Epoxide Hydrolases: Multipotential Biocatalysts
Abstract
:1. Introduction
2. Epoxides and Diols as Chiral Precursors and Their Applications
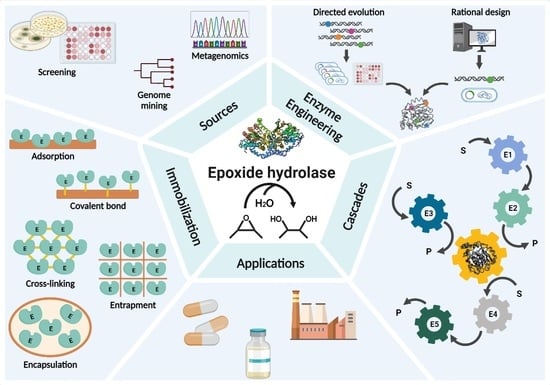
3. Natural and Recombinant EHs
4. Improvement of EHs by Enzyme Engineering
5. Immobilization of EHs
6. Whole-Cell Cascade Biotransformations Using Microbial Epoxide Hydrolases
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choi, W.J.; Choi, C.Y. Production of chiral epoxides: Epoxide hydrolase-catalyzed enantioselective hydrolysis. Biotechnol. Bioprocess. Eng. 2005, 10, 167. [Google Scholar] [CrossRef]
- BRENDA. Available online: www.brenda-enzymes.org (accessed on 20 March 2023).
- Morisseau, C.; Hammock, B.D. Gerry Brooks and epoxide hydrolases: Four decades to a pharmaceutical. Pest Manag. Sci. 2008, 64, 594–609. [Google Scholar] [CrossRef]
- Decker, M.; Arand, M.; Cronin, A. Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch. Toxicol. 2009, 83, 297–318. [Google Scholar] [CrossRef] [Green Version]
- Orru, R.V.A.; Archelas, A.; Furstoss, R.; Faber, K. Epoxide Hydrolases and Their Synthetic Applications. In Biotransformations; Faber, K., Ed.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 145–167. [Google Scholar]
- Archelas, A.; Furstoss, R. Epoxide hydrolases: New tools for the synthesis of fine organic chemicals. Trends Biotechnol. 1998, 16, 108–116. [Google Scholar] [CrossRef]
- Saini, P.; Sareen, D. An Overview on the Enhancement of Enantioselectivity and Stability of Microbial Epoxide Hydrolases. Mol. Biotechnol. 2017, 59, 98–116. [Google Scholar] [CrossRef]
- Choi, W.J. Biotechnological production of enantiopure epoxides by enzymatic kinetic resolution. Appl. Microbiol. Biotechnol. 2009, 84, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Bala, N.; Chimni, S.S. Recent developments in the asymmetric hydrolytic ring opening of epoxides catalysed by microbial epoxide hydrolase. Tetrahedron Asymmetry 2010, 21, 2879–2898. [Google Scholar] [CrossRef]
- Archelas, A.; Iacazio, G.; Kotik, M. Epoxide Hydrolases and their Application in Organic Synthesis. In Green Biocatalysis; Patel, R.N., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 179–229. [Google Scholar]
- Jiang, W.; Fang, B. Synthesizing Chiral Drug Intermediates by Biocatalysis. Appl. Biochem. Biotechnol. 2020, 192, 146–179. [Google Scholar] [CrossRef] [PubMed]
- Research, V.M. Global Chiral Chemicals Market Size by Technology, by Application, by Geographic Scope and Forecast. Available online: https://www.verifiedmarketresearch.com/product/chiral-chemicals-market/ (accessed on 11 April 2023).
- Saini, P.; Sareen, D. Epoxide Hydrolase for the Synthesis of Chiral Drugs. In Nanoscience and Biotechnology for Environmental Applications, 1st ed.; Gothandam, K.M., Ranjan, S., Dasgupta, N., Lichtfouse, E., Eds.; Environmental Chemistry for a Sustainable World; Springer: Cham, Switzerland, 2019; Volume 22, pp. 141–198. [Google Scholar]
- Xuan, J.; Feng, Y. Enantiomeric Tartaric Acid Production Using cis-Epoxysuccinate Hydrolase: History and Perspectives. Molecules 2019, 24, 903. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.J.; Archelas, A.; Furstoss, R. Microbiological transformations. 27. The first examples for preparative-scale enantioselective or diastereoselective epoxide hydrolyses using microorganisms. An unequivocal access to all four bisabolol stereoisomers. J. Org. Chem. 1993, 58, 5528–5532. [Google Scholar] [CrossRef]
- Jacobs, M.H.J.; van den Wijngaard, A.J.; Pentenga, M.; Janssen, D.B. Characterization of the epoxide hydrolase from an epichlorohydrin-degrading Pseudomonas sp. Eur. J. Biochem. 1991, 202, 1217–1222. [Google Scholar] [CrossRef] [PubMed]
- Weijers, C.A.G.M. Enantioselective hydrolysis of aryl, alicyclic and aliphatic epoxides by Rhodotorula glutinis. Tetrahedron Asymmetry 1997, 8, 639–647. [Google Scholar] [CrossRef]
- Chen, L.; Shen, H.; Wei, C.; Zhu, Q. Bioresolution of (R)-glycidyl azide by Aspergillus niger ZJUTZQ208: A new and concise synthon for chiral vicinal amino alcohols. Appl. Microbiol. Biotechnol. 2013, 97, 2609–2616. [Google Scholar] [CrossRef] [PubMed]
- Li, F.-L.; Zheng, Y.-C.; Li, H.; Chen, F.-F.; Yu, H.-L.; Xu, J.-H. Preparing β-blocker (R)-Nifenalol based on enantioconvergent synthesis of (R)-p-nitrophenylglycols in continuous packed bed reactor with epoxide hydrolase. Tetrahedron 2019, 75, 1706–1710. [Google Scholar] [CrossRef]
- Pedragosa-Moreau, S.; Morisseau, C.; Baratti, J.; Zylber, J.; Archelas, A.; Furstoss, R. Microbiological transformations 37. An enantioconvergent synthesis of the β-blocker (R)-Nifénalol® using a combined chemoenzymatic approach. Tetrahedron 1997, 53, 9707–9714. [Google Scholar] [CrossRef]
- Manoj, K.M.; Archelas, A.; Baratti, J.; Furstoss, R. Microbiological transformations. Part 45: A green chemistry preparative scale synthesis of enantiopure building blocks of Eliprodil: Elaboration of a high substrate concentration epoxide hydrolase-catalyzed hydrolytic kinetic resolution process. Tetrahedron 2001, 57, 695–701. [Google Scholar] [CrossRef]
- Zhang, J.; Reddy, J.; Roberge, C.; Senanayake, C.; Greasham, R.; Chartrain, M. Chiral bio-resolution of racemic indene oxide by fungal epoxide hydrolases. J. Ferment. Bioeng. 1995, 80, 244–246. [Google Scholar] [CrossRef]
- Cleij, M.; Archelas, A.; Furstoss, R. Microbiological Transformations 43. Epoxide Hydrolases as Tools for the Synthesis of Enantiopure α-Methylstyrene Oxides: A New and Efficient Synthesis of (S)-Ibuprofen. J. Org. Chem. 1999, 64, 5029–5035. [Google Scholar] [CrossRef]
- Li, C.; Liu, Q.; Song, X.; Ding, D.; Ji, A.; Qu, Y. Epoxide hydrolase-catalyzed resolution of ethyl 3-phenylglycidate using whole cells of Pseudomonas sp. Biotechnol. Lett. 2003, 25, 2113–2116. [Google Scholar] [CrossRef] [PubMed]
- Ueberbacher, B.J.; Osprian, I.; Mayer, S.F.; Faber, K. A Chemoenzymatic, Enantioconvergent, Asymmetric Total Synthesis of(R)-Fridamycin E. Eur. J. Org. Chem. 2005, 2005, 1266–1270. [Google Scholar] [CrossRef]
- Kong, X.-D.; Yuan, S.; Li, L.; Chen, S.; Xu, J.-H.; Zhou, J. Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates. Proc. Natl. Acad. Sci. USA 2014, 111, 15717–15722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, W.J.; Puah, S.M.; Tan, L.L.; Ng, S.S. Production of (R)-ethyl-3,4-epoxybutyrate by newly isolated Acinetobacter baumannii containing epoxide hydrolase. Appl. Microbiol. Biotechnol. 2008, 79, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Wang, Z.; Li, Z. Preparation of (S)-2-, 3-, and 4-chlorostyrene oxides with the epoxide hydrolase from Sphingomonas sp. HXN-200. Tetrahedron Asymmetry 2008, 19, 407–415. [Google Scholar] [CrossRef]
- Monterde, M.I.; Lombard, M.; Archelas, A.; Cronin, A.; Arand, M.; Furstoss, R. Enzymatic transformations. Part 58: Enantioconvergent biohydrolysis of styrene oxide derivatives catalysed by the Solanum tuberosum epoxide hydrolase. Tetrahedron Asymmetry 2004, 15, 2801–2805. [Google Scholar] [CrossRef]
- Monfort, N.; Archelas, A.; Furstoss, R. Enzymatic transformations. Part 53: Epoxide hydrolase-catalysed resolution of key synthons for azole antifungal agents. Tetrahedron Asymmetry 2002, 13, 2399–2401. [Google Scholar] [CrossRef]
- Monfort, N.; Archelas, A.; Furstoss, R. Enzymatic transformations. Part 55: Highly productive epoxide hydrolase catalysed resolution of an azole antifungal key synthon. Tetrahedron 2004, 60, 601–605. [Google Scholar] [CrossRef]
- Fujino, A.; Asano, M.; Yamaguchi, H.; Shirasaka, N.; Sakoda, A.; Ikunaka, M.; Obata, R.; Nishiyama, S.; Sugai, T. Bacillus subtilis epoxide hydrolase-catalyzed preparation of enantiopure 2-methylpropane-1,2,3-triol monobenzyl ether and its application to expeditious synthesis of (R)-bicalutamide. Tetrahedron Lett. 2007, 48, 979–983. [Google Scholar] [CrossRef]
- Roiban, G.-D.; Sutton, P.W.; Splain, R.; Morgan, C.; Fosberry, A.; Honicker, K.; Homes, P.; Boudet, C.; Dann, A.; Guo, J.; et al. Development of an Enzymatic Process for the Production of (R)-2-Butyl-2-ethyloxirane. Org. Process Res. Dev. 2017, 21, 1302–1310. [Google Scholar] [CrossRef]
- Bottalla, A.-L.; Ibrahim-Ouali, M.; Santelli, M.; Furstoss, R.; Archelas, A. Epoxide Hydrolase-Catalysed Kinetic Resolution of a Spiroepoxide, a Key Building Block of Various 11-Heterosteroids. Adv. Synth. Catal. 2007, 349, 1102–1110. [Google Scholar] [CrossRef]
- Goswami, A.; Totleben, M.J.; Singh, A.K.; Patel, R.N. Stereospecific enzymatic hydrolysis of racemic epoxide: A process for making chiral epoxide. Tetrahedron Asymmetry 1999, 10, 3167–3175. [Google Scholar] [CrossRef]
- Patel, R.N. Chapter 11—Applications of Biocatalysis for Pharmaceuticals and Chemicals. In Organic Synthesis Using Biocatalysis; Goswami, A., Stewart, J.D., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 339–411. [Google Scholar]
- Pienaar, D.P.; Mitra, R.K.; Deventer, T.I.v.; Botes, A.L. Synthesis of a variety of optically active hydroxylated heterocyclic compounds using epoxide hydrolase technology. Tetrahedron Lett. 2008, 49, 6752–6755. [Google Scholar] [CrossRef]
- Halama, A.; Kruliš, R.; Rymeš, J. A Convenient Synthesis of Rivaroxaban from (S)-Epichlorohydrin. Org. Prep. Proced. Int. 2020, 52, 201–211. [Google Scholar] [CrossRef]
- Halama, A.; Krulis, R.; Dammer, O.; Kalasek, S. A Process for the Preparation of Rivaroxaban Based on the Use of (S)-Epichlorohydrin. W.O. Patent No. 2013/120465, 22 August 2013. [Google Scholar]
- Kasai, N.; Suzuki, T.; Furukawa, Y. Chiral C3 epoxides and halohydrins: Their preparation and synthetic application. J. Mol. Catal. B Enzym. 1998, 4, 237–252. [Google Scholar] [CrossRef]
- Gu, S.; Wang, X.; Li, Q. A Preparation Method of high-Purity L-Carnitine. W.O. Patent No. 2010/043110, 22 April 2010. [Google Scholar]
- Chang, J.-Y.; Lee, H.S.; Kim, D.J.; Ko, M.-S.; Kim, N.D.; Chang, Y.K.; Kim, M.S. Method for Preparing Sitagliptin and Amine Salt Intermediates Used Therein. W.O. Patent No. 2011/102640, 25 August 2011. [Google Scholar]
- Kitaori, K.; Takehira, Y.; Furukawa, Y.; Yoshimoto, H.; Otera, J. A Pratical Synthesis of Optically Active Atenolol from Chiral Epichlorohydrin. Chem. Pharm. Bull. 1997, 45, 412–414. [Google Scholar] [CrossRef] [Green Version]
- Narina, S.V.; Sudalai, A. Enantioselective synthesis of (S)-timolol via kinetic resolution of terminal epoxides and dihydroxylation of allylamines. Tetrahedron 2007, 63, 3026–3030. [Google Scholar] [CrossRef]
- Ling, X.; Lu, D.; Wang, J.; Chen, J.; Ding, L.; Chen, J.; Chai, H.; Ouyang, P. Kinetic investigation on enantioselective hydrolytic resolution of epichlorohydrin by crude epoxide hydrolase from domestic duck liver. Afr. J. Biotechnol. 2011, 10, 3436–3443. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Sheng, Y.; Jiang, K.; Wang, Z.; Zheng, Y.; Zhu, Q. Bio-resolution of glycidyl (o, m, p)-methylphenyl ethers by Bacillus megaterium. Biotechnol. Lett. 2010, 32, 513–516. [Google Scholar] [CrossRef]
- Kotik, M.; Brichac, J.; Kyslík, P. Novel microbial epoxide hydrolases for biohydrolysis of glycidyl derivatives. J. Biotechnol. 2005, 120, 364–375. [Google Scholar] [CrossRef]
- Zocher, F.; Enzelberger, M.M.; Bornscheuer, U.T.; Hauer, B.; Wohlleben, W.; Schmid, R.D. Epoxide hydrolase activity of Streptomyces strains. J. Biotechnol. 2000, 77, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Moussou, P.; Archelas, A.; Furstoss, R. Microbiological transformations 41.: Screening for novel fungal epoxide hydrolases. J. Mol. Catal. B Enzym. 1998, 5, 447–458. [Google Scholar] [CrossRef]
- Yeates, C.A.; van Dyk, M.S.; Botes, A.L.; Breytenbach, J.C.; Krieg, H.M. Biocatalysis of nitro substituted styrene oxides by non-conventional yeasts. Biotechnol. Lett. 2003, 25, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Osprian, I.; Kroutil, W.; Mischitz, M.; Faber, K. Biocatalytic resolution of 2-methyl-2-(aryl)alkyloxiranes using novel bacterial epoxide hydrolases. Tetrahedron Asymmetry 1997, 8, 65–71. [Google Scholar] [CrossRef]
- Woo, J.-H.; Kang, K.-M.; Kwon, T.-H.; Park, N.-H.; Lee, E.Y. Isolation, identification and characterization of marine bacteria exhibiting complementary enantioselective epoxide hydrolase activity for preparing chiral chlorinated styrene oxide derivatives. J. Ind. Eng. Chem. 2015, 28, 225–228. [Google Scholar] [CrossRef]
- Woo, J.-H.; Kwon, T.-H.; Kim, J.-T.; Kim, C.-G.; Lee, E.Y. Identification and characterization of epoxide hydrolase activity of polycyclic aromatic hydrocarbon-degrading bacteria for biocatalytic resolution of racemic styrene oxide and styrene oxide derivatives. Biotechnol. Lett. 2013, 35, 599–606. [Google Scholar] [CrossRef]
- Woo, J.-H.; Kim, H.-S.; Park, N.-H.; Suk, H.Y. Isolation of a novel strain, Sphingorhabdus sp. YGSMI21 and characterization of its enantioselective epoxide hydrolase activity. J. Microbiol. 2021, 59, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Zocher, F.; Enzelberger, M.M.; Bornscheuer, U.T.; Hauer, B.; Schmid, R.D. A colorimetric assay suitable for screening epoxide hydrolase activity. Anal. Chim. Acta 1999, 391, 345–351. [Google Scholar] [CrossRef]
- Wahler, D.; Reymond, J.-L. The Adrenaline Test for Enzymes. Angew. Chem. Int. Ed. 2002, 41, 1229–1232. [Google Scholar] [CrossRef]
- Mateo, C.; Archelas, A.; Furstoss, R. A spectrophotometric assay for measuring and detecting an epoxide hydrolase activity. Anal. Biochem. 2003, 314, 135–141. [Google Scholar] [CrossRef]
- Saini, P.; Wani, S.I.; Kumar, R.; Chhabra, R.; Chimni, S.S.; Sareen, D. Trigger factor assisted folding of the recombinant epoxide hydrolases identified from C. pelagibacter and S. nassauensis. Protein Expr. Purif. 2014, 104, 71–84. [Google Scholar] [CrossRef]
- van Loo, B.; Kingma, J.; Arand, M.; Wubbolts, M.G.; Janssen, D.B. Diversity and Biocatalytic Potential of Epoxide Hydrolases Identified by Genome Analysis. Appl. Environ. Microbiol. 2006, 72, 2905–2917. [Google Scholar] [CrossRef] [Green Version]
- Stojanovski, G.; Dobrijevic, D.; Hailes, H.C.; Ward, J.M. Identification and catalytic properties of new epoxide hydrolases from the genomic data of soil bacteria. Enzym. Microb. Technol. 2020, 139, 109592. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.L.; Piel, J.; Sunagawa, S. A roadmap for metagenomic enzyme discovery. Nat. Prod. Rep. 2021, 38, 1994–2023. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, D.J.; Dini-Andreote, F.; Ottoni, J.R.; de Oliveira, V.M.; van Elsas, J.D.; Andreote, F.D. Compositional profile of α/β-hydrolase fold proteins in mangrove soil metagenomes: Prevalence of epoxide hydrolases and haloalkane dehalogenases in oil-contaminated sites. Microb. Biotechnol. 2015, 8, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Kotik, M.; Štěpánek, V.; Marešová, H.; Kyslík, P.; Archelas, A. Environmental DNA as a source of a novel epoxide hydrolase reacting with aliphatic terminal epoxides. J. Mol. Catal. B Enzym. 2009, 56, 288–293. [Google Scholar] [CrossRef]
- Kotik, M.; Štěpánek, V.; Grulich, M.; Kyslík, P.; Archelas, A. Access to enantiopure aromatic epoxides and diols using epoxide hydrolases derived from total biofilter DNA. J. Mol. Catal. B Enzym. 2010, 65, 41–48. [Google Scholar] [CrossRef]
- Ferrandi, E.E.; Sayer, C.; Isupov, M.N.; Annovazzi, C.; Marchesi, C.; Iacobone, G.; Peng, X.; Bonch-Osmolovskaya, E.; Wohlgemuth, R.; Littlechild, J.A.; et al. Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. FEBS J. 2015, 282, 2879–2894. [Google Scholar] [CrossRef]
- Ferrandi, E.E.; Sayer, C.; De Rose, S.A.; Guazzelli, E.; Marchesi, C.; Saneei, V.; Isupov, M.N.; Littlechild, J.A.; Monti, D. New Thermophilic α/β Class Epoxide Hydrolases Found in Metagenomes from Hot Environments. Front. Bioeng. Biotechnol. 2018, 6, 144. [Google Scholar] [CrossRef] [Green Version]
- Reetz, M.T. Laboratory Evolution of Stereoselective Enzymes: A Prolific Source of Catalysts for Asymmetric Reactions. Angew. Chem. Int. Ed. 2011, 50, 138–174. [Google Scholar] [CrossRef]
- Basheer, S.M.; Chellappan, S. Enzyme Engineering. In Bioresources and Bioprocess in Biotechnology: Volume 2: Exploring Potential Biomolecules; Sugathan, S., Pradeep, N.S., Abdulhameed, S., Eds.; Springer: Singapore, 2017; pp. 151–168. [Google Scholar]
- Nardini, M.; Rink, R.; Janssen, D.B.; Dijkstra, B.W. Structure and mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J. Mol. Catal. B Enzym. 2001, 11, 1035–1042. [Google Scholar] [CrossRef]
- Zou, J.; Hallberg, B.M.; Bergfors, T.; Oesch, F.; Arand, M.; Mowbray, S.L.; Jones, T.A. Structure of Aspergillus niger epoxide hydrolase at 1.8 Å resolution: Implications for the structure and function of the mammalian microsomal class of epoxide hydrolases. Structure 2000, 8, 111–122. [Google Scholar] [CrossRef] [Green Version]
- Arand, M.; Hallberg, B.M.; Zou, J.; Bergfors, T.; Oesch, F.; van der Werf, M.J.; de Bont, J.A.; Jones, T.A.; Mowbray, S.L. Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site. EMBO J. 2003, 22, 2583–2592. [Google Scholar] [CrossRef] [Green Version]
- Johansson, P.; Unge, T.; Cronin, A.; Arand, M.; Bergfors, T.; Jones, T.A.; Mowbray, S.L. Structure of an atypical epoxide hydrolase from Mycobacterium tuberculosis gives insights into its function. J. Mol. Biol. 2005, 351, 1048–1056. [Google Scholar] [CrossRef]
- Reetz, M.T.; Zonta, A.; Schimossek, K.; Jaeger, K.-E.; Liebeton, K. Creation of Enantioselective Biocatalysts for Organic Chemistry by in vitro Evolution. Angew. Chem. Int. Ed. Engl. 1997, 36, 2830–2832. [Google Scholar] [CrossRef]
- Cedrone, F.; Niel, S.; Roca, S.; Bhatnagar, T.; Ait-abdelkader, N.; Torre, C.; Krumm, H.; Maichele, A.; Reetz, M.T.; Baratti, J.C. Directed Evolution of the Epoxide Hydrolase from Aspergillus niger. Biocatal. Biotransf. 2003, 21, 357–364. [Google Scholar] [CrossRef]
- Reetz, M.T.; Torre, C.; Eipper, A.; Lohmer, R.; Hermes, M.; Brunner, B.; Maichele, A.; Bocola, M.; Arand, M.; Cronin, A.; et al. Enhancing the Enantioselectivity of an Epoxide Hydrolase by Directed Evolution. Org. Lett. 2004, 6, 177–180. [Google Scholar] [CrossRef]
- Rui, L.; Cao, L.; Chen, W.; Reardon, K.F.; Wood, T.K. Active Site Engineering of the Epoxide Hydrolase from Agrobacterium radiobacter AD1 to Enhance Aerobic Mineralization of cis-1,2-Dichloroethylene in Cells Expressing an Evolved Toluene ortho-Monooxygenase. J. Biol. Chem. 2004, 279, 46810–46817. [Google Scholar] [CrossRef] [Green Version]
- Rui, L.; Cao, L.; Chen, W.; Reardon, K.F.; Wood, T.K. Protein Engineering of Epoxide Hydrolase from Agrobacterium radiobacter AD1 for Enhanced Activity and Enantioselective Production of (R)-1-Phenylethane-1,2-Diol. Appl. Environ. Microbiol. 2005, 71, 3995–4003. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Lonsdale, R.; Kong, X.-D.; Xu, J.-H.; Zhou, J.; Reetz, M.T. Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution. Angew. Chem. Int. Ed. 2015, 54, 12410–12415. [Google Scholar] [CrossRef]
- Sun, Z.; Lonsdale, R.; Li, G.; Reetz, M.T. Comparing Different Strategies in Directed Evolution of Enzyme Stereoselectivity: Single- versus Double-Code Saturation Mutagenesis. ChemBioChem 2016, 17, 1865–1872. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Lonsdale, R.; Wu, L.; Li, G.; Li, A.; Wang, J.; Zhou, J.; Reetz, M.T. Structure-Guided Triple-Code Saturation Mutagenesis: Efficient Tuning of the Stereoselectivity of an Epoxide Hydrolase. ACS Catal. 2016, 6, 1590–1597. [Google Scholar] [CrossRef]
- Arabnejad, H.; Bombino, E.; Colpa, D.I.; Jekel, P.A.; Trajkovic, M.; Wijma, H.J.; Janssen, D.B. Computational Design of Enantiocomplementary Epoxide Hydrolases for Asymmetric Synthesis of Aliphatic and Aromatic Diols. ChemBioChem 2020, 21, 1893–1904. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Zong, X.-C.; Xue, F.; Li, C.; Hu, B.-C.; Wu, M.-C. Manipulating regioselectivity of an epoxide hydrolase for single enzymatic synthesis of (R)-1,2-diols from racemic epoxides. Chem. Commun. 2020, 56, 2799–2802. [Google Scholar] [CrossRef]
- Li, G.; Qin, Y.; Fontaine, N.T.; Ng Fuk Chong, M.; Maria-Solano, M.A.; Feixas, F.; Cadet, X.F.; Pandjaitan, R.; Garcia-Borràs, M.; Cadet, F.; et al. Machine Learning Enables Selection of Epistatic Enzyme Mutants for Stability against Unfolding and Detrimental Aggregation. ChemBioChem 2021, 22, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.-D.; Ma, Q.; Zhou, J.; Zeng, B.-B.; Xu, J.-H. A Smart Library of Epoxide Hydrolase Variants and the Top Hits for Synthesis of (S)-β-Blocker Precursors. Angew. Chem. Int. Ed. 2014, 53, 6641–6644. [Google Scholar] [CrossRef] [PubMed]
- van Loo, B.; Spelberg, J.H.L.; Kingma, J.; Sonke, T.; Wubbolts, M.G.; Janssen, D.B. Directed Evolution of Epoxide Hydrolase from A. radiobacter toward Higher Enantioselectivity by Error-Prone PCR and DNA Shuffling. Chem. Biol. 2004, 11, 981–990. [Google Scholar] [CrossRef] [Green Version]
- van Loo, B.; Kingma, J.; Heyman, G.; Wittenaar, A.; Lutje Spelberg, J.H.; Sonke, T.; Janssen, D.B. Improved enantioselective conversion of styrene epoxides and meso-epoxides through epoxide hydrolases with a mutated nucleophile-flanking residue. Enzym. Microb. Technol. 2009, 44, 145–153. [Google Scholar] [CrossRef]
- Xue, F.; Liu, Z.-Q.; Wan, N.-W.; Zhu, H.-Q.; Zheng, Y.-G. Engineering the epoxide hydrolase from Agromyces mediolanus for enhanced enantioselectivity and activity in the kinetic resolution of racemic epichlorohydrin. RSC Adv. 2015, 5, 31525–31532. [Google Scholar] [CrossRef]
- Reetz, M.T.; Wang, L.-W.; Bocola, M. Directed Evolution of Enantioselective Enzymes: Iterative Cycles of CASTing for Probing Protein-Sequence Space. Angew. Chem. Int. Ed. 2006, 45, 1236–1241. [Google Scholar] [CrossRef]
- Reetz, M.T.; Zheng, H. Manipulating the Expression Rate and Enantioselectivity of an Epoxide Hydrolase by Using Directed Evolution. ChemBioChem 2011, 12, 1529–1535. [Google Scholar] [CrossRef]
- Gumulya, Y.; Sanchis, J.; Reetz, M.T. Many Pathways in Laboratory Evolution Can Lead to Improved Enzymes: How to Escape from Local Minima. ChemBioChem 2012, 13, 1060–1066. [Google Scholar] [CrossRef]
- Hu, D.; Hu, B.-C.; Hou, X.-D.; Zhang, D.; Lei, Y.-Q.; Rao, Y.-J.; Wu, M.-C. Structure-Guided Regulation in the Enantioselectivity of an Epoxide Hydrolase to Produce Enantiomeric Monosubstituted Epoxides and Vicinal Diols via Kinetic Resolution. Org. Lett. 2022, 24, 1757–1761. [Google Scholar] [CrossRef]
- Zheng, H.; Reetz, M.T. Manipulating the Stereoselectivity of Limonene Epoxide Hydrolase by Directed Evolution Based on Iterative Saturation Mutagenesis. J. Am. Chem. Soc. 2010, 132, 15744–15751. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Salas, P.T.; Siirola, E.; Lonsdale, R.; Reetz, M.T. Exploring productive sequence space in directed evolution using binary patterning versus conventional mutagenesis strategies. Bioresour. Bioprocess 2016, 3, 44. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Zhang, H.; Sun, Z.; Liu, X.; Reetz, M.T. Multiparameter Optimization in Directed Evolution: Engineering Thermostability, Enantioselectivity, and Activity of an Epoxide Hydrolase. ACS Catal. 2016, 6, 3679–3687. [Google Scholar] [CrossRef]
- Li, A.; Acevedo-Rocha, C.G.; Sun, Z.; Cox, T.; Xu, J.L.; Reetz, M.T. Beating Bias in the Directed Evolution of Proteins: Combining High-Fidelity on-Chip Solid-Phase Gene Synthesis with Efficient Gene Assembly for Combinatorial Library Construction. ChemBioChem 2018, 19, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Wijma, H.J.; Floor, R.J.; Bjelic, S.; Marrink, S.J.; Baker, D.; Janssen, D.B. Enantioselective Enzymes by Computational Design and in silico Screening. Angew. Chem. Int. Ed. 2015, 54, 3726–3730. [Google Scholar] [CrossRef] [PubMed]
- Gurell, A.; Widersten, M. Modification of Substrate Specificity Resulting in an Epoxide Hydrolase with Shifted Enantiopreference for (2,3-Epoxypropyl)benzene. ChemBioChem 2010, 11, 1422–1429. [Google Scholar] [CrossRef]
- Carlsson, Å.J.; Bauer, P.; Ma, H.; Widersten, M. Obtaining Optical Purity for Product Diols in Enzyme-Catalyzed Epoxide Hydrolysis: Contributions from Changes in both Enantio- and Regioselectivity. Biochemistry 2012, 51, 7627–7637. [Google Scholar] [CrossRef]
- Li, Y.; Ou, X.; Guo, Z.; Zong, M.; Lou, W. Using multiple site-directed modification of epoxide hydrolase to significantly improve its enantioselectivity in hydrolysis of rac-glycidyl phenyl ether. Chin. J. Chem. Eng. 2020, 28, 2181–2189. [Google Scholar] [CrossRef]
- Kotik, M.; Zhao, W.; Iacazio, G.; Archelas, A. Directed evolution of metagenome-derived epoxide hydrolase for improved enantioselectivity and enantioconvergence. J. Mol. Catal. B Enzym. 2013, 91, 44–51. [Google Scholar] [CrossRef]
- Kotik, M.; Archelas, A.; Faměrová, V.; Oubrechtová, P.; Křen, V. Laboratory evolution of an epoxide hydrolase—Towards an enantioconvergent biocatalyst. J. Biotechnol. 2011, 156, 1–10. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wu, M.-D.; Zhu, X.-X.; Zhang, X.-D.; Zhang, C.; Xu, Y.-H.; Wu, M.-C. Remarkable improvement in the regiocomplementarity of a Glycine max epoxide hydrolase by reshaping its substrate-binding pocket for the enantioconvergent preparation of (R)-hexane-1,2-diol. Mol. Catal. 2021, 514, 111851. [Google Scholar] [CrossRef]
- Ye, H.-H.; Hu, D.; Shi, X.-L.; Wu, M.-C.; Deng, C.; Li, J.-F. Directed modification of a novel epoxide hydrolase from Phaseolus vulgaris to improve its enantioconvergence towards styrene epoxides. Catal. Commun. 2016, 87, 32–35. [Google Scholar] [CrossRef]
- Zong, X.-C.; Li, C.; Xu, Y.-H.; Hu, D.; Hu, B.-C.; Zang, J.; Wu, M.-C. Substantially improving the enantioconvergence of PvEH1, a Phaseolus vulgaris epoxide hydrolase, towards m-chlorostyrene oxide by laboratory evolution. Microb. Cell Fact. 2019, 18, 202. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.-X.; Hu, B.-C.; Lin, W.-Q.; Zhang, D.; Zhao, J.; Wu, M.-C. Engineering the regiocomplementarity of an epoxide hydrolase from Rhodotorula paludigena by means of computer-aided design for the scale-up enantioconvergent hydrolysis of racemic m-nitrostyrene oxide. Biochem. Eng. J. 2022, 180, 108359. [Google Scholar] [CrossRef]
- Li, F.-L.; Kong, X.-D.; Chen, Q.; Zheng, Y.-C.; Xu, Q.; Chen, F.-F.; Fan, L.-Q.; Lin, G.-Q.; Zhou, J.; Yu, H.-L.; et al. Regioselectivity Engineering of Epoxide Hydrolase: Near-Perfect Enantioconvergence through a Single Site Mutation. ACS Catal. 2018, 8, 8314–8317. [Google Scholar] [CrossRef]
- Li, F.-L.; Qiu, Y.-Y.; Zheng, Y.-C.; Chen, F.-F.; Kong, X.D.; Xu, J.-H.; Yu, H.-L. Reprogramming Epoxide Hydrolase to Improve Enantioconvergence in Hydrolysis of Styrene Oxide Scaffolds. Adv. Synth. Catal. 2020, 362, 4699–4706. [Google Scholar] [CrossRef]
- Wijma, H.J.; Floor, R.J.; Jekel, P.A.; Baker, D.; Marrink, S.J.; Janssen, D.B. Computationally designed libraries for rapid enzyme stabilization. Protein Eng. Des. Sel. 2014, 27, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Gumulya, Y.; Reetz, M.T. Enhancing the Thermal Robustness of an Enzyme by Directed Evolution: Least Favorable Starting Points and Inferior Mutants Can Map Superior Evolutionary Pathways. ChemBioChem 2011, 12, 2502–2510. [Google Scholar] [CrossRef]
- Qiao, P.; Wu, M.; Zhu, L.; Zhang, Y.; Yang, L.; Fei, H.; Lin, J. Enhancing the thermal tolerance of a cis-epoxysuccinate hydrolase via combining directed evolution with various semi-rational redesign methods. J. Mol. Catal. B Enzym. 2015, 121, 96–103. [Google Scholar] [CrossRef]
- Jochens, H.; Stiba, K.; Savile, C.; Fujii, R.; Yu, J.-G.; Gerassenkov, T.; Kazlauskas, R.J.; Bornscheuer, U.T. Converting an Esterase into an Epoxide Hydrolase. Angew. Chem. Int. Ed. 2009, 48, 3532–3535. [Google Scholar] [CrossRef] [PubMed]
- Nagel, B.; Dellweg, H.; Gierasch, L.M. Glossary for chemists of terms used in biotechnology (IUPAC Recommendations 1992). Pure Appl. Chem. 1992, 64, 143–168. [Google Scholar] [CrossRef] [Green Version]
- Hechtberger, P.; Wirnsberger, G.; Mischitz, M.; Klempier, N.; Faber, K. Asymmetric hydrolysis of epoxides using an immobilized enzyme preparation from Rhodococcus sp. Tetrahedron Asymmetry 1993, 4, 1161–1164. [Google Scholar] [CrossRef]
- Lin, H.; Liu, J.-Y.; Wang, H.-B.; Ahmed, A.A.Q.; Wu, Z.-L. Biocatalysis as an alternative for the production of chiral epoxides: A comparative review. J. Mol. Catal. B Enzym. 2011, 72, 77–89. [Google Scholar] [CrossRef]
- Vikartovská, A.; Bučko, M.; Gemeiner, P.; Nahálka, J.; Pätoprstý, V.; Hrabárová, E. Flow Calorimetry—A Useful Tool for Determination of Immobilized cis-Epoxysuccinate Hydrolase Activity from Nocardia tartaricans. Artif. Cells Blood Substit. Biotechnol. 2004, 32, 77–89. [Google Scholar] [CrossRef]
- Bučko, M.; Vikartovská, A.; Lacík, I.; Kolláriková, G.; Gemeiner, P.; Pätoprstý, V.; Brygin, M. Immobilization of a whole-cell epoxide-hydrolyzing biocatalyst in sodium alginate-cellulose sulfate-poly(methylene-co-guanidine) capsules using a controlled encapsulation process. Enzym. Microb. Technol. 2005, 36, 118–126. [Google Scholar] [CrossRef]
- Rosenberg, M.; Miková, H.; Krištofíková, L. Production of L-tartaric acid by immobilized bacterial cells Nocardia tartaricans. Biotechnol. Lett. 1999, 21, 491–495. [Google Scholar] [CrossRef]
- Bučko, M.; Vikartovská, A.; Gemeiner, P.; Lacík, I.; Kolláriková, G.; Marison, I.W. Nocardia tartaricans cells immobilized in sodium alginate–cellulose sulfate–poly (methylene-co-guanidine)capsules: Mechanical resistance and operational stability. J. Chem. Technol. Biotechnol. 2006, 81, 500–504. [Google Scholar] [CrossRef]
- Huang, R.; Wu, M.; Goldman, M.J.; Li, Z. Encapsulation of enzyme via one-step template-free formation of stable organic–inorganic capsules: A simple and efficient method for immobilizing enzyme with high activity and recyclability. Biotechnol. Bioeng. 2015, 112, 1092–1101. [Google Scholar] [CrossRef]
- Zhou, R.; Dong, S.; Feng, Y.; Cui, Q.; Xuan, J. Development of highly efficient whole-cell catalysts of cis-epoxysuccinic acid hydrolase by surface display. Bioresour. Bioprocess 2022, 9, 92. [Google Scholar] [CrossRef]
- Zou, S.-P.; Wang, Z.-C.; Qin, C.; Zheng, Y.-G. Covalent immobilization of Agrobacterium radiobacter epoxide hydrolase on ethylenediamine functionalised epoxy supports for biocatalytical synthesis of (R)-epichlorohydrin. Biotechnol. Lett. 2016, 38, 1579–1585. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.-P.; Xuan, X.-L.; Wang, Z.-J.; Zheng, Y.-G. Conjugation of Agrobacterium radiobacter epoxide hydrolase with ficoll: Catalytic, kinetic and thermodynamic analysis. Int. J. Biol. Macromol. 2018, 119, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Grulich, M.; Maršálek, J.; Kyslík, P.; Štěpánek, V.; Kotik, M. Production, enrichment and immobilization of a metagenome-derived epoxide hydrolase. Process Biochem. 2011, 46, 526–532. [Google Scholar] [CrossRef]
- Mateo, C.; Archelas, A.; Fernandez-Lafuente, R.; Manuel Guisan, J.; Furstoss, R. Enzymatic transformations. Immobilized A. niger epoxide hydrolase as a novel biocatalytic tool for repeated-batch hydrolytic kinetic resolution of epoxides. Org. Biomol. Chem. 2003, 1, 2739–2743. [Google Scholar] [CrossRef]
- Yildirim, D.; Tükel, S.S.; Alagöz, D.; Alptekin, Ö. Preparative-scale kinetic resolution of racemic styrene oxide by immobilized epoxide hydrolase. Enzym. Microb. Technol. 2011, 49, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, D.; Tükel, S.S.; Alptekin, Ö.; Alagöz, D. Immobilized Aspergillus niger epoxide hydrolases: Cost-effective biocatalysts for the prepation of enantiopure styrene oxide, propylene oxide and epichlorohydrin. J. Mol. Catal. B Enzym. 2013, 88, 84–90. [Google Scholar] [CrossRef]
- Petri, A.; Marconcini, P.; Salvadori, P. Efficient immobilization of epoxide hydrolase onto silica gel and use in the enantioselective hydrolysis of racemic para-nitrostyrene oxide. J. Mol. Catal. B Enzym. 2005, 32, 219–224. [Google Scholar] [CrossRef]
- Mateo, C.; Fernandez-Lafuente, R.; Archelas, A.; Guisan, J.M.; Furstoss, R. Preparation of a very stable immobilized Solanum tuberosum epoxide hydrolase. Tetrahedron Asymmetry 2007, 18, 1233–1238. [Google Scholar] [CrossRef]
- Ibrahim, M.; Hubert, P.; Dellacherie, E.; Magdalou, J.; Siest, G. Immobilization of epoxide hydrolase purified from rat liver microsomes. Biotechnol. Lett. 1984, 6, 771–776. [Google Scholar] [CrossRef]
- Ibrahim, M.; Hubert, P.; Dellacherie, E.; Magdalou, J.; Muller, J.; Siest, G. Covalent attachment of epoxide hydrolase to dextran. Enzym. Microb. Technol. 1985, 7, 66–72. [Google Scholar] [CrossRef]
- Kamble, M.; Salvi, H.; Yadav, G.D. Preparation of amino-functionalized silica supports for immobilization of epoxide hydrolase and cutinase: Characterization and applications. J. Porous Mater. 2020, 27, 1559–1567. [Google Scholar] [CrossRef]
- Cassimjee, K.E.; Kourist, R.; Lindberg, D.; Wittrup Larsen, M.; Thanh, N.H.; Widersten, M.; Bornscheuer, U.T.; Berglund, P. One-step enzyme extraction and immobilization for biocatalysis applications. Biotechnol. J. 2011, 6, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Kim, H.S.; Lee, I.S.; Lee, E.Y. Functional expression and magnetic nanoparticle-based Immobilization of a protein-engineered marine fish epoxide hydrolase of Mugil cephalus for enantioselective hydrolysis of racemic styrene oxide. Biotechnol. Lett. 2010, 32, 1685–1691. [Google Scholar] [CrossRef]
- Wang, Z.; Su, M.; Li, Y.; Wang, Y.; Su, Z. Production of tartaric acid using immobilized recominant cis-epoxysuccinate hydrolase. Biotechnol. Lett. 2017, 39, 1859–1863. [Google Scholar] [CrossRef]
- Karboune, S.; Archelas, A.; Baratti, J.C. Free and immobilized Aspergillus niger epoxide hydrolase-catalyzed hydrolytic kinetic resolution of racemic p-chlorostyrene oxide in a neat organic solvent medium. Process Biochem. 2010, 45, 210–216. [Google Scholar] [CrossRef]
- Karboune, S.; Archelas, A.; Furstoss, R.; Baratti, J. Immobilization of epoxide hydrolase from Aspergillus niger onto DEAE-cellulose: Enzymatic properties and application for the enantioselective resolution of a racemic epoxide. J. Mol. Catal. B Enzym. 2005, 32, 175–183. [Google Scholar] [CrossRef]
- Karboune, S.; Amourache, L.; Nellaiah, H.; Morisseau, C.; Baratti, J. Immobilization of the epoxide hydrolase from Aspergillus niger. Biotechnol. Lett. 2001, 23, 1633–1639. [Google Scholar] [CrossRef]
- Kroutil, W.; Genzel, Y.; Pietzsch, M.; Syldatk, C.; Faber, K. Purification and characterization of a highly selective epoxide hydrolase from Nocardia sp. EH1. J. Biotechnol. 1998, 61, 143–150. [Google Scholar] [CrossRef]
- Jin, H.-X.; Liu, Z.-Q.; Hu, Z.-C.; Zheng, Y.-G. Production of (R)-epichlorohydrin from 1,3-dichloro-2-propanol by two-step biocatalysis using haloalcohol dehalogenase and epoxide hydrolase in two-phase system. Biochem. Eng. J. 2013, 74, 1–7. [Google Scholar] [CrossRef]
- Kim, Y.H.; Lee, I.; Choi, S.H.; Lee, O.K.; Shim, J.; Lee, J.; Kim, J.; Lee, E.Y. Enhanced stability and reusability of marine epoxide hydrolase using ship-in-a-bottle approach with magnetically-separable mesoporous silica. J. Mol. Catal. B Enzym. 2013, 89, 48–51. [Google Scholar] [CrossRef]
- Yu, C.-Y.; Li, X.-F.; Lou, W.-Y.; Zong, M.-H. Cross-linked enzyme aggregates of Mung bean epoxide hydrolases: A highly active, stable and recyclable biocatalyst for asymmetric hydrolysis of epoxides. J. Biotechnol. 2013, 166, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-Y.; Wei, P.; Li, X.-F.; Zong, M.-H.; Lou, W.-Y. Using Ionic Liquid in a Biphasic System to Improve Asymmetric Hydrolysis of Styrene Oxide Catalyzed by cross-Linked Enzyme Aggregates (CLEAs) of Mung Bean Epoxide Hydrolases. Ind. Eng. Chem. Res. 2014, 53, 7923–7930. [Google Scholar] [CrossRef]
- Kronenburg, N.A.E.; de Bont, J.A.M.; Fischer, L. Improvement of enantioselectivity by immobilized imprinting of epoxide hydrolase from Rhodotorula glutinis. J. Mol. Catal. B Enzym. 2001, 16, 121–129. [Google Scholar] [CrossRef]
- Salvi, H.M.; Yadav, G.D. Organic-inorganic epoxide hydrolase hybrid nanoflowers with enhanced catalytic activity: Hydrolysis of styrene oxide to 1-phenyl-1,2-ethanediol. J. Biotechnol. 2021, 341, 113–120. [Google Scholar] [CrossRef]
- Cao, S.-L.; Yue, D.-M.; Li, X.-H.; Smith, T.J.; Li, N.; Zong, M.-H.; Wu, H.; Ma, Y.-Z.; Lou, W.-Y. Novel Nano-/Micro-Biocatalyst: Soybean Epoxide Hydrolase Immobilized on UiO-66-NH2 MOF for Efficient Biosynthesis of Enantiopure (R)-1, 2-Octanediol in Deep Eutectic Solvents. ACS Sustain. Chem. Eng. 2016, 4, 3586–3595. [Google Scholar] [CrossRef]
- Onur, H.; Tülek, A.; Aslan, E.S.; Binay, B.; Yildirim, D. A new highly enantioselective stable epoxide hydrolase from Hypsibius dujardini: Expression in Pichia pastoris and immobilization in ZIF-8 for asymmetric hydrolysis of racemic styrene oxide. Biochem. Eng. J. 2022, 189, 108726. [Google Scholar] [CrossRef]
- Maritz, J.; Krieg, H.M.; Yeates, C.A.; Botes, A.L.; Breytenbach, J.C. Calcium alginate entrapment of the yeast Rhodosporidium toruloides for the kinetic resolution of 1,2-epoxyoctane. Biotechnol. Lett. 2003, 25, 1775–1781. [Google Scholar] [CrossRef]
- Bao, W.; Pan, H.; Zhang, Z.; Cheng, Y.; Xie, Z.; Zhang, J. Isolation of the stable strain Labrys sp. BK-8 for l(+)-tartaric acid production. J. Biosci. Bioeng. 2015, 119, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, K.; Kasche, V.; Bornscheuer, U.T. Enzymes in Organic Chemistry. In Biocatalysts and Enzyme Technology, 1st ed.; WILEY-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2005; pp. 141–145. [Google Scholar]
- Faber, K.; Mischitz, M.; Kroutil, W. Microbial Epoxide Hydrolases. Acta Chem. Scand. 1996, 50, 249–258. [Google Scholar] [CrossRef] [Green Version]
- Mayer, S.F.; Steinreiber, A.; Orru, R.V.A.; Faber, K. Enzyme-Triggered Enantioconvergent Transformation of Haloalkyl Epoxides. Eur. J. Org. Chem. 2001, 2001, 4537–4542. [Google Scholar] [CrossRef]
- Xu, Y.; Jia, X.; Panke, S.; Li, Z. Asymmetric dihydroxylation of arylolefins by sequential enantioselective epoxidation and regioselective hydrolysis with tandem biocatalysts. Chem. Commun. 2009, 12, 1481–1483. [Google Scholar] [CrossRef]
- Pandey, R.K.; Fernandes, R.A.; Kumar, P. An asymmetric dihydroxylation route to enantiomerically pure norfluoxetine and fluoxetine. Tetrahedron Lett. 2002, 43, 4425–4426. [Google Scholar] [CrossRef]
- Schrittwieser, J.H.; Velikogne, S.; Hall, M.; Kroutil, W. Artificial Biocatalytic Linear Cascades for Preparation of Organic Molecules. Chem. Rev. 2018, 118, 270–348. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wu, S.; Wu, J.; Li, Z. Enantioselective Cascade Biocatalysis via Epoxide Hydrolysis and Alcohol Oxidation: One-Pot Synthesis of (R)-α-Hydroxy Ketones from Meso- or Racemic Epoxides. ACS Catal. 2015, 5, 51–58. [Google Scholar] [CrossRef]
- Wu, S.; Chen, Y.; Xu, Y.; Li, A.; Xu, Q.; Glieder, A.; Li, Z. Enantioselective trans-Dihydroxylation of Aryl Olefins by Cascade Biocatalysis with Recombinant Escherichia coli Coexpressing Monooxygenase and Epoxide Hydrolase. ACS Catal. 2014, 4, 409–420. [Google Scholar] [CrossRef]
- Xue, F.; Liu, Z.-Q.; Wang, Y.-J.; Zhu, H.-Q.; Wan, N.-W.; Zheng, Y.-G. Efficient synthesis of (S)-epichlorohydrin in high yield by cascade biocatalysis with halohydrin dehalogenase and epoxide hydrolase mutants. Catal. Commun. 2015, 72, 147–149. [Google Scholar] [CrossRef]
- Gao, P.; Wu, S.; Praveen, P.; Loh, K.-C.; Li, Z. Enhancing productivity for cascade biotransformation of styrene to (S)-vicinal diol with biphasic system in hollow fiber membrane bioreactor. Appl. Microbiol. Biotechnol. 2017, 101, 1857–1868. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Parnell, A.; Williams, C.; Bakar, N.A.; Challand, M.R.; van der Kamp, M.W.; Simpson, T.J.; Race, P.R.; Crump, M.P.; Willis, C.L. A Rieske oxygenase/epoxide hydrolase-catalysed reaction cascade creates oxygen heterocycles in mupirocin biosynthesis. Nat. Catal. 2018, 1, 968–976. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Zhang, X.; Liu, F.; Xu, M.; Yang, T.; Long, M.; Zhou, J.; Osire, T.; Yang, S.; Rao, Z. Designing of a Cofactor Self-Sufficient Whole-Cell Biocatalyst System for Production of 1,2-Amino Alcohols from Epoxides. ACS Synth. Biol. 2019, 8, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-D.; Yang, X.-X.; Jia, Q.; Zhao, J.-W.; Gao, L.-L.; Gao, W.-C.; Chang, H.-H.; Wei, W.-L.; Xu, J.-H. Asymmetric ring opening of racemic epoxides for enantioselective synthesis of (S)-β-amino alcohols by a cofactor self-sufficient cascade biocatalysis system. Catal. Sci. Technol. 2019, 9, 70–74. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, X.; Dong, R.; Gao, L.; Li, J.; Li, X.; Huang, S.; Zhang, C.; Chang, H. Cascade Biocatalysis for Regio- and Stereoselective Aminohydroxylation of Styrenyl Olefins to Enantiopure Arylglycinols. ACS Sustain. Chem. Eng. 2020, 8, 18277–18285. [Google Scholar] [CrossRef]
- See, W.W.L.; Choo, J.P.S.; Jung, D.-Y.; Zhi, L. Recent developments in oxidative biocatalytic cascades. Curr. Opin. Green Sustain. Chem. 2022, 38, 100700. [Google Scholar] [CrossRef]



| Chiral Precursor | Final Product | Application of Final Product | Reaction Comment | Ref. |
|---|---|---|---|---|
 |  | Synthetic oxazolidinone antibiotic effective against Gram positive bacteria | Selective hydrolysis of (S)-enantiomer | [18] |
 | Cardio selective β-blocker for treatment of high blood pressure and heart associated chest pain | |||
 | Dietary supplement, involved in long-chain fatty acid transport in cells | |||
 |  | β-adrenergic blocker with antianginal and antiarrhythmic properties. | Chemo-enzymatic enantioconvergent synthesis | [19,20] |
 |  | Neuroprotective agent (aspartate receptor antagonist) | Sequential bi-enzymatic hydrolysis using 2 enantiocomplementary EHs | [21] |
 |  | HIV protease inhibitor MK 639 | Selective hydrolysis of 1(R),2(S)-enantiomer | [22] |
 |  | Non-steroidal anti-inflammatory drug | Selective hydrolysis of (R)-enantiomer | [23] |
 |  | Calcium channel blocker | Kinetic resolution | [24] |
 |  | Anthracycline antibiotic with chemotherapeutic properties | Enzymatic deracemization for production of (S)-diol used for chemical synthesis | [25] |
 |  | Major constituent of Matricaria chamomilla essential oil; ingredient for skin creams, lotions, ointments with anti-inflammatory, bactericidal and antimycotic properties | Chemo-enzymatic process for production of all 4 stereoisomers of bisabolol | [15] |
 |  | β-adrenergic receptor blocking drugs | Selective hydrolysis of (R)-enantiomer | [26] |
 |  | |||
 |  | Neuromediator with antiepileptic and antihypertensive activities | Selective hydrolysis of (S)-enantiomer | [27] |
 | Dietary supplement, involved in long-chain fatty acid transport in cells | |||
 |  | IGF-1R kinase inhibitor | Selective hydrolysis of (R)-enantiomer | [28] |
 | β3-adrenergic receptor agonists | Enantioconvergent hydrolysis of racemic epoxide | [29] | |
 | ||||
 |  | Antifungal triazole drug | Production of optically pure epoxide and diol that can be used for chemical synthesis of optically pure triazole derivatives | [30,31] |
 |  | Non-steroidal antiandrogen drug used for treatment of prostate cancer | Chemo-enzymatic synthesis of optically pure diol | [32] |
 |  | Ileal bile acid transport (iBAT) inhibitor indicated for diabetes type II | Kinetic resolution | [33] |
 |  | Chiral precursors for synthesis of various steroidal compounds | Kinetic resolution to produce both enantiomers of spiroepoxide, using 2 different EHs | [34] |
 |  | Melatonin receptor agonist used for treatment of sleep disorders | Selective hydrolysis of (R)-enantiomer | [35,36] |
 |  | Calcium channel blocker used for treatment of hypertension | Selective hydrolysis of (R)-enantiomer | [37] |
 | Chiral chemical building block with broad applications in chemical, pharmaceutical, food industries | Asymmetric hydrolysis to produce optically pure diol | [14] | |
 |  | Anticoagulant; direct factor Xa inhibitor developed by Bayer and marketed as Xarelto | - 1 | [38,39] |
 | Dietary supplement, involved in long-chain fatty acid transport in cells | - | [40,41] | |
 | Antidiabetic drug | - | [42] | |
 | β-adrenergic receptor blocking drug | - | [43] | |
 | β-adrenergic antagonist drug | - | [44,45] | |
| Modified Property | Source of EH Used as Template for Mutagenesis | Enzyme Engineering Method | Mutant | Substrate | E (-) | eeP (%) | C (%) | Ref. | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutant | WT | Mutant | WT | Mutant | WT | ||||||
| Enantioselectivity | Agrobacterium radiobacter AD1 | Directed evolution—Error-prone PCR and DNA shuffling | F108I/P205H/Y215H/E271V | styrene oxide | >50 | 16 | NI 1 | NI | [85] | ||
| p-nitrostyrene oxide | 81 | 56 | NI | NI | |||||||
| p-nitrophenyl glycidyl ether | 32 | 3.4 | NI | NI | |||||||
| epichlorohydrin | 40 | <2 | NI | NI | |||||||
| 1,2-epoxyhexane | 27 | 3.6 | NI | NI | |||||||
| Directed evolution—DNA shuffling and site-saturation mutagenesis | I219F | styrene oxide | 91 (R) | 17 (R) | NI | NI | [77] | ||||
| Rational design—Site-saturation mutagenesis | F108I | p-nitrophenyl glycidyl ether | 20 (S) | 3.4 (S) | NI | NI | [86] | ||||
| F108T | 22 (S) | 3.4 (S) | NI | NI | |||||||
| Agromyces mediolanus ZJB120203 | Rational design—Structure-based site saturation and site-directed mutagenesis | W182F/S207V/N240D | epichlorohydrin | 90.0 (R) | 12.9 (R) | NI | NI | [87] | |||
| Aspergillus niger LCP 521 | Directed evolution—one round of error-prone PCR | A217V/K332E/A390E | phenyl glycidyl ether | 10.8 (S) | 4.6 (S) | 74 (S) | 56 (S) | 39 | 33 | [75] | |
| Semi-rational design —Directed evolution using ISM with combinatorial active site saturation (CASTing) | L215F/A217N/R219S/L249Y/T317W/T318V/M329P/L330Y/C350V | phenyl glycidyl ether | 115 (S) | 4.6 (S) | 95 (S) | 56 (S) | 48 | 33 | [88] | ||
| Semi-rational design—Directed evolution using ISM and optimalization of expression in recombinant cells | P221S/F244C/L249F/L215F/T317F/T318V(ProThrAlaSerAlaProHisThrTyrArgGluPheIle)-L349V 2 | phenyl glycidyl ether | 160 (S) | 4.6 (S) | 97 (S) | 56 (S) | 45 | 33 | [89] | ||
| Semi-rational design—Directed evolution using all 24 possible pathways using 4 randomization sites for ISM | L215F/R219V/L249F/T317F/T318C/L349D/C350Y | phenyl glycidyl ether | 158 (S) | 4 (S) | 98 (S) | 61 (S) | 30 | 28 | [90] | ||
| Aspergillus usamii (AuEH2) | Semi-rational design—Microtuning of the substrate-binding pocket | A214C/A250I | styrene oxide | 202 | 16 | >99 (R) | NI | 50.2 | 40 | [91] | |
| A250I | o-nitrostyrene oxide | 341 | 96 | 98.0 (R) | NI | 50.5 | NI | ||||
| A250W | isopropyl glycidyl ether | 204 | 6.8 | 80.2 (S) | NI | 55.2 | NI | ||||
| Rhodococcus erythropolis DCL14 | Directed evolution—ISM using NDT codon degeneracy | M32C/I80F/L114C/I116V | cyclopentene oxide | NI | 93 (S,S) | 13 (R,R) | 65 | 72 | [92] | ||
| cyclohexene oxide | NI | 97 (S,S) | 4 (S,S) | 94 | 84 | ||||||
| cycloheptene oxide | NI | 98 (S,S) | 17 (S,S) | 75 | 74 | ||||||
| cis-2,3-butene oxide | NI | 93 (R,R) | 5 (S,S) | NI | |||||||
| phenyl glycidyl ether | 32 (R) | 2.6 (R) | 92 (R) | 37 (R) | 31 | 33 | |||||
| styrene oxide | 44 (S) | 2.8 (R) | 91 (S) | 40 (R) | 43 | 36 | |||||
| M32C/L74I/M78F/I80C/V83I | cyclopentene oxide | NI | 80 (R,R) | 13 (R,R) | 81 | 72 | |||||
| cyclohexene oxide | NI | 90 (R,R) | 4 (S,S) | 74 | 84 | ||||||
| cycloheptene oxide | NI | 77 (R,R) | 17 (S,S) | 84 | 74 | ||||||
| cis-2,3-butene oxide | NI | 83 (R,R) | 5 (S,S) | NI | |||||||
| styrene oxide | 36 (R) | 2.8 (R) | 91 (R) | 40 (R) | 30 | 36 | |||||
| Directed evolution—ISM using a single-code saturation mutagenesis (SCSM) | L74F/M78F/L103V/L114V/I116V/F139V/L147V | cyclohexene oxide | NI | 92 (S,S) | 4(S,S) | >99 | 84 | [78] | |||
| cycloheptene oxide | NI | 94 (S,S) | 17 (S,S) | 52 | 97 | ||||||
| L74F/M78F/I80V/L114F | cyclohexene oxide | NI | 96 (R,R) | 4 (S,S) | 83 | 84 | |||||
| cycloheptene oxide | NI | 94 (R,R) | 17 (S,S) | 66 | 97 | ||||||
| Directed evolution—ISM using double-code saturation mutagenesis (DCSM) | L74F/M78F/I80F/L114V/I116V/F138V | cyclopentene oxide | NI | 85 (S,S) | 13 (R,R) | 13 | 84 | [79] | |||
| cyclohexene oxide | NI | 97 (S,S) | 4 (S,S) | 98 | >99 | ||||||
| cycloheptene oxide | NI | 97 (S,S) | 17 (S,S) | 73 | 97 | ||||||
| M78V/I80V/L114F | cyclohexene oxide | NI | 92 (R,R) | 13 (R,R) | 99 | >99 | |||||
| cycloheptene oxide | NI | 85 (R,R) | 4 (S,S) | 40 | 97 | ||||||
| styrene oxide | NI | 57 (S) | 21 (R) | 7 | 46 | ||||||
| Directed evolution—ISM using triple-code saturation mutagenesis (TCSM) | I80Y/L114V/I116V | cyclohexene oxide | NI | 99 (S,S) | 4 (S,S) | 97 | >99 | [80] | |||
| cycloheptene oxide | NI | 98 (S,S) | 17 (S,S) | 81 | 97 | ||||||
| styrene oxide | 28.0 | 1.8 | 92 (S) | 26 (R) | 15 | 17 | |||||
| M32V/M78V/I80V/L114F | cyclohexene oxide | NI | 97 (R,R) | 4 (S,S) | >99 | >99 | |||||
| cycloheptene oxide | NI | 94 (R,R) | 17 (S,S) | 83 | 97 | ||||||
| Semi-rational design—Directed evolution using ISM with reduced AA alphabets using binary pattern based on choosing hydrophobic and hydrophilic amino acids | I80F/V83I/L114 V/I116V | cyclopentene oxide | NI | 94 (S,S) | 7 (R,R) | 34 | 69 | [93] | |||
| cyclohexene oxide | NI | 97 (S,S) | 3 (S,S) | 93 | 87 | ||||||
| cycloheptene oxide | NI | 97 (S,S) | 22 (S,S) | 96 | 99 | ||||||
| I80V/V83I/L114 V | cyclopentene oxide | NI | 51 (R,R) | 7 (R,R) | 48 | 69 | |||||
| cyclohexene oxide | NI | 79 (R,R) | 3 (S,S) | 97 | 87 | ||||||
| cycloheptene oxide | NI | 53 (R,R) | 22 (S,S) | 99 | 99 | ||||||
| Semi-rational design—Directed evolution using ISM with the aim to improve thermostability, enantioselectivity and activity | T76K/L114V/I116V/N92K/F139V/L147F/S15D/A19K/L74F/M78F/E45D | cyclohexene oxide | NI | 94 (S,S) | 2 (S,S) | 100 | 100 | [94] | |||
| S15P/M78F/N92K/F139V/T76K/T85K/E45D/I80V/E124D | cyclohexene oxide | NI | 80 (R,R) | 2 (S,S) | 100 | 100 | |||||
| Semi-rational design—Directed evolution using saturation mutagenesis, mutants were prepared by high-fidelity solid-phase chemical gene synthesis on silicon chips followed by efficient gene assembly instead of PCR to overcome AA bias | M78F/I80Y/L114V/I116V | cyclohexene oxide | NI | >98 (S,S) | NI | >98 | NI | [95] | |||
| R. erythropolis DCL14 (mutant LEH-P) 3 | Rational design—Computational design of mutant library using CASCO strategy | M32L/L74I/I80V/L103F/F139L | cyclopentene oxide | NI | 85.5 (R,R) | 23.9 (R,R) | NI | [96] | |||
| M32L/L35W/L74F/M78F/I80A/I116V/F139L | NI | 90.2 (S,S) | 23.9 (R,R) | NI | |||||||
| Rational design—Use of Rosetta enzyme design to computationally predict enantioselective mutants and high-throughput-multiple independent molecular docking simulations for in silico screening of the generated mutant libraries | M32A/M78I/I80F/L103I/I116V/F139L | cyclopentene oxide | NI | 85 (S,S) | 14 (R,R) | NI | [81] | ||||
| L35W/L74F/I80G/I116V/F139L | cis-2,3-butane oxide | NI | 82 (S,S) | 2 (S,S) | NI | ||||||
| M32L/L35G/I80W/L103V/F139L | cis-stilbene oxide | NI | >99 (R,R) | 92 (R,R) | 98 | NI | |||||
| M32L/L35M/L103I/L114M/I116F/F139L | NI | 88 (S,S) | 92 (R,R) | 63 | NI | ||||||
| Solanum tuberosum (StEH1) | Semi-rational design—Directed evolution—ISM targeting AA residues around active site of enzyme | W106L/L109Y/V141K/I155V | (2,3-epoxypropyl)benzene | 15 (R) | 0.4 (R) | 60 (R) | 32 (S) | NI | [97] | ||
| Semi-rational design—Directed evolution with 2 rounds of iterative saturation mutagenesis | W106L/L109Y/V141K/I155W/F189C | styrene oxide | 5800 (S) | 69 (S) | NI | NI | [98] | ||||
| trans-2-methylstyrene oxide | 770 (S) | 84 (S) | NI | NI | |||||||
| Sphingomonas sp. HXN-200 | Semi-rational design—Site-directed mutagenesis of selected AA residues in active site based on homology modelling | V196A/N226A/M332A | phenyl glycidyl ether | 21.2 (R) | 2.2 (R) | 79.2 (S) | 61.9 (S) | 50 | 50 | [99] | |
| metagenomic DNA (Kau2EH) | Semi-rational design—Directed evolution by randomizing selected sites within substrate binding pocket | V290Y | p-chlorostyrene oxide | 130 | NI | 97 (R) | NI | 50 | NI | [100] | |
| Enantioconvergence | A. niger M200 | Semi-rational design—ISM, mutated sites were chosen on structural similarity with EH from A. niger LCP 521 | L349V/C350W/T317W/T318V/M218W/R219E/L215M/A217G/M245A | styrene oxide | 22 | 10 | 70.1 (R) | 3.0 (R) | 100 | 100 | [101] |
| p-chlorostyrene oxide | 20 | 40 | 70.5 (R) | 4.4 (R) | 100 | 100 | |||||
| Glycine max (GmEH3) | Semi-rational design —Site-saturation and combinatorial mutagenesis used for reshaping substrate-binding pocket | W102V/P187F | 1,2-epoxyhexane | NI | 83.8 (R) | 47.2 (R) | >99 | >99 | [102] | ||
| Phaseolus vulgaris (PvEH1) | Rational design—Site-directed mutagenesis based on molecular docking simulations and multiple alignment | L105I/M160A/M175I | styrene oxide | 3.6 | 1.5 | 87.8 (R) | 33.6 (R) | NI | [103] | ||
| m-chlorostyrene oxide | NI | 69.7 (R) | 1.0 (R) | NI | |||||||
| p-nitrostyrene oxide | NI | 64.7 (R) | 50.3 (R) | NI | |||||||
| m-nitrostyrene oxide | NI | 52.3 (R) | 14.7 (R) | NI | |||||||
| p-chlorostyrene oxide | NI | 70.9 (R) | 51.4 (R) | NI | |||||||
| Rational design—Leucine scanning used for identification of AA residues at sites lining the enzyme’s binding pocket responsible for enantioconvergence and subsequent saturation mutagenesis | L105I/M160A/M175I/Y149L/P184W | m-chlorostyrene oxide | NI | 96.1 (R) | 1.0 (R) | >99 | >99 | [104] | |||
| Rational design—Reshaping of substrate binding pocket | L105I/V106I/M160A/M175I/S178T/P184W | styrene oxide | NI | 90.3 (R,R) | 33.6 (R,R) | >99.9 | 99.1 | [82] | |||
| p-nitrostyrene oxide | NI | 86.7 (R,R) | 50.3 (R,R) | 84.2 | 99.3 | ||||||
| m-nitrostyrene oxide | NI | 85.1 (R,R) | 14.7 (R,R) | >99.9 | 99.7 | ||||||
| p-fluorostyrene oxide | NI | 90.6 (R,R) | 13.6 (R,R) | >99.9 | 98.7 | ||||||
| m-chlorostyrene oxide | 6 | 2 | 96.2 (R,R) | 1.0 (R,R) | 99.2 | 99.9 | |||||
| Rhodotorula paludigena JNU001 | Rational design—Microtuning substrate-binding pocket of EH by computer-aided design using valine scanning mutagenesis | L360C | m-nitrostyrene oxide | NI | 93.4 (R) | 85.7 (R) | 99 | >99 | [105] | ||
| Vigna radiata (VrEH2) | Rational design—Creation of smart library by site-directed mutagenesis using reduced AA alphabet to prepare enantioconvergent EH | M263N | p-nitrostyrene oxide | NI | 98 (R) | 84 (R) | 99.5 | NI | [106] | ||
| m-nitrostyrene oxide | NI | 90 (R) | 20 (R) | >99 | >99 | ||||||
| Rational design—Creation of smart library by site-directed mutagenesis using reduced AA alphabet to prepare enantioconvergent EH | M263Q | m-chlorostyrene oxide | NI | 90 (R) | 20 (R) | NI | [107] | ||||
| M263V | 2-naphthyloxirane | NI | 90 | 60 | NI | ||||||
| metagenomic DNA (Kau2EH) | Semi-rational design —Directed evolution by randomizing selected sites within substrate binding pocket | W110L/F113L/F161Y/P193G/V290W | p-chlorostyrene oxide | 17 | 23 | 93 (R) | 84 (R) | 100 | 100 | [100] | |
| Immobilization Technique | EH (Source) | Immobilized Biocatalyst | Support | Benefit of Immobilization | Ref. |
|---|---|---|---|---|---|
| Covalent bond | ArEH (Agrobacterium radiobacter AD1) | Crude enzyme extract | LX-1000EP modified by EDA LX-1000EP | Operational stability, reusability, increased thermal stability as compared to free enzyme | [121] |
| Purified enzyme | Dextran activated with NaIO4 and ethylene glycol Ficoll activated with NaIO4 and ethylene glycol Amylopectin activated with NaIO4 and ethylene glycol Carboxymethyl cellulose activated with NaIO4 and ethylene glycol | Improved tolerance to the inhibitory effects of Co2+, Fe3+ and EDTA | [122] | ||
| Kau2EH (metagenomic DNA) | Purified enzyme | Eupergit C 250L Eupergit C Eupergit C modified by IDA and CuSO4 Sepabeads EC-EP Sepabeads EC-EP modified by IDA and CuSO4 | Significantly higher thermal stability as compared to free enzyme | [123] | |
| VrEH2M263N (Vigna radiata) | Purified enzyme | ECR8205F (Epoxy) ECR4204F (Epoxy) ECR8215F (Epoxy) ES-103 (Epoxy) ESR-1 (Amino) ESQ-1 (Amino) ECR8405F (Amino) | Improvement of thermal and operational stability as compared to free enzyme | [19] | |
| AnEH (Aspergillus niger LCP 521) | Purified enzyme (lyophilized powder) | Eupergit C Eupergit C 250L Eupergit C 250L modified by EDA Eupergit C modified by IDA and CuSO4 | Improvement of enzyme stability and enantioselectivity | [124] | |
| Eupergit C 250L modified by EDA and glutaraldehyde | Improvement of enzyme storage and thermal stability and enantioselectivity | [125] | |||
| Eupergit C modified by EDA and glutaraldehyde; Florisil® silanized with 3-APTES and activated with glutaraldehyde | Improvement of enzyme reusability and enantioselectivity | [126] | |||
| Epoxide-derived silica gel | Enhancement of enzyme stability in the presence of DMSO | [127] | |||
| StEH (Solanum tuberosum) | Crude enzyme extract (lyophilized powder) | Sepabeads EP—Epoxy modified by IDA and CuSO4 Glyoxyl–agarose (agarose modified by glycidol and oxidized by NaIO4) | Stabilization of enzyme | [128] | |
| mEH (rat liver) | Purified enzyme | Sephadex G-150 activated by 1,1′-carbonyldiimidazole | Enhancement of stability and repeated use of the enzyme | [129] | |
| Dextran activated by 1,1′-carbonyldiimidazole | Increasement of enzyme stability | [130] | |||
| VaEH (Vigna angularis) | Partially purified enzyme | Mesocellular foam silica (MCF) amino modified and activated by glutaraldehyde; Santa Barbara Amorphous (SBA-15) amino modified and activated by glutaraldehyde | Enhancement of enzyme operational stability and thermal stability | [131] | |
| Ionic bond/Affinity bond (His-tag) | StEH (Solanum tuberosum) | Crude enzyme extract | Silica oxide powder modified by resacetophenone and Co2+ | Observation of enzyme activity in organic solvents | [132] |
| mMcEH (triple mutant) (Mugil cephalus) | Purified enzyme | NiO presenting magnetic nanoparticles | Reusability of enzyme | [133] | |
| CESH (Nocardia tartaricans CAS-52) | Purified enzyme | Metal ion affinity chromatography media Ni-IDA QZT 6FF | Enhancement of enzyme activity | [134] | |
| Adsorption | AnEH (Aspergillus niger LCP 521) | Purified enzyme | Accurel EP 100 (polypropylene resin) | Enhancement of enzyme operational stability using nonporous DEAE-cellulose | [135] |
| DEAE cellulose (ionic bond) | Reusability of enzyme | [135,136] | |||
| Porous polypropylene | Immobilized for preparative purposes (reuse, continuous reactor) | [137] | |||
| Lewatit® VP OC 1600 | Enzyme reusability, enhancement of enantioselectivity | [126] | |||
| Nsp.EH (Nocardia sp. EH1) | Partially purified enzyme | DEAE cellulose (ionic bond) | Enzyme stabilization | [138] | |
| ArEH (Agrobacterium radiobacter AD1 expressed in E. coli) | Whole cells | Perlite | Immobilized for preparative purposes | [139] | |
| McEH (Mugil cephalus) | Purified enzyme | Magnetically separable silica Mag-MSU-F (adsorption) + cross-linking with glutaraldehyde | Enhancement of enzyme stability and reusability | [140] | |
| CLEA | VrEH (Vigna radiata) | Partially purified enzyme extract | Cross-linker: glutaraldehyde | Enhancement of catalytic efficiency, enantioselectivity and product yield | [141] |
| Enhancement of initial reaction rate, product yield, enantioselectivity, operational stability | [142] | ||||
| Co-polymerization | RgEH (Rhodotorula glutinis CIMW 147 (ATCC 201718)) | Partially purified enzyme | Acylation of enzyme by itaconic acid, bio-imprinted with substrate and copolymerized with ethylene glycol dimethacrylate | Enzyme stabilization, reusability and product separation, improvement of enantioselectivity | [143] |
| Nanoflowers | GmEH (Glycine max) | Purified enzyme | Organic–inorganic nanoflowers formed with Ca2+ ions | High catalytic activity and stability | [144] |
| Metal–organic framework (MOFs) | GmEH (Glycine max) | Crude enzyme preparation (extract) | UiO-66-NH2 metal−organic framework (MOF) cross-linked with glutaraldehyde | Higher enzyme pH stability, thermostability and tolerance to organic solvents as compared to free enzyme | [145] |
| HdEH (Hypsibius dujardini) | Purified enzyme | Zeolitic imidazole frameworks (ZIF-8) Zeolitic imidazole frameworks treated with glutaraldehyde (Glu/ZIF-8) | Enhancement of stability, enantioselectivity, reusability of enzyme | [146] | |
| Encapsulation | CESH (Nocardia tartaricans ATCC 31191) | Whole cells | Polyelectrolyte complex microcapsules from sodium alginate−cellulose sulfate−poly(methylene-co-guanidine) | Enhanced enzyme activity, storage stability and decreased reaction time using immobilized whole cells as compared to free cells | [116] |
| Enhancement of operational stability | [118] | ||||
| Entrapment | RtEH (Rhodosporidium toruloides UOFS Y-0471) | Whole cells | Calcium alginate | Stabilization of cells | [147] |
| CESH (Labrys sp. BK-8) | Whole cells | κ-carrageenan | Stabilization of cells | [148] | |
| not mentioned | NOVO SP409 (Rhodococcus sp. commercial preparation) | Crude enzyme | Not mentioned | Preparative purposes | [113] |
| Enzymes in the Cascade including EH and Enzyme Source/GMO Cells | Substrate(s) | Product(s) | Note to the Role of EH in the Cascade | Ref. |
|---|---|---|---|---|
Epoxide hydrolase SpEH from Sphingomonas sp. HXN-200 and butanediol dehydrogenase BDHA from Bacillus subtilis BGSC1A1 and NADH oxidase NOX from Lactobacillus brevis DSM 20054/
| Meso- or racemic epoxides | R-(α)-hydroxyketones | No significant influence of using separately expressed vs. co-expressed enzymes of the cascade on ee and conversion | [155] |
Epoxide hydrolase SpEH from Sphingomonas sp. HXN-200 or Epoxide hydrolase StEH from Solanum tuberosum and styrene monooxygenase SMO/
| Aryl olefins | Chiral vicinal diols | The first enzyme cascade which enabled reversing enantioselectivity of dihydroxylation using StEH instead of SpEH | [156] |
Epoxide hydrolase AmEH from Agromyces mediolanus and halohydrin dehalogenase HheC from Agrobacterium radiobacter AD1/
| 1,3-dichloro-2-propanol | Chiral epichlorohydrin | Effect of co-expressed vs. separately expressed enzymes on the enantioselectivity of the cascade | [157] |
Epoxide hydrolase SpEH from Sphingomonas sp. and styrene monooxygenase SMO from Pseudomonas sp./
| Styrene | (S)-1-phenyl-1,2-ethanediol | Aqueous/organic biphasic reaction system was used for the first time for cascade biotransformation to enhance productivity | [158] |
Epoxide hydrolase MupZ from Pseudomonas fluorescens NCIMB 10586 and Rieske non-heme oxygenase MupW Pseudomonas fluorescens NCIMB 10586/
| Mupirocins | Hydroxylated tetrahydropyrans and tetrahydrofurans | Cascade containing epoxide hydrolase and Rieske non-heme oxygenase enabled formation of heterocyclic THP ring, which is difficult to achieve biosynthetically | [159] |
| Epoxide hydrolase SpEH from Sphingomonas sp. HXN-200, alcohol dehydrogenase MnADH from Mycobacterium neoaurum VKM AC-1815D, ω-transaminase PAKω-TA from Pseudomonas aeruginosa and glutamate dehydrogenase GluDH from E. coli BL21/ E. coli BL21 (SGMP) co-expressing 4-enzyme self-sufficient cascade system SpEH-MnADH-PAKω-TA-GluDH | (S)-epoxides | Chiral 1,2-aminoalcohols | The first one-step synthesis of optically pure 1,2-amino alcohols from (S)-epoxides employing a synthetic redox-self-sufficient enzyme cascade in recombinant cells | [160] |
Epoxide hydrolase SpEH from Sphingomonas sp. HXN-200, 2,3-butanediol dehydrogenase BDHA from Bacillus subtilis, polyol dehydrogenase GoSCR from Gluconobacter oxydans, (R)-ω-transaminase MVTA from Mycobacterium vanbaalenii/
| Racemic epoxides | Enantiopure β-amino alcohols | General access to variety of chiral β-amino alcohols starting from inexpensive racemic epoxides using designed enzyme cascade process in recombinant cells | [161] |
Styrene monooxygenase SMO from Pseudomonas sp., epoxide hydrolase SpEH from Sphingomonas sp. HXN-200, polyol dehydrogenase GoSCR from Gluconobacter oxydans, (R)-ω-transaminase MVTA from Mycobacterium vanbaalenii or transaminase BMTA from Bacillus megaterium SC6394/
| Styrenyl olefins | 2-amino-2-phenyl ethanols | Challenging direct regio- and stereoselective aminohydroxylation of olefins to unprotected enantioenriched β-amino alcohols was enabled by novel one-pot four-enzyme biocatalytic cascade in good yields and excellent enantioselectivity | [162] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bučko, M.; Kaniaková, K.; Hronská, H.; Gemeiner, P.; Rosenberg, M. Epoxide Hydrolases: Multipotential Biocatalysts. Int. J. Mol. Sci. 2023, 24, 7334. https://doi.org/10.3390/ijms24087334
Bučko M, Kaniaková K, Hronská H, Gemeiner P, Rosenberg M. Epoxide Hydrolases: Multipotential Biocatalysts. International Journal of Molecular Sciences. 2023; 24(8):7334. https://doi.org/10.3390/ijms24087334
Chicago/Turabian StyleBučko, Marek, Katarína Kaniaková, Helena Hronská, Peter Gemeiner, and Michal Rosenberg. 2023. "Epoxide Hydrolases: Multipotential Biocatalysts" International Journal of Molecular Sciences 24, no. 8: 7334. https://doi.org/10.3390/ijms24087334
APA StyleBučko, M., Kaniaková, K., Hronská, H., Gemeiner, P., & Rosenberg, M. (2023). Epoxide Hydrolases: Multipotential Biocatalysts. International Journal of Molecular Sciences, 24(8), 7334. https://doi.org/10.3390/ijms24087334