.; Portmann, R.; Menard, O.; Marze, S.; Minekus, M.; Le Feunteun, S.; Sarkar, A.; Grundy, M.M.; Carriere, F.; et al. A standardised semi-dynamic in vitro digestion method suitable for food—An international consensus. Food Funct. 2020, 11, 1702–1720. [Google Scholar] [CrossRef] [Green Version]
- Cuc, N.T.; Anh, L.T.; Cuong, N.T.; Anh, N.H.; Nhiem, N.X.; Tai, B.H.; Hoai, N.T.; Yen, P.H.; Van Minh, C.; Van Kiem, P. Flavonol glycosides and their α-glucosidase inhibitory effect from Camellia sinensis. Phytochem. Lett. 2020, 35, 68–72. [Google Scholar] [CrossRef]
- Kong, F.; Kang, S.; Tian, J.; Li, M.; Liang, X.; Yang, M.; Zheng, Y.; Pi, Y.; Cao, X.; Liu, Y.; et al. Interaction of xylitol with whey proteins: Multi-spectroscopic techniques and docking studies. Food Chem. 2020, 326, 126804. [Google Scholar] [CrossRef] [PubMed]
- Lang, Y.; Li, E.; Meng, X.; Tian, J.; Ran, X.; Zhang, Y.; Zang, Z.; Wang, W.; Li, B. Protective effects of bovine serum albumin on blueberry anthocyanins under illumination conditions and their mechanism analysis. Food Res. Int. 2019, 122, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, I.; Nie, R.; Ge, Z.; Li, K.; Li, C. Understanding the shielding effects of whey protein on mulberry anthocyanins: Insights from multispectral and molecular modelling investigations. Int. J. Biol. Macromol. 2018, 119, 116–124. [Google Scholar] [CrossRef]
- Tang, C.-H. Nanocomplexation of proteins with curcumin: From interaction to nanoencapsulation (A review). Food Hydrocoll. 2020, 109, 106106. [Google Scholar] [CrossRef]





| Compounds | Class | Relative Quantitative (%) Mean ± SD |
|---|---|---|
| Aureusidin-4-O-glucoside | Aurones | 6.88 ± 0.11 |
| Kaempferol-3-O-glucoside | Flavonols | 4.62 ± 0.16 |
| Luteolin-7-O-rutinoside | Flavones | 4.22 ± 0.26 |
| Kaempferol-3-O-neohesperidoside | Flavonols | 4.04 ± 0.10 |
| Luteolin-7-O-neohesperidoside | Flavones | 4.04 ± 0.01 |
| Kaempferol-3-O-glucorhamnoside | Flavonols | 3.98 ± 0.08 |
| Quercetin-5-O-β-D-glucoside | Flavonols | 3.58 ± 0.11 |
| 6-C-MethylKaempferol-3-glucoside | Flavones | 3.51 ± 0.02 |
| Diosmetin-7-O-glucoside | Flavones | 3.51 ± 0.04 |
| Diosmetin-7-O-galactoside | Flavones | 3.68 ± 0.05 |
| Hispidulin-7-O-Glucoside | Flavones | 3.42 ± 0.02 |
| Diosmetin-7-O-rutinoside (Diosmin) | Flavones | 2.10 ± 0.08 |
| Luteolin-4′-O-glucoside | Flavones | 2.36 ± 0.23 |
| Quercetin-3-O-galactoside (Hyperin) | Flavonols | 2.28 ± 0.08 |
| Luteolin-7,3′-di-O-glucoside | Flavones | 1.78 ± 0.07 |
| Quercetin-3-O-glucoside (Isoquercitrin) | Flavonols | 2.15 ± 0.05 |
| Luteolin-3′-O-glucoside | Flavones | 1.92 ± 0.02 |
| Yuanhuanin | Flavones | 1.55 ± 0.18 |
| Isobavachalcone | Chalcones | 1.73 ± 0.03 |
| Xanthoangelol F | Chalcones | 1.63 ± 0.05 |
| Quercetin-7-O-glucoside | Flavonols | 1.49 ± 0.02 |
| Quercetin-4′-O-glucoside (Spiraeoside) | Flavonols | 1.49 ± 0.03 |
| Compounds | Docking Energy (kJ mol–1) | Docking Interaction Energy (kJ mol−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 3VO3 | 1HFZ | 3NPO | 1O5D | 3AUP | 3VO3 | 1HFZ | 3NPO | 1O5D | 3AUP | |
| Aureusidin-4-O-glucoside | −11.6258 | −7.5076 | −1.8926 | −16.3452 | −15.0624 | −50.1247 | −52.6416 | −46.8467 | −68.3019 | −53.0224 |
| Kaempferol-3-O-glucoside | −14.2774 | −26.1547 | −29.1191 | −22.3671 | −13.7369 | −53.6227 | −50.7946 | −57.8633 | −65.2053 | −55.6249 |
| Luteolin-7-O-rutinoside | −10.6571 | −28.2081 | −25.8475 | −23.1766 | −12.6838 | −50.6169 | −59.0243 | −51.0303 | −79.6111 | −70.0672 |
| Kaempferol-3-O-glucorhamnoside | −4.8362 | −16.1509 | −20.2018 | −23.4815 | −6.8266 | −57.3834 | −63.6072 | −62.1672 | −75.4202 | −57.5002 |
| Kaempferol-3-O-neohesperidoside | −6.8933 | −14.0788 | −15.8156 | −20.6677 | −8.4797 | −58.3429 | −57.0833 | −65.0644 | −70.7281 | −63.8887 |
| Luteolin-7-O-neohesperidoside | −0.4392 | −15.3671 | −16.6973 | −19.2924 | −0.2389 | −41.7946 | −51.7348 | −64.2016 | −70.7572 | −57.5002 |
| Protein | T | Kq | Ka | n | ΔH | ΔS | ΔG |
|---|---|---|---|---|---|---|---|
| (°C) | (1012 L·mol−1s−1) | (105L·mol−1) | (kJ·mol−1) | (J·mol−1·K−1) | (kJ·mol−1) | ||
| 18 | 1.23 ± 0.66 | 0.1486 ± 0.01 | 0.95 ± 0.14 | 104.38 ± 4.39 | −23.23 ± 0.14 | ||
| SPI | 23 | 1.10 ± 0.94 | 0.1866 ± 0.01 | 1.15 ± 0.22 | 7.13 ± 0.71 | 110.26 ± 2.18 | −25.50 ± 0.09 |
| 28 | 1.03 ± 0.10 | 0.2245 ± 0.02 | 1.16 ± 0.13 | 110.40 ± 2.75 | −26.09 ± 0.15 | ||
| 18 | 1.21 ± 0.21 | 1.6867 ± 0.11 | 1.35 ± 0.05 | 178.16 ± 2.79 | −28.25 ± 0.17 | ||
| WPI | 23 | 1.23 ± 0.12 | 2.8689 ± 0.02 | 1.36 ± 0.18 | 26.59 ± 0.63 | 188.19 ± 3.75 | −29.11 ± 0.17 |
| 28 | 1.19 ± 0.27 | 5.9921 ± 0.10 | 1.42 ± 0.29 | 198.94 ± 2.14 | −33.29 ± 0.04 | ||
| 18 | 1.85 ± 0.19 | 1.0525 ± 0.10 | 0.98 ± 0.18 | 133.91 ± 0.92 | −28.73 ± 0.16 | ||
| BSA | 23 | 2.90 ± 0.30 | 1.4437 ± 0.22 | 1.13 ± 0.13 | 10.23 ± 0.37 | 132.31 ± 2.29 | −28.93 ± 0.17 |
| 28 | 1.43 ± 0.18 | 3.3640 ± 0.07 | 1.21 ± 0.18 | 138.02 ± 1.55 | −31.31 ± 0.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Tu, L.; Pan, D.; Gao, X.; Du, L.; Cai, Z.; Wu, J.; Dang, Y. A Comparative Study of Binding Interactions between Proteins and Flavonoids in Angelica Keiskei: Stability, α-Glucosidase Inhibition and Interaction Mechanisms. Int. J. Mol. Sci. 2023, 24, 6582. https://doi.org/10.3390/ijms24076582
Wang R, Tu L, Pan D, Gao X, Du L, Cai Z, Wu J, Dang Y. A Comparative Study of Binding Interactions between Proteins and Flavonoids in Angelica Keiskei: Stability, α-Glucosidase Inhibition and Interaction Mechanisms. International Journal of Molecular Sciences. 2023; 24(7):6582. https://doi.org/10.3390/ijms24076582
Chicago/Turabian StyleWang, Rui, Lanlan Tu, Daodong Pan, Xinchang Gao, Lihui Du, Zhendong Cai, Jinhong Wu, and Yali Dang. 2023. "A Comparative Study of Binding Interactions between Proteins and Flavonoids in Angelica Keiskei: Stability, α-Glucosidase Inhibition and Interaction Mechanisms" International Journal of Molecular Sciences 24, no. 7: 6582. https://doi.org/10.3390/ijms24076582
APA StyleWang, R., Tu, L., Pan, D., Gao, X., Du, L., Cai, Z., Wu, J., & Dang, Y. (2023). A Comparative Study of Binding Interactions between Proteins and Flavonoids in Angelica Keiskei: Stability, α-Glucosidase Inhibition and Interaction Mechanisms. International Journal of Molecular Sciences, 24(7), 6582. https://doi.org/10.3390/ijms24076582