DNA Mimics for the Rapid Identification of Microorganisms by Fluorescence in situ Hybridization (FISH)
Abstract
:1. FISH for Microbial Detection
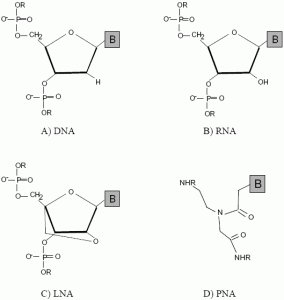
2. Emergence of DNA Mimics
2.1. Peptide nucleic acids (PNA)
2.2. Locked nucleic acids (LNA)
2.3. Other mimics
3. Conclusions and Future Work
Acknowledgments
References and Notes
- Kricka, LJ. Stains, labels and detection strategies for nucleic acids assays. Ann. Clin. Biochem. 2002, 39, 114–129. [Google Scholar]
- Amann, R; Fuchs, BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol 2008, 6, 339–348. [Google Scholar]
- Michalet, X; Pinaud, FF; Bentolila, LA; Tsay, JM; Doose, S; Li, JJ; Sundaresan, G; Wu, AM; Gambhir, SS; Weiss, S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science 2005, 307, 538–544. [Google Scholar]
- Berlier, JE; Rothe, A; Buller, G; Bradford, J; Gray, DR; Filanoski, BJ; Telford, WG; Yue, S; Liu, J; Cheung, CY; Chang, W; Hirsch, JD; Beechem, JM; Haugland, RP. Quantitative comparison of long-wavelength Alexa Fluor dyes to Cy dyes: Fluorescence of the dyes and their bioconjugates. J. Histochem. Cytochem. 2003, 51, 1699–1712. [Google Scholar]
- Barken, KB; Haagensen, JA; Tolker-Nielsen, T. Advances in nucleic acid-based diagnostics of bacterial infections. Clin. Chim. Acta 2007, 384, 1–11. [Google Scholar]
- Guimaraes, N; Azevedo, NF; Figueiredo, C; Keevil, CW; Vieira, MJ. Development and application of a novel peptide nucleic acid probe for the specific detection of Helicobacter pylori in gastric biopsy specimens. J. Clin. Microbiol 2007, 45, 3089–3094. [Google Scholar]
- Bayani, J; Squire, JA. Application and interpretation of FISH in biomarker studies. Cancer Lett 2007, 249, 97–109. [Google Scholar]
- Arentsen, HC; de la Rosette, JJ; de Reijke, TM; Langbein, S. Fluorescence in situ hybridization: A multitarget approach in diagnosis and management of urothelial cancer. Expert. Rev. Mol. Diagn. 2007, 7, 11–19. [Google Scholar]
- Sanz, JL; Kochling, T. Molecular biology techniques used in wastewater treatment: An overview. Process Biochem 2007, 42, 119–133. [Google Scholar]
- Rogers, SW; Moorman, TB; Ong, SK. Fluorescent in situ hybridization and micro-autoradiography applied to ecophysiology in soil. Soil Sci. Soc. Amer. J. 2007, 71, 620–631. [Google Scholar]
- Dmochowski, IJ; Tang, XJ. Taking control of gene expression with light-activated oligonucleotides. Biotechniques 2007, 43, 161–171. [Google Scholar]
- Miyauchi, R; Oki, K; Aoi, Y; Tsuneda, S. Diversity of nitrite reductase genes in “Candidatus Accumulibacter phosphatis”-dominated cultures enriched by flow-cytometric sorting. Appl. Environ. Microbiol. 2007, 73, 5331–5337. [Google Scholar]
- Catalina, P; Cobo, F; Cortes, JL; Nieto, AI; Cabrera, C; Montes, R; Concha, A; Menendez, P. Conventional and molecular cytogenetic diagnostic methods in stem cell research: A concise review. Cell Biol. Int. 2007, 31, 861–869. [Google Scholar]
- Wagner, M; Horn, M; Daims, H. Fluorescence in situ hybridisation for the identification and characterisation of prokaryotes. Curr. Opin. Microbiol. 2003, 6, 302–309. [Google Scholar]
- Silverman, AP; Kool, ET. Oligonucleotide probes for RNA-targeted fluorescence in situ hybridization. Adv. Clin. Chem 2007, 43, 79–115. [Google Scholar]
- Stender, H; Fiandaca, M; Hyldig-Nielsen, JJ; Coull, J. PNA for rapid microbiology. J. Microbiol. Methods 2002, 48, 1–17. [Google Scholar]
- Delong, EF; Wickham, GS; Pace, NR. Phylogenetic Stains - Ribosomal RNA-Based Probes for the Identification of Single Cells. Science 1989, 243, 1360–1363. [Google Scholar]
- Pernthaler, A; Pernthaler, J; Amann, R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 2002, 68, 3094–3101. [Google Scholar]
- Wagner, M; Schmid, M; Juretschko, S; Trebesius, KH; Bubert, A; Goebel, W; Schleifer, KH. In situ detection of a virulence factor mRNA and 16S rRNA in Listeria monocytogenes. FEMS Microbiol. Lett 1998, 160, 159–168. [Google Scholar]
- Yilmaz, LS; Okten, HE; Noguera, DR. Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides. Appl. Environ. Microbiol. 2006, 72, 733–744. [Google Scholar]
- Yazbeck, DR; Min, KL; Damha, MJ. Molecular requirements for degradation of a modified sense RNA strand by Escherichia coli ribonuclease H1. Nucleic Acids Res 2002, 30, 3015–3025. [Google Scholar]
- Cummins, LL; Owens, SR; Risen, LM; Lesnik, EA; Freier, SM; Mcgee, D; Guinosso, CJ; Cook, PD. Characterization of fully 2’-modified oligoribonucleotide heteroduplex and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995, 23, 2019–2024. [Google Scholar]
- Shakeel, S; Karim, S; Ali, A. Peptide nucleic acid (PNA) - a review. J. Chem. Technol. Biotechnol. 2006, 81, 892–899. [Google Scholar]
- Yilmaz, LS; Noguera, DR. Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization. Appl. Environ. Microbiol. 2004, 70, 7126–7139. [Google Scholar]
- Nielsen, PE; Egholm, M; Berg, RH; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar]
- Petersen, M; Wengel, J. LNA: A versatile tool for therapeutics and genomics. Trends Biotechnol 2003, 21, 74–81. [Google Scholar]
- Hanvey, JC; Peffer, NJ; Bisi, JE; Thomson, SA; Cadilla, R; Josey, JA; Ricca, DJ; Hassman, CF; Bonham, MA; Au, KG; Carter, SG; Bruckenstein, DA; Boyd, AL; Noble, SA; Babiss, LE. Antisense and Antigene Properties of Peptide Nucleic-Acids. Science 1992, 258, 1481–1485. [Google Scholar]
- Wahlestedt, C; Salmi, P; Good, L; Kela, J; Johnsson, T; Hokfelt, T; Broberger, C; Porreca, F; Lai, J; Ren, KK; Ossipov, M; Koshkin, A; Jakobsen, N; Skouv, J; Oerum, H; Jacobsen, MH; Wengel, J. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. USA 2000, 97, 5633–5638. [Google Scholar]
- Forrest, GN; Mehta, S; Weekes, E; Lincalis, DP; Johnson, JK; Venezia, RA. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J. Antimicrob. Chemother 2006, 58, 154–158. [Google Scholar]
- Forrest, GN; Mankes, K; Jabra-Rizk, MA; Weekes, E; Johnson, JK; Lincalis, DP; Venezia, RA. Peptide nucleic acid fluorescence in situ hybridization-based identification of Candida albicans and its impact on mortality and antifungal therapy costs. J. Clin. Microbiol. 2006, 44, 3381–3383. [Google Scholar]
- Shepard, JR; Addison, RM; Alexander, BD; Della-Latta, P; Gherna, M; Haase, G; Hall, G; Johnson, JK; Merz, WG; Peltroche-Llacsahuanga, H; Stender, H; Venezia, RA; Wilson, D; Procop, GW; Wu, F; Fiandaca, MJ. Multicenter evaluation of the Candida albicans/Candida glabrata peptide nucleic acid fluorescent in situ hybridization method for simultaneous dual-color identification of C. albicans and C. glabrata directly from blood culture bottles. J. Clin. Microbiol 2008, 46, 50–55. [Google Scholar]
- Koshkin, AA; Nielsen, P; Meldgaard, M; Rajwanshi, VK; Singh, SK; Wengel, J. LNA (locked nucleic acid): An RNA mimic forming exceedingly stable LNA : LNA duplexes. J. Am. Chem. Soc. 1998, 120, 13252–13253. [Google Scholar]
- Shirude, PS; Kumar, VA; Ganesh, KN. (2S,5R/2R,5S)-aminoethylpipecolyl aepip-aegPNA chimera: synthesis and duplex/triplex stability. Tetrahedron 2004, 60, 9485–9491. [Google Scholar]
- Corradini, R; Sforza, S; Tedeschi, T; Totsingan, F; Marchelli, R. Peptide nucleic acids with a structurally biased backbone: Effects of conformational constraints and stereochemistry. Curr. Top. Med. Chem. 2007, 7, 681–694. [Google Scholar]
- Jensen, J; Sjogren, G; Hansen, JB; Rosenbohm, C; Koch, T. Synthesis and biological evaluation of LNA phosphoramidates. Nucleos. Nucleot. Nucleic Acids 2008, 27, 37–42. [Google Scholar]
- Sharma, PK; Kumar, S; Nielsen, P. Synthesis of a branched locked nucleic acid (LNA) analogue. Nucleos. Nucleot. Nucleic Acids 2007, 26, 1505–1508. [Google Scholar]
- Kubota, K; Ohashi, A; Imachi, H; Harada, H. Improved in situ hybridization efficiency with locked-nucleic-acid-incorporated DNA probes. Appl. Environ. Microbiol 2006, 72, 5311–5317. [Google Scholar]
- Stender, H; Mollerup, TA; Lund, K; Petersen, KH; Hongmanee, P; Godtfredsen, SE. Direct detection and identification of Mycobacterium tuberculosis in smear-positive sputum samples by fluorescence in situ hybridization (FISH) using peptide nucleic acid (PNA) probes. Int. J. Tuberc. Lung Dis 1999, 3, 830–837. [Google Scholar]
- Stender, H; Broomer, A; Oliveira, K; Perry-O’Keefe, H; Hyldig-Nielsen, JJ; Sage, A; Young, B; Coull, J. Rapid detection, identification, and enumeration of Pseudomonas aeruginosa in bottled water using peptide nucleic acid probes. J. Microbiol. Methods 2000, 42, 245–253. [Google Scholar]
- Drobniewski, FA; More, PG; Harris, GS. Differentiation of Mycobacterium tuberculosis complex and nontuberculous mycobacterial liquid cultures by using peptide nucleic acid-fluorescence in situ hybridization probes. J. Clin. Microbiol 2000, 38, 444–447. [Google Scholar]
- Prescott, AM; Fricker, CR. Use of PNA oligonucleotides for the in situ detection of Escherichia coli in water. Mol. Cell. Probes 1999, 13, 261–268. [Google Scholar]
- Nielsen, PE. Peptide nucleic acid: a versatile tool in genetic diagnostics and molecular biology. Curr. Opin. Biotechnol. 2001, 12, 16–20. [Google Scholar]
- Perry-O’Keefe, H; Rigby, S; Oliveira, K; Sorensen, D; Stender, H; Coull, J; Hyldig-Nielsen, JJ. Identification of indicator microorganisms using a standardized PNA FISH method. J. Microbiol. Methods 2001, 47, 281–292. [Google Scholar]
- Lomakin, A; Frank-Kamenetskii, MD. A theoretical analysis of specificity of nucleic acid interactions with oligonucleotides and peptide nucleic acids (PNAs). J. Mol. Biol. 1998, 276, 57–70. [Google Scholar]
- Orum, H; Nielsen, PE; Jorgensen, M; Larsson, C; Stanley, C; Koch, T. Sequence-Specific Purification of Nucleic-Acids by PNA-Controlled Hybrid Selection. Biotechniques 1995, 19, 472–480. [Google Scholar]
- Fuchs, BM; Syutsubo, K; Ludwig, W; Amann, R. In situ accessibility of Escherichia coli 23S rRNA to fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol 2001, 67, 961–968. [Google Scholar]
- Fuchs, BM; Wallner, G; Beisker, W; Schwippl, I; Ludwig, W; Amann, R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol 1998, 64, 4973–4982. [Google Scholar]
- Demidov, VV; Potaman, VN; Frank-Kamenetskii, MD; Egholm, M; Buchard, O; Sonnichsen, SH; Nielsen, PE. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 1994, 48, 1310–1313. [Google Scholar]
- Cole, JR; Chai, B; Farris, RJ; Wang, Q; Kulam, SA; McGarrell, DM; Garrity, GM; Tiedje, JM. The Ribosomal Database Project (RDP-II): Sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 2005, 33, D294–D296. [Google Scholar]
- McGinnis, S; Madden, TL. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 2004, 32, W20–W25. [Google Scholar]
- Azevedo, NF; Vieira, MJ; Keevil, CW. Establishment of a continuous model system to study Helicobacter pylori survival in potable water biofilms. Water Sci. Technol 2003, 47, 155–160. [Google Scholar]
- Vester, B; Wengel, J. LNA (Locked nucleic acid): High-affinity targeting of complementary RNA and DNA. Biochemistry 2004, 43, 13233–13241. [Google Scholar]
- Demidov, VV. PNA and LNA throw light on DNA. Trends Biotechnol 2003, 21, 4–7. [Google Scholar]
- Kauppinen, S; Vester, B; Wengel, J. Locked nucleic acid: high-affinity targeting of complementary RNA for RNomics. Handb. Exp. Pharmacol. 2006, 173, 405–422. [Google Scholar]
- Darfeuille, F; Hansen, JB; Orum, H; Primo, CD; Toulme, JJ. LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1. Nucleic Acids Res 2004, 32, 3101–3107. [Google Scholar]
- Kloosterman, WP; Wienholds, E; de Bruijn, E; Kauppinen, S; Plasterk, RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods 2006, 3, 27–29. [Google Scholar]
- Silahtaroglu, AN; Tommerup, N; Vissing, H. FISHing with locked nucleic acids (LNA): evaluation of different LNA/DNA mixmers. Mol. Cell. Probes 2003, 17, 165–169. [Google Scholar]
- Thomsen, R; Nielsen, PS; Jensen, TH. Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA 2005, 11, 1745–1748. [Google Scholar]
- Pitts, AE; Corey, DR. Inhibition of human telomerase by 2 ‘-O-methyl-RNA. Proc. Natl. Acad. Sci. USA 1998, 95, 11549–11554. [Google Scholar]
- Majlessi, M; Nelson, NC; Becker, MM. Advantages of 2 ‘-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998, 26, 2224–2229. [Google Scholar]
- Tsourkas, A; Behlke, MA; Bao, G. Hybridization of 2 ‘-O-methyl and 2 ‘-deoxy molecular beacons to RNA and DNA targets. Nucleic Acids Res. 2002, 30, 5168–5174. [Google Scholar]
- Lamond, AI; Sproat, BS. Antisense oligonucleotides made of 2’-o-alkylrna - their properties and applications in rna biochemistry. FEBS Lett 1993, 325, 123–127. [Google Scholar]
- Sproat, BS. Chemistry and Applications of Oligonucleotide Analogs. J. Biotechnol. 1995, 41, 221–238. [Google Scholar]
- Molenaar, C; Marras, SA; Slats, JCM; Truffert, JC; Lemaitre, M; Raap, AK; Dirks, RW; Tanke, HJ. Linear 2’ O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001, 29, E89–E99. [Google Scholar]
- Monia, BP; Lesnik, EA; Gonzalez, C; Lima, WF; Mcgee, D; Guinosso, CJ; Kawasaki, AM; Cook, PD; Freier, SM. Evaluation of 2’-modified oligonucleotides containing 2’-deoxy gaps as antisense inhibitors of gene-expression. J. Biol. Chem. 1993, 268, 14514–14522. [Google Scholar]
- Kawasaki, AM; Casper, MD; Freier, SM; Lesnik, EA; Zounes, MC; Cummins, LL; Gonzalez, C; Cook, PD. Uniformly modified 2’-deoxy-2’-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high-affinity and specificity for rna targets. J. Med. Chem. 1993, 36, 831–841. [Google Scholar]
- Summerton, J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta 1999, 1489, 141–158. [Google Scholar]
- Ge, Q; Pastey, M; Kobasa, D; Puthavathana, P; Lupfer, C; Bestwick, RK; Iversen, PL; Chen, J; Stein, DA. Inhibition of multiple subtypes of influenza A virus in cell cultures with morpholino oligomers. Antimicrob. Agents Chemother. 2006, 50, 3724–3733. [Google Scholar]
- Deas, TS; Bennett, CJ; Jones, SA; Tilgner, M; Ren, P; Behr, MJ; Stein, DA; Iversen, PL; Kramer, LD; Bernard, KA; Shi, PY. In vitro resistance selection and in vivo efficacy of morpholino oligomers against West Nile virus. Antimicrob. Agents Chemother. 2007, 51, 2470–2482. [Google Scholar]
- Tilley, LD; Hine, OS; Kellogg, JA; Hassinger, JN; Weller, DD; Iversen, PL; Geller, BL. Gene-specific effects of antisense phosphorodiamidate morpholino oligomer-peptide conjugates on Escherichia coli and Salmonella enterica serovar typhimurium in pure culture and in tissue culture. Antimicrob. Agents Chemother 2006, 50, 2789–2796. [Google Scholar]
- Klee, EW; Shim, KJ; Pickart, MA; Ekker, SC; Ellis, LB. AMOD: A morpholino oligonucleotide selection tool. Nucleic Acids Res 2005, 33, W506–W511. [Google Scholar]
- Efimov, VA; Birikh, KR; Staroverov, DB; Lukyanov, SA; Tereshina, MB; Zaraisky, AG; Chakhmakhcheva, OG. Hydroxyproline-based DNA mimics provide an efficient gene silencing in vitro and in vivo. Nucleic Acids Res. 2006, 34, 2247–2257. [Google Scholar]
- Efimov, VA; Buryakova, AA; Chakhmakhcheva, OG. Synthesis of polyacrylamides N-substituted with PNA-like oligonucleotide mimics for molecular diagnostic applications. Nucleic Acids Res. 1999, 27, 4416–4426. [Google Scholar]
- Efimov, VA; Choob, MV; Buryakova, AA; Kalinkina, AL; Chakhmakhcheva, OG. Synthesis and evaluation of some properties of chimeric oligomers containing PNA and phosphono-PNA residues. Nucleic Acids Res 1998, 26, 566–575. [Google Scholar]
- Fluiter, K; Frieden, M; Vreijling, J; Rosenbohm, C; De Wissel, MB; Christensen, SM; Koch, T; Orum, H; Baas, F. On the in vitro and in vivo properties of four locked nucleic acid nucleotides incorporated into an anti-H-Ras antisense oligonucleotide. Chembiochem 2005, 6, 1104–1109. [Google Scholar]
- Rosenbohm, C; Christensen, SM; Sorensen, MD; Pedersen, DS; Larsen, LE; Wengel, J; Koch, T. Synthesis of 2’-amino-LNA: a new strategy. Org. Biomol. Chem. 2003, 1, 655–663. [Google Scholar]
- Sorensen, MD; Kvaerno, L; Bryld, T; Hakansson, AE; Verbeure, B; Gaubert, G; Herdewijn, P; Wengel, J. Alpha-L-ribo-configured locked nucleic acid (alpha-L-LNA): Synthesis and properties. J. Am. Chem. Soc. 2002, 124, 2164–2176. [Google Scholar]
- Kumar, R; Singh, SK; Koshkin, AA; Rajwanshi, VK; Meldgaard, M; Wengel, J. The first analogues of LNA (locked nucleic acids): phosphorothioate-LNA and 2’-thio-LNA. Bioorg. Med. Chem. Lett. 1998, 8, 2219–2222. [Google Scholar]
- Hakansson, AE; Koshkin, AA; Sorensen, MD; Wengel, J. Convenient syntheses of 7-hydroxy-1-(hydroxymethyl)-3-(thymin-1-yl)-2,5-dioxabicyclo. J. Org. Chem 2000, 65, 5161–5166. [Google Scholar]
- SantaLucia, J; Hicks, D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 415–440. [Google Scholar]
- SantaLucia, J; Allawi, HT; Seneviratne, A. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochem 1996, 35, 3555–3562. [Google Scholar]
- Giesen, U; Kleider, W; Berding, C; Geiger, A; Orum, H; Nielsen, PE. A formula for thermal stability (Tm) prediction of PNA/DNA duplexes. Nucleic Acids Res 1998, 26, 5004–5006. [Google Scholar]
- Silverman, AP; Kool, ET. Quenched autoligation probes allow discrimination of live bacterial species by single nucleotide differences in rRNA. Nucleic Acids Res 2005, 33, 4978–4986. [Google Scholar]
- Zwirglmaier, K; Ludwig, W; Schleifer, KH. Recognition of individual genes in a single bacterial cell by fluorescence in situ hybridization - RING-FISH. Mol. Microbiol 2004, 51, 89–96. [Google Scholar]
- Smolina, I; Lee, C; Frank-Kamenetskii, M. Detection of low-copy-number genomic DNA sequences in individual bacterial cells by using peptide nucleic acid-assisted rolling-circle amplification and fluorescence in situ hybridization. Appl. Environ. Microbiol 2007, 73, 2324–2328. [Google Scholar]
- Rigby, S; Procop, GW; Haase, G; Wilson, D; Hall, G; Kurtzman, C; Oliveira, K; Von Oy, S; Hyldig-Nielsen, JJ; Coull, J; Stender, H. Fluorescence in situ hybridization with peptide nucleic acid probes for rapid identification of Candida albicans directly from blood culture bottles. J. Clin. Microbiol. 2002, 40, 2182–2186. [Google Scholar]
- Oliveira, K; Haase, G; Kurtzman, C; Hyldig-Nielsen, JJ; Stender, H. Differentiation ofCandida albicans and Candida dubliniensis by fluorescent in situ hybridization with peptide nucleic acid probes. J. Clin. Microbiol. 2001, 39, 4138–4141. [Google Scholar]
- Stender, H; Kurtzman, C; Hyldig-Nielsen, JJ; Sorensen, D; Broomer, A; Oliveira, K; Perry-O’ Keefe, H; Sage, A; Young, B; Coull, J. Identification of Dekkera bruxellensis (Brettanomyces) from wine by fluorescence in situ hybridization using peptide nucleic acid probes. Appl. Environ. Microbiol. 2001, 67, 938–941. [Google Scholar]
- Perry-O’ Keefe, H; Stender, H; Broomer, A; Oliveira, K; Coull, J; Hyldig-Nielsen, JJ. Filter-based PNA in situ hybridization for rapid detection, identification and enumeration of specific microorganisms. J. Appl. Microbiol 2001, 90, 180–189. [Google Scholar]
- Radwanska, M; Magez, S; Perry-O’Keefe, H; Stender, H; Coull, J; Sternberg, JM; Buscher, P; Hyldig-Nielsen, JJ. Direct detection and identification of African trypanosomes by fluorescence in situ hybridization with peptide nucleic acid probes. J. Clin. Microbiol 2002, 40, 4295–4297. [Google Scholar]
- Lehtola, MJ; Loades, CJ; Keevil, CW. Advantages of peptide nucleic acid oligonucleotides for sensitive site directed 16S rRNA fluorescence in situ hybridization (FISH) detection of Campylobacter jejuni, Campylobacter coli and Campylobacter lari. J. Microbiol. Methods 2005, 62, 211–219. [Google Scholar]
- Juhna, T; Birzniece, D; Larsson, S; Zulenkovs, D; Sharipo, A; Azevedo, NF; Menard-Szczebara, F; Castagnet, S; Feliers, C; Keevil, CW. Detection of Escherichia coli in biofilms from pipe samples and coupons in drinking water distribution networks. Appl. Environ. Microbiol. 2007, 73, 7456–7464. [Google Scholar]
- Bragança, SM; Azevedo, NF; Simoes, LC; Keevil, CW; Vieira, MJ. Use of fluorescent in situ hybridisation for the visualisation of Helicobacter pylori in real drinking water biofilms. Water Sci. Technol. 2007, 55, 387–393. [Google Scholar]
- Sogaard, M; Hansen, DS; Fiandaca, MJ; Stender, H; Schonheyder, HC. Peptide nucleic acid fluorescence in situ hybridization for rapid detection of Klebsiella pneumoniae from positive blood cultures. J. Med. Microbiol 2007, 56, 914–917. [Google Scholar]
- Wilks, SA; Keevil, CW. Targeting species-specific low-affinity 16S rRNA binding sites by using peptide nucleic acids for detection of legionellae in biofilms. Appl. Environ. Microbiol 2006, 72, 5453–5462. [Google Scholar]
- Brehm-Stecher, BF; Hyldig-Nielsen, JJ; Johnson, EA. Design and evaluation of 16S rRNA-targeted peptide nucleic acid probes for whole-cell detection of members of the genus Listeria. Appl. Environ. Microbiol. 2005, 71, 5451–5457. [Google Scholar]
- Lefmann, M; Schweickert, B; Buchholz, P; Gobel, UB; Ulrichs, T; Seiler, P; Theegarten, D; Moter, A. Evaluation of peptide nucleic acid-fluorescence in situ hybridization for identification of clinically relevant mycobacteria in clinical specimens and tissue sections. J. Clin. Microbiol. 2006, 44, 3760–3767. [Google Scholar]
- Lehtola, MJ; Torvinen, E; Miettinen, LT; Keevil, CW. Fluorescence in situ hybridization using peptide nucleic acid probes for rapid detection of Mycobacterium avium subsp avium and Mycobacterium avium subsp paratuberculosis in potable-water biofilms. Appl. Environ. Microbiol 2006, 72, 848–853. [Google Scholar]

| Microorganism | Sequence (5'–3') | Target | GC Content (%) | Hybridization temperature (°C)/solvent concentration | Specificity (%) | Sensitivity (%) | Ref. |
|---|---|---|---|---|---|---|---|
| Eucarya | |||||||
| C. albicans | AGAGAGCAGCATGCA | 26S | 53 | 55°C / 30% DMF | 96 | 46 | [86] |
| C. albicans | ACAGCAGAAGCCGTG | 26S | 60 | 50°C / 30% DMF | 91 | 70 | [87] |
| C. dubliniensis | TAGCCAGAAGAAAGG | 18S | 47 | 50°C / 30% DMF | 100 | 4 | [87] |
| D. bruxellensis | CGGTCTCCAGCGATT | 26S | 60 | 50°C / 50% DMF or 50°C / 30% DMF | 100 | 85 | [88] |
| Eucarya | ACCAGACTTGCCCTC | 18S | 60 | 55°C/ 0.5% (w/v) SDS 50°C/50% DMF | N.D. | N.D. | [43, 89] |
| S. cerevisae | TTACCGAGGCAAGCT | 18S | 53 | 50°C / 50% DMF | N.D. | N.D. | [43] |
| T. subgenus A | CGGAACCCAGCCA | 18S | 69 | 45°C / 30% DMF | N.D. | N.D. | [90] |
| T. subgenus A | GTTGCCACCAGCAGT | 18S | 60 | 45°C / 30% DMF | N.D. | N.D. | [90] |
| T. genus B | GCCCTAACAGGTGTG | 18S | 60 | 55°C / 30% DMF | N.D. | N.D. | [90] |
| Z. bailii | CGAGCGAAACGCCTG | 18S | 67 | 50°C / 50% DMF | 5 | 50 | [89] |
| Bacteria | |||||||
| C. coli, C. jejuni and C. lari | CCCTACTCAACTTGT | 16S | 47 | 50°C / 30% DMF | 100 | 91 | [91] |
| E. coli | TCAATGAGCAAAGGT | 16S | 40 | 55°C/ 0.5%(w/v) SDS or 50°C/50% DMF or 57°C / 30% DMF | 59 | 10 | [43, 89, 91, 92] |
| E. coli | GCAAAGCAGCAAGCTC | 16S | 56 | 50°C/ 0.01% SDS | 100 | 1 | [41] |
| Eubacteria | CTGCCTCCCGTAGGA | 16S | 67 | 55°C/ 0.5% (w/v) SDS or 50°C/50% DMF | N.D. | 93 | [89] |
| H. pylori | GAGACTAAGCCCTCC | 16S | 60 | 59°C / 30% DMF | 96 | 90 | [6] |
| H. pylori | TAATCAGCACTCTAGCAA | 16S | 39 | 55°C / 30% DMF | 100 | 24 | [51, 93] |
| K. pneumoniae | CACCTACACACCAGC | 23S | 60 | 55°C | 100 | 92 | [94] |
| L. brevis | CTCTAAGATTGGCAG | 16S | 47 | 50°C / 50% DMF | 81 | 97 | [89] |
| Legionella genus | GACGCAGGCTAATCT | 16S | 53 | 55°C to 65°C/ 30% DMF | 88 | 68 | [95] |
| L. pneumophila | CTGACCGTCCCAGGT | 16S | 67 | 55°C to 65°C / 30% DMF | 92 | 100 | [95] |
| Listeria genus | CCCCAACTTACAGGC | 16S | 60 | 55°C /0.5% SDS | 98 | 91 | [96] |
| Listeria genus | AAGGGACAAGCAGT | 16S | 50 | 55°C /0.5% SDS | 97 | 97 | [96] |
| M. avium | ATGCGTCTTGAGGTC | 16S | 53 | 55°C / 40% DMF | 95 | 91 | [97] |
| M. avium subsp. avium and M. avium subsp. paratuberculosis | TGCGTCTTGAGGTCC | 16S | 60 | 59°C / 30% DMF | 100 | 89 | [98] |
| M. kansasii | TATCCCGGTGTGCAG | 16S | 60 | 55°C / 40% DMF | 57 | 100 | [97] |
| M. leprae | CGCCTTGAAGTCCTA | 16S | 53 | 55°C / 40% DMF | 100 | 100 | [97] |
| M. tuberculosis complex (MTC) species | GCATCCCGTGGTCCT | 16S | 67 | 60°C / 50% DMF | 76 | 100 | [97] |
| M. tuberculosis complex (MTC) species | GGTTTTAAGGATTC | 16S | 40 | 55°C / 30% DMF | 62 | 100 | [38] |
| Nontuberculous (NTM) mycobacteria species | GCATTACCCGCTGGC | 16S | 67 | 55°C / 30% DMF | 34 | 34 | [38] |
| P. aeruginosa | CTGAATCCAGGAGCA | 16S | 53 | 55°C/ 0.5% (w/v) SDS or 50°C/50% DMF | 80 | 87 | [43, 89] |
| Salmonella | TAAGCCGGGATGGC | 23S | 64 | 55°C/ 0.5% (w/v) SDS or 50°C/50% DMF | 41 | 60 | [43, 89] |
| S. aureus | GCTTCTCGTCCGTTC | 16S | 60 | 55°C/ 0.5%(w/v) SDS or 50°C/50% DMF | 100 | 92 | [43, 89] |
© 2008 by MDPI This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Cerqueira, L.; Azevedo, N.F.; Almeida, C.; Jardim, T.; Keevil, C.W.; Vieira, M.J. DNA Mimics for the Rapid Identification of Microorganisms by Fluorescence in situ Hybridization (FISH). Int. J. Mol. Sci. 2008, 9, 1944-1960. https://doi.org/10.3390/ijms9101944
Cerqueira L, Azevedo NF, Almeida C, Jardim T, Keevil CW, Vieira MJ. DNA Mimics for the Rapid Identification of Microorganisms by Fluorescence in situ Hybridization (FISH). International Journal of Molecular Sciences. 2008; 9(10):1944-1960. https://doi.org/10.3390/ijms9101944
Chicago/Turabian StyleCerqueira, Laura, Nuno F. Azevedo, Carina Almeida, Tatiana Jardim, Charles William Keevil, and Maria J. Vieira. 2008. "DNA Mimics for the Rapid Identification of Microorganisms by Fluorescence in situ Hybridization (FISH)" International Journal of Molecular Sciences 9, no. 10: 1944-1960. https://doi.org/10.3390/ijms9101944
APA StyleCerqueira, L., Azevedo, N. F., Almeida, C., Jardim, T., Keevil, C. W., & Vieira, M. J. (2008). DNA Mimics for the Rapid Identification of Microorganisms by Fluorescence in situ Hybridization (FISH). International Journal of Molecular Sciences, 9(10), 1944-1960. https://doi.org/10.3390/ijms9101944