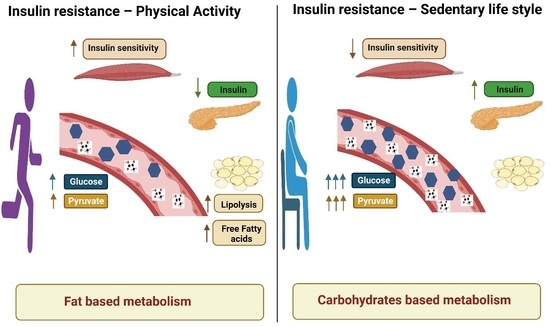
The Metabolic Switch of Physical Activity in Non-Obese Insulin Resistant Individuals
Abstract
:1. Introduction
2. Results
2.1. Comparing the Effect of Physical Activity on Clinical Traits between IS and IR Subjects
2.2. Multivariate Analysis of Metabolites Differentiating IS and IR in Sedentary and Active Non-Obese Subjects
2.3. Univariate Analysis of Metabolites Differentiating Sedentary Insulin Sensitive and Insulin Resistant Individuals
2.4. Univariate Analysis of Metabolites Differentiating Physically Active Insulin Sensitive and Insulin Resistant Individuals
2.5. Common Metabolites That Are Significantly Different between IS and IR Individuals in Active and Sedentary Groups
2.6. Spearman’s Correlation of Clinical Traits and Top Metabolite Hits from the Linear Regression Analysis in Active and Sedentary IR Individuals
3. Discussion
4. Materials and Methods
4.1. The Data Source and Study Participants
4.2. Metabolomics
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cornier, M.; Dabelea, D.; Hernandez, T.; Lindstrom, R.; Steig, A.; Stob, N.; Van Pelt, R.; Wang, H.; Eckel, R. The metabolic syndrome. Endocr. Rev. 2008, 29, 777–822. [Google Scholar] [CrossRef]
- Kramer, C.; Zinman, B.; Retnakaran, R. Are metabolically healthy overweight and obesity benign conditions? A systematic review and meta-analysis. Ann. Intern. Med. 2013, 159, 758–769. [Google Scholar] [CrossRef]
- Dvorak, R.V.; DeNino, W.F.; Ades, P.A.; Poehlman, E.T. Phenotypic characteristics associated with insulin resistance in metabolically obese but normal-weight young women. Diabetes 1999, 48, 2210–2214. [Google Scholar] [CrossRef]
- Ruderman, N.; Chisholm, D.; Pi-Sunyer, X.; Schneider, S. The metabolically obese, normal-weight individual revisited. Diabetes 1998, 47, 699–713. [Google Scholar] [CrossRef]
- McLaughlin, T.; Allison, G.; Abbasi, F.; Lamendola, C.; Reaven, G. Prevalence of insulin resistance and associated cardiovascular disease risk factors among normal weight, overweight, and obese individuals. Metabolism 2004, 53, 495–499. [Google Scholar] [CrossRef]
- St-Onge, M.; Janssen, I.; Heymsfield, S. Metabolic syndrome in normal-weight Americans: New definition of the metabolically obese, normal-weight individual. Diabetes Care 2004, 27, 2222–2228. [Google Scholar] [CrossRef]
- Owei, I.; Umekwe, N.; Provo, C.; Wan, J.; Dagogo-Jack, S. Insulin-sensitive and insulin-resistant obese and non-obese phenotypes: Role in prediction of incident pre-diabetes in a longitudinal biracial cohort. BMJ Open Diabetes Res. Care 2017, 5, e000415. [Google Scholar] [CrossRef] [PubMed]
- DeFronzo, R.A.; Ferrannini, E. Insulin Resistance: A Multifaceted Syndrome Responsible for NIDDM, Obesity, Hypertension, Dyslipidemia, and Atherosclerotic Cardiovascular Disease. Diabetes Care 1991, 14, 173–194. [Google Scholar] [CrossRef] [PubMed]
- Kahn, S.E.; Hull, R.L.; Utzschneider, K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006, 444, 840–846. [Google Scholar] [CrossRef]
- Grundy, S.; Brewer, H., Jr.; Cleeman, J.; Smith, S., Jr.; Lenfant, C.; Heart, A.A.; Heart, L.N.; Blood, I. Definition of metabolic syndrome: Report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Circulation 2004, 109, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Al Dubayee, M.; Alshahrani, A.; Masood, A.; Benabdelkamel, H.; Zahra, M.; Li, L.; Rahman, A.M.A.; Aljada, A. Distinctive Metabolomics Patterns Associated With Insulin Resistance and Type 2 Diabetes Mellitus. Front. Mol. Biosci. 2020, 7, 609806. [Google Scholar] [CrossRef]
- Ahola-Olli, A.V.; Mustelin, L.; Kalimeri, M.; Kettunen, J.; Jokelainen, J.; Auvinen, J.; Puukka, K.; Havulinna, A.S.; Lehtimäki, T.; Kähönen, M.; et al. Circulating metabolites and the risk of type 2 diabetes: A prospective study of 11,896 young adults from four Finnish cohorts. Diabetologia 2019, 62, 2298–2309. [Google Scholar] [CrossRef]
- Anton, S.; Moehl, K.; Donahoo, W.; Marosi, K.; Lee, S.; Mainous, A., III; Leeuwenburgh, C.; Mattson, M. Flipping the Metabolic Switch: Understanding and Applying the Health Benefits of Fasting. Obesity 2018, 26, 254–268. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.; Moehl, K.; Ghena, N.; Schmaedick, M.; Cheng, A. Intermittent metabolic switching, neuroplasticity and brain health. Nat. Rev. Neurosci. 2018, 19, 63–80. [Google Scholar] [CrossRef]
- Luc, K.; Schramm-Luc, A.; Guzik, T.J.; Mikolajczyk, T.P. Oxidative stress and inflammatory markers in prediabetes and diabetes. J. Physiol. Pharmacol. 2019, 70. [Google Scholar] [CrossRef]
- Horton, E.S. Effects of Lifestyle Changes to Reduce Risks of Diabetes and Associated Cardiovascular Risks: Results from Large Scale Efficacy Trials. Obesity 2009, 17 (Suppl. S3), S43–S48. [Google Scholar] [CrossRef]
- Stanford, K.I.; Goodyear, L.J. Exercise and type 2 diabetes: Molecular mechanisms regulating glucose uptake in skeletal muscle. Adv. Physiol. Educ. 2014, 38, 308–314. [Google Scholar] [CrossRef]
- Tuomilehto, J.; Lindström, J.; Eriksson, J.G.; Valle, T.T.; Hämäläinen, H.; Ilanne-Parikka, P.; Keinänen-Kiukaanniemi, S.; Laakso, M.; Louheranta, A.; Rastas, M.; et al. Prevention of Type 2 Diabetes Mellitus by Changes in Lifestyle among Subjects with Impaired Glucose Tolerance. N. Engl. J. Med. 2001, 344, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Blonde, L. Current Antihyperglycemic Treatment Guidelines and Algorithms for Patients with Type 2 Diabetes Mellitus. Am. J. Med. 2010, 123, S12–S18. [Google Scholar] [CrossRef]
- James, D.; Kraegen, E.W.; Chisholm, D.J. Effects of exercise training on in vivo insulin action in individual tissues of the rat. J. Clin. Investig. 1985, 76, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Kido, K.; Sase, K.; Yokokawa, T.; Fujita, S. Enhanced skeletal muscle insulin sensitivity after acute resistance-type exercise is upregulated by rapamycin-sensitive mTOR complex 1 inhibition. Sci. Rep. 2020, 10, 8509. [Google Scholar] [CrossRef]
- Eriksson, J.; Taimela, S.; Koivisto, V. Exercise and the metabolic syndrome. Diabetologia 1997, 40, 125–135. [Google Scholar]
- AlMuraikhy, S.; Anwardeen, N.; Naeem, A.; Sellami, M.; Domling, A.; Agouni, A.; Elrayess, M.A. Comparing the Metabolic Profiles Associated with Fitness Status between Insulin-Sensitive and Insulin-Resistant Non-Obese Individuals. Int. J. Environ. Res. Public Health 2022, 19, 12169. [Google Scholar] [CrossRef]
- Stefanov, T.; Temelkova-Kurktschiev, T.; Koehler, C.; Henkel, E.; Schaper, F.; Hanefeld, M. Association of physical activity with insulin resistance, subclinical inflammation, coagulation, and fibrinolytic biomarkers among population at high risk for type 2 diabetes. Folia Med. 2012, 54, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Brozek, J. Changes of body composition in man during maturity and their nutritional implications. Fed. Proc. 1952, 11, 784–793. [Google Scholar] [PubMed]
- Goodpaster, B.H.; Sparks, L.M. Metabolic Flexibility in Health and Disease. Cell Metab. 2017, 25, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Meigs, J.B.; D’Agostino, R.B., Sr.; Wilson, P.W.; Cupples, L.A.; Nathan, D.M.; Singer, D.E. Risk variable clustering in the insulin resistance syndrome. The Framingham Offspring Study. Diabetes 1997, 46, 1594–1600. [Google Scholar] [CrossRef]
- Beck, E.; Esser, N.; Paquot, N.; Scheen, A.J. Metabolically obese normal-weight individuals and metabolically healthy, but obese, subjects. Rev. Med. Suisse 2009, 5, 1644–1646, 1648–1649. [Google Scholar]
- Oliveros, E.; Somers, V.K.; Sochor, O.; Goel, K.; Lopez-Jimenez, F. The Concept of Normal Weight Obesity. Prog. Cardiovasc. Dis. 2013, 56, 426–433. [Google Scholar] [CrossRef]
- Hernandes, V.V.; Dordevic, N.; Hantikainen, E.M.; Sigurdsson, B.B.; Smárason, S.V.; Garcia-Larsen, V.; Gögele, M.; Caprioli, G.; Bozzolan, I.; Pramstaller, P.P.; et al. Age, Sex, Body Mass Index, Diet and Menopause Related Metabolites in a Large Homogeneous Alpine Cohort. Metabolites 2022, 12, 205. [Google Scholar] [CrossRef]
- Al-Khelaifi, F.; Diboun, I.; Donati, F.; Botrè, F.; Alsayrafi, M.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; Elrayess, M.A. A pilot study comparing the metabolic profiles of elite-level athletes from different sporting disciplines. Sport. Med.—Open 2018, 4, 2. [Google Scholar] [CrossRef]
- Halama, A.; Kulinski, M.; Kader, S.A.; Satheesh, N.J.; Abou-Samra, A.B.; Suhre, K.; Mohammad, R.M. Measurement of 1,5-anhydroglucitol in blood and saliva: From non-targeted metabolomics to biochemical assay. J. Transl. Med. 2016, 14, 140. [Google Scholar] [CrossRef] [PubMed]
- Wang-Sattler, R.; Yu, Z.; Herder, C.; Messias, A.; Floegel, A.; He, Y.; Heim, K.; Campillos, M.; Holzapfel, C.; Thorand, B.; et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol. 2012, 8, 615. [Google Scholar] [CrossRef]
- Al-Sulaiti, H.; Diboun, I.; Agha, M.V.; Mohamed, F.F.S.; Atkin, S.; Dömling, A.S.; Elrayess, M.A.; Mazloum, N.A. Metabolic signature of obesity-associated insulin resistance and type 2 diabetes. J. Transl. Med. 2019, 17, 348. [Google Scholar] [CrossRef] [PubMed]
- Arneth, B.; Arneth, R.; Shams, M. Metabolomics of Type 1 and Type 2 Diabetes. Int. J. Mol. Sci. 2019, 20, 2467. [Google Scholar] [CrossRef] [PubMed]
- Gray, L.R.; Tompkins, S.C.; Taylor, E.B. Regulation of pyruvate metabolism and human disease. Cell. Mol. Life Sci. 2014, 71, 2577–2604. [Google Scholar] [CrossRef]
- Nakai, N.; Sato, Y.; Oshida, Y.; Fujitsuka, N.; Yoshimura, A.; Shimomura, Y. Insulin activation of pyruvate dehydrogenase complex is enhanced by exercise training. Metabolism 1999, 48, 865–869. [Google Scholar] [CrossRef] [PubMed]
- Ranallo, R.F.; Rhodes, E.C.; Buchanan, J.M. Lipid Metabolism During Exercise. Sport. Med. 1998, 26, 29–42. [Google Scholar] [CrossRef]
- Purdom, T.; Kravitz, L.; Dokladny, K.; Mermier, C. Understanding the factors that effect maximal fat oxidation. J. Int. Soc. Sport. Nutr. 2018, 15, 3. [Google Scholar] [CrossRef]
- Smith, R.L.; Soeters, M.R.; Wüst, R.C.I.; Houtkooper, R.H. Metabolic Flexibility as an Adaptation to Energy Resources and Requirements in Health and Disease. Endocr. Rev. 2018, 39, 489–517. [Google Scholar] [CrossRef] [PubMed]
- Iwayama, K.; Ogawa, A.; Tanaka, Y.; Yajima, K.; Park, I.; Ando, A.; Ogata, H.; Kayaba, M.; Zhang, S.; Tanji, F.; et al. Effects of exercise before breakfast on plasma free fatty acid profile and 24-h fat oxidation. Metab. Open 2020, 8, 100067. [Google Scholar] [CrossRef]
- Diboun, I.; Al-Mansoori, L.; Al-Jaber, H.; Albagha, O.; Elrayess, M.A. Metabolomics of Lean/Overweight Insulin-Resistant Females Reveals Alterations in Steroids and Fatty Acids. J. Clin. Endocrinol. Metab. 2020, 106, e638–e649. [Google Scholar] [CrossRef] [PubMed]
- Güttler, F.; Kühl, C.; Pedersen, L.; Påby, P. Effects of oral phenylalanine load on plasma glucagon, insulin, amino acid and glucose concentrations in man. Scand. J. Clin. Lab. Investig. 1978, 38, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, F.; Schweim, K.; Gannon, M. Effect of orally administered phenylalanine with and without glucose on insulin, glucagon and glucose concentrations. Horm. Metab. Res. 2006, 38, 518–523. [Google Scholar] [CrossRef]
- Zhou, Q.; Sun, W.; Chen, J.; Zhang, H.; Liu, J.; Lin, Y.; Lin, P.; Wu, B.; An, Y.; Huang, L.; et al. Phenylalanine impairs insulin signaling and inhibits glucose uptake through modification of IRbeta. Nat. Commun. 2022, 13, 4291. [Google Scholar] [CrossRef] [PubMed]
- Zaghlool, S.B.; Halama, A.; Stephan, N.; Gudmundsdottir, V.; Gudnason, V.; Jennings, L.L.; Thangam, M.; Ahlqvist, E.; Malik, R.A.; Albagha, O.M.E.; et al. Metabolic and proteomic signatures of type 2 diabetes subtypes in an Arab population. Nat. Commun. 2022, 13, 7121. [Google Scholar] [CrossRef]
- Yu, D.; Richardson, N.; Green, C.; Spicer, A.; Murphy, M.; Flores, V.; Jang, C.; Kasza, I.; Nikodemova, M.; Wakai, M.; et al. The adverse metabolic effects of branched-chain amino acids are mediated by isoleucine and valine. Cell Metab. 2021, 33, 905–922 e6. [Google Scholar] [CrossRef]
- Lei, Y.; Yang, J.; Li, H.; Zhong, H.; Wan, Q. Changes in glucose-lipid metabolism, insulin resistance, and inflammatory factors in patients with autoimmune thyroid disease. J. Clin. Lab. Anal. 2019, 33, e22929. [Google Scholar] [CrossRef]
- Evans, A.; Bridgewater, B.; Liu, Q.; Mitchell, M.; Robinson, R.; Dai, H.; Stewart, S.; DeHaven, C.; Miller, L. High Resolution Mass Spectrometry Improves Data Quantity and Quality as Compared to Unit Mass Resolution Mass Spectrometry in High-Throughput Profiling Metabolomics. Metabolomics 2014, 4, 1000132. [Google Scholar]







| Physically Active | Sedentary | |||||
|---|---|---|---|---|---|---|
| ISA (78) | IRA (50) | p | ISS (87) | IRS (90) | p | |
| Gender | ||||||
| Male | 44 (56.41%) | 26 (52%) | 0.624 | 33 (37.93%) | 56 (62.23%) | 0.001 |
| Female | 34 (43.58%) | 24 (48%) | 54 (62.06%) | 34 (37.78%) | ||
| Vital Signs | ||||||
| Age (years) | 25.7 (2.7) | 26.0 (2.9) | 0.562 | 25.8 (3.0) | 25.8 (3.0) | 0.881 |
| BMI (kg/m2) | 24.4 (22.9–26.9) | 26.1 (23.4–27.8) | 0.174 | 23.7 (22.2–26.0) | 24.9 (23.4–27.5) | 1.3 × 10−3 |
| Average systolic BP (mmHg) | 106.0 (101.0–113.7) | 108.5 (101.0–116.0) | 0.355 | 102.0 (96.0–111.0) | 110.0 (101.0–115.7) | 2.3 × 10−3 |
| Average diastolic BP (mmHg) | 67.8 (7.4) | 67.4 (7.6) | 0.957 | 67.5 (8.1) | 69.74 (7.30) | 0.043 |
| Pulse rate (beats/min) | 65.0 (59.0–72.0) | 69.0 (63.0–76.0) | 0.062 | 68.0 (62.0–72.0) | 70.0 (65.0–78.0) | 0.048 |
| Blood sugar | ||||||
| HOMA-IR | 1.2 (1.0–1.5) | 2.9 (2.3–5.0) | 1.7 × 10−21 | 1.3 (1.0–1.5) | 3.2 (2.2–5.2) | 3.2 × 10−30 |
| C Peptide (ng/mL) | 1.4 (1.2–1.7) | 2.6 (2.1–4.0) | 9.6 × 10−16 | 1.5 (1.3–1.7) | 2.9 (2.1–4.2) | 4.8 × 10−18 |
| Insulin (uu/mL) | 6.0 (4.9–7.0) | 13.2 (10.2–21.9) | 1.0 × 10−20 | 6.0 (4.8–7.0) | 14.5 (10.0–23.1) | 6.9 × 10−27 |
| Hemoglobin A1c (HbA1c) % | 5.2 (5.0–5.3) | 5.3 (5.0–5.4) | 0.182 | 5.2 (5.0–5.4) | 5.3 (5.1–5.5) | 0.124 |
| Glucose (mmol/L) | 4.8 (4.5–5.0) | 5.1 (4.8–5.4) | 1.1 × 10−4 | 4.7 (4.5–4.9) | 5.2 (4.8–5.4) | 6.2 × 10−11 |
| Physical tests | ||||||
| Sitting height (cm) | 90.7 (85.2–134.3) | 90.8 (85.5–134.1) | 0.778 | 91.7 (86.4–134.8) | 91.4 (87.2–133.6) | 0.783 |
| Weight (kg) | 69.2 (61.3–76.2) | 73.4 (63.2–79.8) | 0.134 | 65.4 (58.6–72.2) | 70.4 (63.9–78.7) | 2.1 × 10−3 |
| Waist size (cm) | 78.5 (73.0–84.0) | 82.5 (75.0–89.0) | 0.044 | 75.0 (71.0–80.0) | 82.0 (75.0–89.0) | 4.0 × 10−6 |
| Hip size (cm) | 100.2 ± 6.8 | 102.3 ± 6.1 | 0.072 | 100.5 ± 6.8 | 101.1 ± 5.8 | 0.463 |
| Waist to hip ratio | 0.79 ± 0.07 | 0.80 ± 0.08 | 0.305 | 0.76 ± 0.08 | 0.81 ± 0.08 | 2.2 × 10−5 |
| Handgrip (left) | 33.0 (22.0–44.0) | 33.0 (28.0–43.0) | 0.925 | 26.0 (20.0–38.0) | 31.5 (22.0–40.0) | 0.046 |
| Handgrip (right) | 35.0 (26.0–48.0) | 32.0 (26.0–45.5) | 0.739 | 29.0 (22.0–39.5) | 34.0 (23.2–42.0) | 0.114 |
| Maximum heart rate (beats/min) | 120.0 (106.5–133.0) | 122.0 (111.5–132.0) | 0.485 | 129.0 (112.0–142.0) | 126.5 (114.2–139.0) | 0.893 |
| Run time (seconds) | 764.0 (742.0–764.0) | 764.0 (742.0–764.0) | 0.981 | 764.0 (684.0–764.0) | 764.0 (742.0–764.0) | 0.12 |
| Kidney profile | ||||||
| Sodium (mmol/L) | 140.5 (139.0–141.8) | 141.0 (139.0–142.0) | 0.739 | 140.0 (138.5–141.0) | 140.0 (139.0–142.0) | 0.133 |
| Potassium (mmol/L) | 4.3 (4.1–4.5) | 4.3 (4.1–4.5) | 0.787 | 4.3 (4.1–4.5) | 4.2 (4.0–4.4) | 0.037 |
| Chloride (mmol/L) | 101.0 (100.0–102.0) | 102.0 (99.2–102.7) | 0.398 | 101.0 (100.0–102.0) | 101.0 (100.0–102.0) | 0.794 |
| Bicarbonate (mmol/L) | 27.0 (25.0–28.0) | 26.0 (25.2–28.0) | 0.41 | 26.0 (25.0–27.0) | 26.0 (25.0–27.0) | 0.826 |
| Urea (mmol/L) | 4.3 (3.4–5.3) | 4.1 (3.4–5.1) | 0.606 | 4.2 (3.3–4.9) | 4.2 (3.4–4.8) | 0.822 |
| Creatinine (mmol/L) | 69.0 (52.7–78.7) | 67.0 (54.7–80.0) | 0.743 | 61.0 (54.0–76.5) | 70.5 (58.2–77.0) | 0.023 |
| Calcium (mmol/L) | 2.4 ± 0.1 | 2.4 ± 0.1 | 0.571 | 2.4 ± 0.1 | 2.4 ± 0.1 | 0.264 |
| Calcium Corrected (mmol/L) | 2.3 ± 0.1 | 2.3 ± 0.1 | 0.382 | 2.3 ± 0.1 | 2.3 ± 0.1 | 0.146 |
| Phosphorus (mmol/L) | 1.2 ± 0.1 | 1.2 ± 0.2 | 0.752 | 1.2 ± 0.1 | 1.1 ± 0.2 | 1.1 × 10−3 |
| Uric Acid (umol/L) | 296.5 (240.2–335.5) | 282.0 (234.5–361.7) | 0.625 | 257.0 (221.5–305.0) | 308.5 (242.0–349.7) | 6.1 × 10−4 |
| Creatine kinase (U/L) | 86.5 (69.2–130.0) | 99.0 (60.0–159.5) | 0.746 | 67.0 (55.0–105.0) | 89.0 (62.0–117.0) | 0.037 |
| Creatine Kinase 1 (ng/mL) | 1.1 (0.9–1.5) | 1.1 (0.7–1.5) | 0.771 | 1.0 (0.7–2.0) | 1.31 (1.0–1.6) | 1 |
| Creatine kinase 2 (U/L) | 88.0 (72.0–124.0) | 71.0 (52.2–239.0) | 0.758 | 86.5 (60.0–127.0) | 75 (47.5–109) | 0.478 |
| Magnesium(µmol/L) | 0.8 ± 0.05 | 0.8 ± 0.05 | 0.887 | 0.8 ± 0.05 | 0.84 ± 0.05 | 0.621 |
| Total Protein (g/L) | 74.0 (72.0–76.0) | 75.0 (72.0–77.0) | 0.737 | 73.0 (70.0–76.0) | 74.0 (72.0–76.0) | 0.049 |
| Homocysteine (µmol/L) | 8.7 (6.8–10.6) | 8.0 (6.6–9.5) | 0.142 | 8.3 (6.2–9.9) | 8.0 (6.6–9.5) | 0.952 |
| Liver profile | ||||||
| Total Bilirubin (µmol/L) | 7.0 (5.2–10.0) | 5.9 (4.6–7.85) | 0.045 | 7.6 (6.0–9.0) | 6.0 (4.3–9.0) | 0.031 |
| Albumin (g/L) | 47.0 (45.0–48.0) | 47.0 (45.0–48.0) | 0.63 | 46.0 (45.0–48.0) | 46.0 (44.2–49.0) | 0.992 |
| Alkaline phosphatase (U/L) | 61.0 (48.5–72.0) | 64.5 (57.2–79.7) | 0.054 | 60.0 (52.5–70.5) | 68.5 (60.0–77.7) | 1.1 × 10−3 |
| Alanine Transaminase (U/L) | 17.0 (12.0–22.7) | 19.0 (13.0–31.5) | 0.292 | 14.0 (10.0–19.5) | 21.5 (14.0–32.0) | 8.5 × 10−6 |
| Aspartate aminotransferase (U/L) | 19.0 (16.0–21.7) | 18.0 (15.0–23.7) | 0.835 | 17.0 (14.0–19.0) | 18.5 (15.0–21.0) | 8.3 × 10−3 |
| GGT (U/L) | 13.0 (10.5–18.5) | 13.0 (10.5–17.5) | 0.788 | 13.0 (8.2–17.0) | 16.0 (13.0–23.0) | 0.037 |
| GGT 2 (U/L) | 14.0 (10.0–19.0) | 17.5 (12–25.5) | 0.069 | 14.0 (12.0–25.0) | 23.0 (16.0–35.0) | 2.7 × 10−4 |
| Lipid profile | ||||||
| HDL Cholesterol (mmol/L) | 1.5 ± 0.4 | 1.4 ± 0.4 | 0.356 | 1.5 ± 0.3 | 1.2 ± 0.3 | 1.5 × 10−5 |
| LDL Cholesterol Calc (mmol/L) | 2.8 (2.1–3.0) | 2.9 (2.1–3.3) | 0.413 | 2.6 (2.1–3.0) | 2.9 (2.5–3.2) | 0.082 |
| Triglyceride (mmol/L) | 0.8 (0.6–1.0) | 1.0 (0.7–1.4) | 0.041 | 0.8 (0.5–1.1) | 1.1 (0.8–1.6) | 3.7 × 10−6 |
| Iron profile | ||||||
| Iron (µmol/L) | 15.5 (11.5–20.0) | 14.0 (10.5–17.0) | 0.143 | 16.0 (10.5–20.0) | 15.15 (11.21–19.1) | 0.971 |
| TIBC (µmol/L) | 56.0 (52.0–63.0) | 60.0 (56.0–67.0) | 0.004 | 60.5 (54.2–66.5) | 59.0 (52.0–64.0) | 0.443 |
| UIBC (µmol/L) | 41.0 (34.0–48.0) | 46.0 (39.0–55.0) | 0.014 | 43.0 (36.0–53.6) | 42.0 (34.5–49.8) | 0.583 |
| Ferritin (µg/L) | 54.0 (22.0–110.5) | 34.0 (17.0–105.5) | 0.332 | 34.0 (10.0–70.0) | 63.5 (20.5–119.0) | 0.014 |
| Vitamins | ||||||
| Folate (nmol/L) | 22.0 ± 7.4 | 22.6 ± 8.2 | 0.673 | 24.0 ± 8.9 | 21.6 ± 7.0 | 0.157 |
| Vitamin B12 (pmol/L) | 288.0 (232.2–418.0) | 271.5 (208.0–356.2) | 0.194 | 283.0 (222.0–374.0) | 295.0 (229.0–377.0) | 0.935 |
| Dihydroxyvitamin D Total (ng/mL) | 15.0 (11.2–18.0) | 13.5 (11.0–24.0) | 0.46 | 16.5 (12.0–24.7) | 13.0 (10.2–17.7) | 4.2 × 103 |
| Hormones | ||||||
| Total Testosterone (nmol/L) | 15.9 (1.4–25.4) | 4.14 (1.2–17.6) | 0.047 | 1.85 (1.1–20.4) | 14.2 (1.4–19.0) | 0.383 |
| Estradiol (pmol/L) | 124.0 (88.0–237.0) | 122.0 (84.0–234.0) | 0.499 | 146 (86.7–349.7) | 105 (77.2–169.7) | 0.022 |
| SHBG (nmol/L) | 44.3 (30.8–64.7) | 37.1 (23.5–60.0) | 0.1 | 52.8 (36.7–79.2) | 31.5 (22.5–46.8) | 4.1 × 10−6 |
| Free Thyroxine (pmol/L) | 13.7 (12.5–14.5) | 13.3 (12.5–14.1) | 0.469 | 13.3 (12.4–14.6) | 13.4 (12.6–14.6) | 0.63 |
| Free triiodothyronine (nmol/L) | 4.6 (4.1–4.9) | 4.4 (4.0–4.85) | 0.701 | 4.4 (4.14–4.8) | 4.7 (4.3–5.1) | 0.007 |
| TSH (mU/L) | 1.3 (1.0–2.0) | 1.3 (1.0–1.6) | 0.241 | 1.6 (1.1–2.5) | 1.3 (0.9–2.2) | 0.079 |
| Blood inflammatory markers | ||||||
| Hemoglobin (g/dL) | 13.7 (12.6–15.1) | 13.7 (12.5–14.9) | 0.655 | 13.3 (11.9–14.56) | 14.4 (13.1–15.3) | 8.6 × 10−4 |
| Estimated hemoglobin * | 18.5 ± 3.3 | 19.1 ± 3.3 | 0.527 | 17.7 ± 3.8 | 18.3 ± 3.9 | 0.236 |
| Hematocrit % | 42.1 (37.6–45.0) | 41.9 (38.5–44.9) | 0.843 | 40.0 (36.1–42.78) | 42.7 (39.4–45.7) | 4.2 × 10−4 |
| Red Blood Cell count (×106/µL) | 5 (4.4–5.3) | 5.1 (4.6–5.5) | 0.095 | 4.7 (4.5–5.1) | 5.1 (4.7–5.6) | 3.5 × 10−5 |
| White Blood Cell count (×103/µL) | 6.5 (5.4–7.5) | 6.5 (5.5–8.1) | 0.696 | 6.4 (5.4–7.6) | 6.6 (5.1–7.8) | 0.984 |
| Monocyte % | 7 (5.9–8.2) | 7.3 (6.2–8.6) | 0.506 | 6.8 (5.6–8.2) | 8.0 (6.5–9.3) | 2.8 x 10−3 |
| Monocyte count (×103/µL) | 0.5 (0.4–0.6) | 0.5 (0.4–0.6) | 0.473 | 0.4 (0.3–0.5) | 0.5 (0.4–0.6) | 8.7 × 10−4 |
| Absolute neutrophil count (×103/µL) | 3.3 (2.7–4.3) | 3.6 (2.8–4.5) | 0.535 | 3.4 (2.7–4.7) | 3.5 (2.6–4.4) | 0.901 |
| Neutrophil % | 53.2 ± 9.6 | 55.0 ± 10.3 | 0.325 | 54.2 ± 9.0 | 54.6 ± 9.6 | 0.847 |
| Lymphocyte count (×103/µL) | 2.3 (1.9–2.8) | 2.1 (1.9–2.5) | 0.386 | 2.2 (1.8–2.6) | 2.2 (1.7–2.6) | 0.477 |
| Lymphocyte % | 36.5 ± 8.7 | 34.1 ± 8.7 | 0.134 | 35.6 ± 7.6 | 34.4 ± 7.8 | 0.291 |
| Eosinophil % | 2.3 (1.3–3.8) | 2.3 (1.5–3.475) | 0.966 | 1.8 (1.2–3.3) | 2.5 (1.6–3.5) | 0.049 |
| Eosinophils count (×103/µL) | 0.1 (0.1–0.2) | 0.1 (0.1–0.3) | 0.911 | 0.1 (0.1–0.2) | 0.1 (0.1–0.2) | 0.327 |
| Basophil % | 0.6 (0.3–0.7) | 0.6 (0.4–0.7) | 0.601 | 0.7 (0.5–0.9) | 0.6 (0.4–0.8) | 0.231 |
| Basophils count (×103/µL) | 0.0 (0.0–0.1) | 0.0 (0.0–0.03) | 0.864 | 0.0 (0.0–0.1) | 0.0 (0.0–0.1) | 0.589 |
| Platelet count (×103/µL) | 233 (207.0–263.0) | 235 (208.8–275.8) | 0.674 | 237 (195.0–284.0) | 244 (209.0–276.5) | 0.805 |
| MCH (pg) | 28.6 (26.6–29.9) | 27.5 (25.7–28.8) | 0.013 | 28.6 (26.1–29.8) | 28.2 (26.6–29.6) | 0.743 |
| MCHC (g/dL) | 33.4 (32.7–33.9) | 33 (32.4–33.6) | 0.184 | 33.3 (32.5–34.0) | 33.6 (32.8–34.2) | 0.185 |
| C Reactive Protein (mg/L) | 5 (5–5) | 5 (5–5) | 0.659 | 5 (5–5) | 5 (5–5) | 0.023 |
| Metabolite | Super-Pathway | Sub-Pathway | Estimate | SE | p-Value | FDR |
|---|---|---|---|---|---|---|
| Pyruvate | Carbohydrate | Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | −0.182 | 0.033 | 7.22 × 10−8 | 6.11 × 10−5 |
| 1-carboxyethylphenylalanine | Amino Acid | Phenylalanine Metabolism | −0.372 | 0.079 | 3.76 × 10−6 | 0.001 |
| Gamma-glutamylcitrulline | Peptide | Gamma-glutamyl Amino Acid | 0.214 | 0.051 | 3.59 × 10−5 | 0.010 |
| N-acetylglycine | Amino Acid | Glycine, Serine and Threonine Metabolism | 0.268 | 0.068 | 1.01 × 10−4 | 0.021 |
| Glucose | Carbohydrate | Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | −0.103 | 0.028 | 2.31 × 10−4 | 0.039 |
| Metabolite | Super-Pathway | Sub-Pathway | Estimate | SE | p-Value | FDR |
|---|---|---|---|---|---|---|
| Myristoleate (14:1n5) | Lipid | Long Chain Monounsaturated Fatty Acid | 0.684 | 0.138 | 1.38 × 10−6 | 0.001 |
| Branched-chain, straight-chain, or cyclopropyl 12:1 fatty acid | Partially Characterized Molecules | Partially Characterized Molecules | 0.597 | 0.136 | 1.59 × 10−5 | 0.006 |
| Dodecadienoate (12:2) | Lipid | Fatty Acid, Dicarboxylate | 0.415 | 0.098 | 2.67 × 10−5 | 0.006 |
| 1-carboxyethylphenylalanine | Amino Acid | Phenylalanine Metabolism | −0.395 | 0.093 | 2.90 × 10−5 | 0.006 |
| Palmitoleate (16:1n7) | Lipid | Long Chain Monounsaturated Fatty Acid | 0.584 | 0.141 | 4.58 × 10−5 | 0.007 |
| Isoleucine | Amino Acid | Leucine, Isoleucine and Valine Metabolism | −0.119 | 0.029 | 5.59 × 10−5 | 0.007 |
| Tetradecadienoate (14:2) | Lipid | Long Chain Polyunsaturated Fatty Acid (n3 and n6) | 0.499 | 0.123 | 6.40 × 10−5 | 0.007 |
| Myristate (14:0) | Lipid | Long Chain Saturated Fatty Acid | 0.407 | 0.102 | 8.04 × 10−5 | 0.008 |
| 7-alpha-hydroxy-3-oxo-4-cholestenoate (7-Hoca) | Lipid | Sterol | 0.194 | 0.05 | 1.57 × 10−4 | 0.015 |
| 5-dodecenoate (12:1n7) | Lipid | Medium Chain Fatty Acid | 0.42 | 0.112 | 2.16 × 10−4 | 0.017 |
| 10-heptadecenoate (17:1n7) | Lipid | Long Chain Monounsaturated Fatty Acid | 0.439 | 0.117 | 2.24 × 10−4 | 0.017 |
| Hexadecadienoate (16:2n6) | Lipid | Long Chain Polyunsaturated Fatty Acid (n3 and n6) | 0.43 | 0.12 | 4.21 × 10−4 | 0.029 |
| Prolylglycine | Peptide | Dipeptide | −0.375 | 0.106 | 4.87 × 10−4 | 0.031 |
| Stearidonate (18:4n3) | Lipid | Long Chain Polyunsaturated Fatty Acid (n3 and n6) | 0.448 | 0.13 | 6.55 × 10−4 | 0.036 |
| 16-hydroxypalmitate | Lipid | Fatty Acid, Monohydroxy | 0.227 | 0.066 | 6.61 × 10−4 | 0.036 |
| Gamma-glutamylmethionine | Peptide | Gamma-glutamyl Amino Acid | −0.195 | 0.057 | 8.04 × 10−4 | 0.041 |
| Oleate/vaccenate (18:1) | Lipid | Long Chain Monounsaturated Fatty Acid | 0.369 | 0.109 | 8.29 × 10−4 | 0.041 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almuraikhy, S.; Anwardeen, N.; Doudin, A.; Sellami, M.; Domling, A.; Agouni, A.; Al Thani, A.A.; Elrayess, M.A. The Metabolic Switch of Physical Activity in Non-Obese Insulin Resistant Individuals. Int. J. Mol. Sci. 2023, 24, 7816. https://doi.org/10.3390/ijms24097816
Almuraikhy S, Anwardeen N, Doudin A, Sellami M, Domling A, Agouni A, Al Thani AA, Elrayess MA. The Metabolic Switch of Physical Activity in Non-Obese Insulin Resistant Individuals. International Journal of Molecular Sciences. 2023; 24(9):7816. https://doi.org/10.3390/ijms24097816
Chicago/Turabian StyleAlmuraikhy, Shamma, Najeha Anwardeen, Asmma Doudin, Maha Sellami, Alexander Domling, Abdelali Agouni, Asmaa A. Al Thani, and Mohamed A. Elrayess. 2023. "The Metabolic Switch of Physical Activity in Non-Obese Insulin Resistant Individuals" International Journal of Molecular Sciences 24, no. 9: 7816. https://doi.org/10.3390/ijms24097816
APA StyleAlmuraikhy, S., Anwardeen, N., Doudin, A., Sellami, M., Domling, A., Agouni, A., Al Thani, A. A., & Elrayess, M. A. (2023). The Metabolic Switch of Physical Activity in Non-Obese Insulin Resistant Individuals. International Journal of Molecular Sciences, 24(9), 7816. https://doi.org/10.3390/ijms24097816