Impact of Adipose Tissue Depot Harvesting Site on the Multilineage Induction Capacity of Male Rat Adipose-Derived Mesenchymal Stem Cells: An In Vitro Study
Abstract
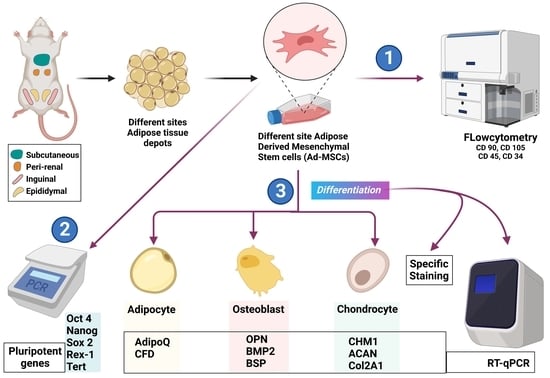
:1. Introduction
2. Results
2.1. Morphologic and Immunophenotypic Characterization of Different Sites’ r-AdMSCs
2.2. Potential to Express the Pluripotency Genes via Different Sites’ r-AdMSCs
2.3. Qualitative and Quantitative Assessment of the Multilineage Differentiation Capacity of Different Sites’ r-AdMSCs
2.3.1. Adipogenic Differentiation Potential
2.3.2. Osteogenic Differentiation Potential
2.3.3. Chondrogenic Differentiation Potential
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. Isolation and In Vitro Culture of Different Sites’ r-AdMSCs
4.3. Characterization of r-AdMSCs’ Morphology and Immunophenotypes
4.4. Expression of Pluripotent Genes in r-AdMSCs
4.5. In Vitro Assessment of Cell Heterogeneity and Multilineage Differentiation Capacity
4.5.1. Adipogenic Differentiation
4.5.2. Osteogenic Differentiation
4.5.3. Chondrogenic Induction
4.5.4. Quantitative Assessment of the Differentiation Capacity Using RT-qPCR
4.6. Statistical Analysis
4.7. Limitations of the Present Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ji, S.; Xiong, M.; Chen, H.; Liu, Y.; Zhou, L.; Hong, Y.; Wang, M.; Wang, C.; Fu, X.; Sun, X. Cellular rejuvenation: Molecular mechanisms and potential therapeutic interventions for diseases. Signal Transduct. Target. Ther. 2023, 8, 116. [Google Scholar] [CrossRef]
- El-Husseiny, H.M.; Mady, E.A.; Radwan, Y.; Nagy, M.; Abugomaa, A.; Elbadawy, M.; Tanaka, R. Hybrid Biodegradable Polymeric Scaffolds for Cardiac Tissue Engineering. In Handbook of Biodegradable Materials; Ali, G.A.M., Makhlouf, A.S.H., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 1045–1092. [Google Scholar]
- Prakoso, A.T.; Basri, H.; Adanta, D.; Yani, I.; Ammarullah, M.I.; Akbar, I.; Ghazali, F.A.; Syahrom, A.; Kamarul, T. The Effect of Tortuosity on Permeability of Porous Scaffold. Biomedicines 2023, 11, 427. [Google Scholar] [CrossRef]
- Doghish, A.S.; Ismail, A.; El-Mahdy, H.A.; Elkhawaga, S.Y.; Elsakka, E.G.E.; Mady, E.A.; Elrebehy, M.A.; Khalil, M.A.F.; El-Husseiny, H.M. miRNAs insights into rheumatoid arthritis: Favorable and detrimental aspects of key performers. Life Sci. 2023, 314, 121321. [Google Scholar] [CrossRef]
- El-Husseiny, M. Platelet rich fibrin augmented versus non-augmented glycerolized bovine pericardium and polypropylene mesh for repairing of large abdominal wall defects. Eur. J. Med. Nat. Sci. 2019, 3, 33–48. [Google Scholar] [CrossRef]
- Sharun, K.; Chandran, D.; Manjusha, K.M.; Mankuzhy, P.D.; Kumar, R.; Pawde, A.M.; Dhama, K.; El-Husseiny, H.M. Advances and prospects of platelet-rich plasma therapy in veterinary ophthalmology. Vet. Res. Commun. 2023. [Google Scholar] [CrossRef]
- El-Husseiny, H. Evaluation of Some Prosthetic Implants for Surgical Management of Different Varieties of Hernias in Domestic Animals; Department of Surgery, Anesthesiology, and Radiology, Faculty of Veterinary Medicine, Benha University: Benha, Egypt, 2017; pp. 42–43. [Google Scholar]
- El-Husseiny, H.M.; Mady, E.A.; El-Dakroury, W.A.; Doghish, A.S.; Tanaka, R. Stimuli-Responsive Hydrogels: Smart State Of-the-Art Platforms for Cardiac Tissue Engineering. 2022. Available online: https://www.researchsquare.com/article/rs-2011475/v1 (accessed on 10 September 2022).
- El-Husseiny, H.M.; Mady, E.A.; Helal, M.A.; Tanaka, R. The Pivotal Role of Stem Cells in Veterinary Regenerative Medicine and Tissue Engineering. Vet. Sci. 2022, 9, 648. [Google Scholar] [CrossRef]
- Hendawy, H.; Uemura, A.; Ma, D.; Namiki, R.; Samir, H.; Ahmed, M.F.; Elfadadny, A.; El-Husseiny, H.M.; Chieh-Jen, C.; Tanaka, R. Tissue Harvesting Site Effect on the Canine Adipose Stromal Vascular Fraction Quantity and Quality. Animals 2021, 11, 460. [Google Scholar] [CrossRef]
- Farashah, M.S.G.; Mohammadi, A.; Javadi, M.; Rad, J.S.; Shakouri, S.K.; Meshgi, S.; Roshangar, L. Bone marrow mesenchymal stem cells’ osteogenic potential: Superiority or non-superiority to other sources of mesenchymal stem cells? Cell Tissue Bank. 2023. [Google Scholar] [CrossRef]
- Yasumura, Y.; Teshima, T.; Nagashima, T.; Takano, T.; Michishita, M.; Taira, Y.; Suzuki, R.; Matsumoto, H. Immortalized Canine Adipose-Derived Mesenchymal Stem Cells as a Novel Candidate Cell Source for Mesenchymal Stem Cell Therapy. Int. J. Mol. Sci. 2023, 24, 2250. [Google Scholar] [CrossRef]
- Wen, W.; Pang, Y.; Tian, Y.; Xu, C.; Wang, J.; Wu, Y.; Xie, X. Osteogenic mesenchymal stem cells/progenitors in the periodontium. Oral Dis. 2023, 1–7. [Google Scholar] [CrossRef]
- Chamberlain, G.; Fox, J.; Ashton, B.; Middleton, J. Concise Review: Mesenchymal Stem Cells: Their Phenotype, Differentiation Capacity, Immunological Features, and Potential for Homing. Stem Cells 2007, 25, 2739–2749. [Google Scholar] [CrossRef]
- Gaur, M.; Dobke, M.; Lunyak, V.V. Methods and Strategies for Procurement, Isolation, Characterization, and Assessment of Senescence of Human Mesenchymal Stem Cells from Adipose Tissue. In Stem Cells and Aging: Methods and Protocols; Turksen, K., Ed.; Springer: New York, NY, USA, 2019; pp. 37–92. [Google Scholar]
- Pak, J.; Lee, J.H.; Pak, N.; Pak, Y.; Park, K.S.; Jeon, J.H.; Jeong, B.C.; Lee, S.H. Cartilage Regeneration in Humans with Adipose Tissue-Derived Stem Cells and Adipose Stromal Vascular Fraction Cells: Updated Status. Int. J. Mol. Sci. 2018, 19, 2146. [Google Scholar] [CrossRef]
- Yang, Z.; Huang, C.-Y.C.; Candiotti, K.A.; Zeng, X.; Yuan, T.; Li, J.; Yu, H.; Abdi, S. Sox-9 facilitates differentiation of adipose tissue-derived stem cells into a chondrocyte-like phenotype in vitro. J. Orthop. Res. 2011, 29, 1291–1297. [Google Scholar] [CrossRef]
- De Girolamo, L.; Stanco, D.; Salvatori, L.; Coroniti, G.; Arrigoni, E.; Silecchia, G.; Russo, M.A.; Niada, S.; Petrangeli, E.; Brini, A.T. Stemness and Osteogenic and Adipogenic Potential are Differently Impaired in Subcutaneous and Visceral Adipose Derived Stem Cells (ASCs) Isolated from Obese Donors. Int. J. Immunopathol. Pharmacol. 2013, 26, 11–21. [Google Scholar] [CrossRef]
- Scioli, M.G.; Bielli, A.; Gentile, P.; Mazzaglia, D.; Cervelli, V.; Orlandi, A. The Biomolecular Basis of Adipogenic Differentiation of Adipose-Derived Stem Cells. Int. J. Mol. Sci. 2014, 15, 6517. [Google Scholar] [CrossRef]
- Young, D.A.; Choi, Y.S.; Engler, A.J.; Christman, K.L. Stimulation of adipogenesis of adult adipose-derived stem cells using substrates that mimic the stiffness of adipose tissue. Biomaterials 2013, 34, 8581–8588. [Google Scholar] [CrossRef]
- Choi, Y.S.; Dusting, G.J.; Stubbs, S.; Arunothayaraj, S.; Han, X.L.; Collas, P.; Morrison, W.A.; Dilley, R.J. Differentiation of human adipose-derived stem cells into beating cardiomyocytes. J. Cell. Mol. Med. 2010, 14, 878–889. [Google Scholar] [CrossRef]
- Si, Z.; Wang, X.; Sun, C.; Kang, Y.; Xu, J.; Wang, X.; Hui, Y. Adipose-derived stem cells: Sources, potency, and implications for regenerative therapies. Biomed. Pharmacother. 2019, 114, 108765. [Google Scholar] [CrossRef]
- Arévalo-Turrubiarte, M.; Olmeo, C.; Accornero, P.; Baratta, M.; Martignani, E. Analysis of mesenchymal cells (MSCs) from bone marrow, synovial fluid and mesenteric, neck and tail adipose tissue sources from equines. Stem Cell Res. 2019, 37, 101442. [Google Scholar] [CrossRef]
- Reumann, M.K.; Linnemann, C.; Aspera-Werz, R.H.; Arnold, S.; Held, M.; Seeliger, C.; Nussler, A.K.; Ehnert, S. Donor Site Location Is Critical for Proliferation, Stem Cell Capacity, and Osteogenic Differentiation of Adipose Mesenchymal Stem/Stromal Cells: Implications for Bone Tissue Engineering. Int. J. Mol. Sci. 2018, 19, 1868. [Google Scholar] [CrossRef]
- Arnhold, S.; Elashry, M.I.; Klymiuk, M.C.; Geburek, F. Investigation of stemness and multipotency of equine adipose-derived mesenchymal stem cells (ASCs) from different fat sources in comparison with lipoma. Stem Cell Res. Ther. 2019, 10, 309. [Google Scholar] [CrossRef]
- Rebelatto, C.K.; Aguiar, A.M.; Moretão, M.P.; Senegaglia, A.C.; Hansen, P.; Barchiki, F.; Oliveira, J.; Martins, J.; Kuligovski, C.; Mansur, F.; et al. Dissimilar Differentiation of Mesenchymal Stem Cells from Bone Marrow, Umbilical Cord Blood, and Adipose Tissue. Exp. Biol. Med. 2008, 233, 901–913. [Google Scholar] [CrossRef]
- Barberini, D.J.; Freitas, N.P.P.; Magnoni, M.S.; Maia, L.; Listoni, A.J.; Heckler, M.C.; Sudano, M.J.; Golim, M.A.; da Cruz Landim-Alvarenga, F.; Amorim, R.M. Equine mesenchymal stem cells from bone marrow, adipose tissue and umbilical cord: Immunophenotypic characterization and differentiation potential. Stem Cell Res. Ther. 2014, 5, 25. [Google Scholar] [CrossRef]
- Boeuf, S.; Richter, W. Chondrogenesis of mesenchymal stem cells: Role of tissue source and inducing factors. Stem Cell Res. Ther. 2010, 1, 31. [Google Scholar] [CrossRef]
- Tholpady, S.S.; Katz, A.J.; Ogle, R.C. Mesenchymal stem cells from rat visceral fat exhibit multipotential differentiation in vitro. Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol. 2003, 272A, 398–402. [Google Scholar] [CrossRef]
- Hendawy, H.; Kaneda, M.; Metwally, E.; Shimada, K.; Tanaka, T.; Tanaka, R. A Comparative Study of the Effect of Anatomical Site on Multiple Differentiation of Adipose-Derived Stem Cells in Rats. Cells 2021, 10, 2469. [Google Scholar] [CrossRef]
- Song, N.; Scholtemeijer, M.; Shah, K. Mesenchymal Stem Cell Immunomodulation: Mechanisms and Therapeutic Potential. Trends Pharmacol. Sci. 2020, 41, 653–664. [Google Scholar] [CrossRef]
- Russo, V.; Yu, C.; Belliveau, P.; Hamilton, A.; Flynn, L.E. Comparison of Human Adipose-Derived Stem Cells Isolated from Subcutaneous, Omental, and Intrathoracic Adipose Tissue Depots for Regenerative Applications. Stem Cells Transl. Med. 2014, 3, 206–217. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Discher, D.E.; Péault, B.M.; Phinney, D.G.; Hare, J.M.; Caplan, A.I. Mesenchymal stem cell perspective: Cell biology to clinical progress. NPJ Regen. Med. 2019, 4, 22. [Google Scholar] [CrossRef]
- He, Q.; Ye, Z.; Zhou, Y.; Tan, W.-S. Comparative study of mesenchymal stem cells from rat bone marrow and adipose tissue. Turk. J. Biol. 2018, 42, 477–489. [Google Scholar] [CrossRef]
- Liu, B.-H.; Yeh, H.-Y.; Lin, Y.-C.; Wang, M.-H.; Chen, D.C.; Lee, B.-H.; Hsu, S.-h. Spheroid Formation and Enhanced Cardiomyogenic Potential of Adipose-Derived Stem Cells Grown on Chitosan. BioRes. Open Access 2012, 2, 28–39. [Google Scholar] [CrossRef]
- Barzilay, R.; Sadan, O.; Melamed, E.; Offen, D. Comparative characterization of bone marrow-derived mesenchymal stromal cells from four different rat strains. Cytotherapy 2009, 11, 435–442. [Google Scholar] [CrossRef]
- Schäffler, A.; Büchler, C. Concise Review: Adipose Tissue-Derived Stromal Cells—Basic and Clinical Implications for Novel Cell-Based Therapies. Stem Cells 2007, 25, 818–827. [Google Scholar] [CrossRef]
- Taha, M.F.; Javeri, A.; Rohban, S.; Mowla, S.J. Upregulation of Pluripotency Markers in Adipose Tissue-Derived Stem Cells by miR-302 and Leukemia Inhibitory Factor. BioMed Res. Int. 2014, 2014, 941486. [Google Scholar] [CrossRef]
- Pierantozzi, E.; Gava, B.; Manini, I.; Roviello, F.; Marotta, G.; Chiavarelli, M.; Sorrentino, V. Pluripotency Regulators in Human Mesenchymal Stem Cells: Expression of NANOG But Not of OCT-4 and SOX-2. Stem Cells Dev. 2010, 20, 915–923. [Google Scholar] [CrossRef]
- Gao, Q.; Zhao, L.; Song, Z.; Yang, G. Expression pattern of embryonic stem cell markers in DFAT cells and ADSCs. Mol. Biol. Rep. 2012, 39, 5791–5804. [Google Scholar] [CrossRef]
- Casella, S. Gene Expression Analysis of Rat Adi-pose Tissue-Derived Stem Cells. Int. J. Stem Cell Res. Transpl. 2015, 3, 120–124. [Google Scholar]
- Scotland, K.B.; Chen, S.; Sylvester, R.; Gudas, L.J. Analysis of Rex1 (zfp42) function in embryonic stem cell differentiation. Dev. Dyn. 2009, 238, 1863–1877. [Google Scholar] [CrossRef]
- Hidema, S.; Fukuda, T.; Date, S.; Tokitake, Y.; Matsui, Y.; Sasaki, H.; Nishimori, K. Transgenic expression of Telomerase reverse transcriptase (Tert) improves cell proliferation of primary cells and enhances reprogramming efficiency into the induced pluripotent stem cell. Biosci. Biotechnol. Biochem. 2016, 80, 1925–1933. [Google Scholar] [CrossRef]
- Dani, C.; Foissac, R.; Ladoux, A.; Chignon-Sicard, B. Autologous Fat Grafts: Can We Match the Donor Fat Site and the Host Environment for Better Postoperative Outcomes and Safety? Curr. Surg. Rep. 2017, 5, 14. [Google Scholar] [CrossRef]
- Yang, L.; Chen, L.; Lu, X.; Tan, A.; Chen, Y.; Li, Y.; Peng, X.; Yuan, S.; Cai, D.; Yu, Y. Peri-ovarian adipose tissue contributes to intraovarian control during folliculogenesis in mice. Reproduction 2018, 156, 133–144. [Google Scholar] [CrossRef]
- Foissac, R.; Villageois, P.; Chignon-Sicard, B.; Georgiou, C.; Camuzard, O.; Dani, C. Homeotic and Embryonic Gene Expression in Breast Adipose Tissue and in Adipose Tissues Used as Donor Sites in Plastic Surgery. Plast. Reconstr. Surg. 2017, 139, 685e–692e. [Google Scholar] [CrossRef]
- Kouidhi, M.; Villageois, P.; Mounier, C.M.; Ménigot, C.; Rival, Y.; Piwnica, D.; Aubert, J.; Chignon-Sicard, B.; Dani, C. Characterization of Human Knee and Chin Adipose-Derived Stromal Cells. Stem Cells Int. 2015, 2015, 592090. [Google Scholar] [CrossRef]
- Charbord, P.; Livne, E.; Gross, G.; Häupl, T.; Neves, N.M.; Marie, P.; Bianco, P.; Jorgensen, C. Human Bone Marrow Mesenchymal Stem Cells: A Systematic Reappraisal Via the Genostem Experience. Stem Cell Rev. Rep. 2011, 7, 32–42. [Google Scholar] [CrossRef]
- Satomura, K.; Krebsbach, P.; Bianco, P.; Gehron Robey, P. Osteogenic imprinting upstream of marrow stromal cell differentiation. J. Cell. Biochem. 2000, 78, 391–403. [Google Scholar] [CrossRef]
- Wang, Q.A.; Tao, C.; Gupta, R.K.; Scherer, P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 2013, 19, 1338–1344. [Google Scholar] [CrossRef]
- Pellegrinelli, V.; Carobbio, S.; Vidal-Puig, A. Adipose tissue plasticity: How fat depots respond differently to pathophysiological cues. Diabetologia 2016, 59, 1075–1088. [Google Scholar] [CrossRef]
- Joe, A.W.B.; Yi, L.; Even, Y.; Vogl, A.W.; Rossi, F.M.V. Depot-Specific Differences in Adipogenic Progenitor Abundance and Proliferative Response to High-Fat Diet. Stem Cells 2009, 27, 2563–2570. [Google Scholar] [CrossRef]
- Pan, Z.; Zhou, Z.; Zhang, H.; Zhao, H.; Song, P.; Wang, D.; Yin, J.; Zhao, W.; Xie, Z.; Wang, F.; et al. CD90 serves as differential modulator of subcutaneous and visceral adipose-derived stem cells by regulating AKT activation that influences adipose tissue and metabolic homeostasis. Stem Cell Res. Ther. 2019, 10, 355. [Google Scholar] [CrossRef]
- Arrigoni, E.; Lopa, S.; de Girolamo, L.; Stanco, D.; Brini, A.T. Isolation, characterization and osteogenic differentiation of adipose-derived stem cells: From small to large animal models. Cell Tissue Res. 2009, 338, 401–411. [Google Scholar] [CrossRef]
- Fathi, E.; Farahzadi, R. Enhancement of osteogenic differentiation of rat adipose tissue-derived mesenchymal stem cells by zinc sulphate under electromagnetic field via the PKA, ERK1/2 and Wnt/β-catenin signaling pathways. PLoS ONE 2017, 12, e0173877. [Google Scholar] [CrossRef]
- Tang, Y.; Pan, Z.-y.; Zou, Y.; He, Y.; Yang, P.-y.; Tang, Q.-q.; Yin, F. A comparative assessment of adipose-derived stem cells from subcutaneous and visceral fat as a potential cell source for knee osteoarthritis treatment. J. Cell. Mol. Med. 2017, 21, 2153–2162. [Google Scholar] [CrossRef]
- Habib, S.A.; Kamal, M.M.; El-Maraghy, S.A.; Senousy, M.A. Exendin-4 enhances osteogenic differentiation of adipose tissue mesenchymal stem cells through the receptor activator of nuclear factor-kappa B and osteoprotegerin signaling pathway. J. Cell. Biochem. 2022, 123, 906–920. [Google Scholar] [CrossRef]
- Ogawa, R.; Mizuno, H.; Watanabe, A.; Migita, M.; Shimada, T.; Hyakusoku, H. Osteogenic and chondrogenic differentiation by adipose-derived stem cells harvested from GFP transgenic mice. Biochem. Biophys. Res. Commun. 2004, 313, 871–877. [Google Scholar] [CrossRef]
- Baer, P.C.; Koch, B.; Hickmann, E.; Schubert, R.; Cinatl, J.; Hauser, I.A.; Geiger, H. Isolation, Characterization, Differentiation and Immunomodulatory Capacity of Mesenchymal Stromal/Stem Cells from Human Perirenal Adipose Tissue. Cells 2019, 8, 1346. [Google Scholar] [CrossRef]
- Cassis, L.A.; Police, S.B.; Yiannikouris, F.; Thatcher, S.E. Local adipose tissue renin-angiotensin system. Curr. Hypertens. Rep. 2008, 10, 93–98. [Google Scholar] [CrossRef]
- Hall, J.E.; do Carmo, J.M.; da Silva, A.A.; Wang, Z.; Hall, M.E. Obesity-Induced Hypertension: Interaction of neurohumoral and renal mechanisms. Circ. Res. 2015, 116, 991–1006. [Google Scholar] [CrossRef]
- Wei, K.; Yin, Z.; Xie, Y. Roles of the kidney in the formation, remodeling and repair of bone. J. Nephrol. 2016, 29, 349–357. [Google Scholar] [CrossRef]
- Huang, J.I.; Beanes, S.R.; Zhu, M.; Lorenz, P.H.; Hedrick, M.H.; Benhaim, P. Rat Extramedullary Adipose Tissue as a Source of Osteochondrogenic Progenitor Cells. Plast. Reconstr. Surg. 2002, 109, 1033–1041. [Google Scholar] [CrossRef]
- Zheng, B.; Cao, B.; Li, G.; Huard, J. Mouse Adipose-Derived Stem Cells Undergo Multilineage Differentiation in Vitro but Primarily Osteogenic and Chondrogenic Differentiation in Vivo. Tissue Eng. 2006, 12, 1891–1901. [Google Scholar] [CrossRef]
- Ni, X.; Shan, X.; Xu, L.; Yu, W.; Zhang, M.; Lei, C.; Xu, N.; Lin, J.; Wang, B. Adipose-derived stem cells combined with platelet-rich plasma enhance wound healing in a rat model of full-thickness skin defects. Stem Cell Res. Ther. 2021, 12, 226. [Google Scholar] [CrossRef]
- Tapp, H.; Deepe, R.; Ingram, J.A.; Kuremsky, M.; Hanley, E.N.; Gruber, H.E. Adipose-derived mesenchymal stem cells from the sand rat: Transforming growth factor beta and 3D co-culture with human disc cells stimulate proteoglycan and collagen type I rich extracellular matrix. Arthritis Res. Ther. 2008, 10, R89. [Google Scholar] [CrossRef]
- Khalilzadeh, M.; Panahi, G.; Rashidian, A.; Hadian, M.R.; Abdollahi, A.; Afshari, K.; Shakiba, S.; Norouzi-Javidan, A.; Rahimi, N.; Momeny, M.; et al. The protective effects of sumatriptan on vincristine—Induced peripheral neuropathy in a rat model. NeuroToxicology 2018, 67, 279–286. [Google Scholar] [CrossRef]
- Lotfy, A.; Salama, M.; Zahran, F.; Jones, E.; Badawy, A.; Sobh, M. Characterization of Mesenchymal Stem Cells Derived from Rat Bone Marrow and Adipose Tissue: A Comparative Study. Int. J. Stem Cells 2014, 7, 135–142. [Google Scholar] [CrossRef]
- Luo, Y.; Mohsin, A.; Xu, C.; Wang, Q.; Hang, H.; Zhuang, Y.; Chu, J.; Guo, M. Co-culture with TM4 cells enhances the proliferation and migration of rat adipose-derived mesenchymal stem cells with high stemness. Cytotechnology 2018, 70, 1409–1422. [Google Scholar] [CrossRef]
- Parvaneh, M.; Karimi, G.; Jamaluddin, R.; Ng, M.H.; Zuriati, I.; Muhammad, S.I. Lactobacillus helveticus (ATCC 27558) upregulates Runx2 and Bmp2 and modulates bone mineral density in ovariectomy-induced bone loss rats. Clin. Interv. Aging 2018, 13, 1555–1564. [Google Scholar] [CrossRef]
- Li, J.; Zhao, Z.; Liu, J.; Huang, N.; Long, D.; Wang, J.; Li, X.; Liu, Y. MEK/ERK and p38 MAPK regulate chondrogenesis of rat bone marrow mesenchymal stem cells through delicate interaction with TGF-β1/Smads pathway. Cell Prolif. 2010, 43, 333–343. [Google Scholar] [CrossRef]
- Zhang, M.; Zhou, Q.; Liang, Q.Q.; Li, C.G.; Holz, J.D.; Tang, D.; Sheu, T.J.; Li, T.F.; Shi, Q.; Wang, Y.J. IGF-1 regulation of type II collagen and MMP-13 expression in rat endplate chondrocytes via distinct signaling pathways. Osteoarthr. Cartil. 2009, 17, 100–106. [Google Scholar] [CrossRef]







| Antigen | Primary Antibody | Secondary Antibody | Isotype Control Antibody |
|---|---|---|---|
| CD 90 | FITC anti-rat CD90 (206105, BioLegend, San Diego, CA, USA) | ---- | FITC Mouse IgG1, κ (981802, BioLegend, San Diego, CA, USA) |
| CD 105 | Mouse anti-CD105 (OTI8A1) (ab156756, Abcam, Cambridge, UK) | Goat anti-mouse IgG H&L (Alexa Fluor® 488) (ab150113, Abcam, Cambridge, UK) | Purified Mouse IgG2b, κ (402202, BioLegend, San Diego, CA, USA) |
| CD 45 | FITC anti-rat CD45 (202205, BioLegend, San Diego, CA, USA) | ---- | FITC Mouse IgG1, κ (981802, BioLegend, San Diego, CA, USA) |
| CD 34 | Mouse anti CD34 (ICO115) (SC-7324, Santa Cruz Biotechnology, Inc., CA, USA) | Goat anti-mouse IgG H&L (Alexa Fluor® 488) (ab150113, Abcam, Cambridge, UK) | Purified Mouse IgG1, κ (401402, BioLegend, San Diego, CA, USA) |
| Name | Direction | Primer Sequence (5′-3′) | Accession Number |
|---|---|---|---|
| Oct 4 | F | CGAACCTGGCTAAGCTTCCA | NM_001009178.2 |
| R | GCCATCCCTCCACAGAACTC | ||
| Nanog | F | TACCTCAGCCTCCAGCAGAT | XM_006237310.3 |
| R | CATTGGTTTTTCTGCCACCT | ||
| Sox 2 | F | CTCGCAGACCTACATGAAC | NM_001109181.1 |
| R | TCGGACTTGACCACAGAG | ||
| Rex-1 | F | GCTCCGGCGGAATCGAGTGG | XM_032907726.1 |
| R | GCACGTGTTGCTTGGCGACC | ||
| Tert | F | CCCGAGTATGGCTGCATGAT | NM_053423.1 |
| R | AAAGTCCGAGTGTCCAGCAG | ||
| β-actin | F | GCAGGAGTACGATGAGTCCG | [67] |
| R | ACGCAG CTCAGTAACAGTCC |
| Purpose | Name | Direction | Primer Sequence (5′-3′) | Refs. |
|---|---|---|---|---|
| Adipogenic induction | ADIPOQ | F | TAATTCAGAGCAGCCCGTAG | [69] |
| R | TGGGGATAACACTCAGAACC | |||
| CFD | F | GGAGTGACCAAGGATGAGG | [69] | |
| R | ACCCAGTGAGGCATTGTG | |||
| Osteogenic induction | BMP2 | F | CAGGTCTTTGCACCAAGATG | [70] |
| R | GCTGGACTTAAGACGCTTCC | |||
| OPN | F | GAAGAGCCAGGAGTCCGATG | [30] | |
| R | CTTCCCGTTGCTGTCCTGAT | |||
| BSP | F | AGGCTACGAGGGTCAGGATT | [30] | |
| R | CTCTGCCTCCCGTGAAAC | |||
| Chondrogenic induction | ACAN | F | CTCTGCCTCCCGTGAAAC | [71] |
| R | TGAAGTGCCTGCATCTATGT | |||
| COL2A1 | F | TCCTAAGGGTGCCAATGGTGA | [72] | |
| R | AGGACCAACTTTGCCTTGAGGAC | |||
| CHM1 | F | GAGAACTGTGAGGGCTGTCA | [30] | |
| R | GATACCTCGGGCCAGAAGTG | |||
| Internal control | β-actin | F | GCAGGAGTACGATGAGTCCG | [67] |
| R | ACGCAGCTCAGTAACAGTCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Husseiny, H.M.; Kaneda, M.; Mady, E.A.; Yoshida, T.; Doghish, A.S.; Tanaka, R. Impact of Adipose Tissue Depot Harvesting Site on the Multilineage Induction Capacity of Male Rat Adipose-Derived Mesenchymal Stem Cells: An In Vitro Study. Int. J. Mol. Sci. 2023, 24, 7513. https://doi.org/10.3390/ijms24087513
El-Husseiny HM, Kaneda M, Mady EA, Yoshida T, Doghish AS, Tanaka R. Impact of Adipose Tissue Depot Harvesting Site on the Multilineage Induction Capacity of Male Rat Adipose-Derived Mesenchymal Stem Cells: An In Vitro Study. International Journal of Molecular Sciences. 2023; 24(8):7513. https://doi.org/10.3390/ijms24087513
Chicago/Turabian StyleEl-Husseiny, Hussein M., Masahiro Kaneda, Eman A. Mady, Tadashi Yoshida, Ahmed S. Doghish, and Ryou Tanaka. 2023. "Impact of Adipose Tissue Depot Harvesting Site on the Multilineage Induction Capacity of Male Rat Adipose-Derived Mesenchymal Stem Cells: An In Vitro Study" International Journal of Molecular Sciences 24, no. 8: 7513. https://doi.org/10.3390/ijms24087513
APA StyleEl-Husseiny, H. M., Kaneda, M., Mady, E. A., Yoshida, T., Doghish, A. S., & Tanaka, R. (2023). Impact of Adipose Tissue Depot Harvesting Site on the Multilineage Induction Capacity of Male Rat Adipose-Derived Mesenchymal Stem Cells: An In Vitro Study. International Journal of Molecular Sciences, 24(8), 7513. https://doi.org/10.3390/ijms24087513