Dielectrophoretic Separation of Live and Dead Monocytes Using 3D Carbon-Electrodes
Abstract
:1. Introduction
2. Theory
3. Materials and Methods
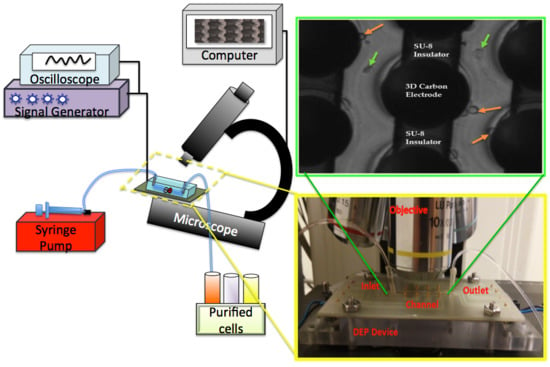
3.1. 3D Carbon-DEP Device Fabrication
3.2. Low Conductive DEP Buffer Preparation
3.3. Cell Preparation
3.4. Experimental Setup
3.5. Experimental Procedure
3.6. Downstream Analysis
3.7. Image Acquisition and Analysis
3.8. Statistical Analysis
4. Results
4.1. Dielectrophoretic Characterization of the Live and Dead U937 Monocytes
4.2. Dielectrophoretic Separation of the Live and Dead U937 Monocytes
5. Discussion
6. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
References
- Masuda, T.; Hayashi, N.; Iguchi, T.; Ito, S.; Eguchi, H.; Mimori, K. Clinical and biological significance of circulating tumor cells in cancer. Mol. Oncol. 2016, 10, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, D.T.; Ting, D.T.; Toner, M.; Maheswaran, S.; Haber, D.A. Single-Cell Analysis of Circulating Tumor Cells as a Window into Tumor Heterogeneity. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Alt, J.R.; Hunsley, B.A.; Williams, T.L.; Fernando, M.R. Stabilization of circulating tumor cells in blood using a collection device with a preservative reagent. Cancer Cell Int. 2014, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Fritsche, H.; Müller, V.; Rau, T.; Schindlbeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Jänicke, F.; et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the Cell Search system. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Gascoyne, P.R.C.; Wang, X.B.; Huang, Y.; Becker, F.F. Dielectrophoretic separation of cancer cells from blood. IEEE Trans. Ind. Appl. 1997, 33, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Sueblinvong, T.; Ghebre, R.; Iizuka, Y.; Pambuccian, S.E.; Vogel, R.I.; Skubitz, A.P.N.; Bazzaro, M. Establishment, characterization and downstream application of primary ovarian cancer cells derived from solid tumors. PLoS ONE 2012, 7, e50519. [Google Scholar] [CrossRef] [PubMed]
- Bøyum, A. [9] Separation of lymphocytes, granulocytes, and monocytes from human blood using iodinated density gradient media. Methods Enzymol. 1984, 108, 88–102. [Google Scholar]
- Bøyum, A.; Løvhaug, D.; Tresland, L.; Nordlie, E.M. Separation of leucocytes: Improved cell purity by fine adjustments of gradient medium density and osmolality. Scand. J. Immunol. 1991, 34, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Thöne, F.; Schwanhäusser, B.; Becker, D.; Ballmaier, M.; Bumann, D. FACS-isolation of Salmonella-infected cells with defined bacterial load from mouse spleen. J. Microbiol. Methods 2007, 71, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Wigzell, H.; Sundqvist, K.G.; Yoshida, T.O. Separation of Cells According to Surface Antigens by the Use of Antibody-Coated Columns. Fractionation of Cells Carrying Immunoglobulins and Blood Group Antigen. Scand. J. Immunol. 1972, 1, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Quirici, N.; Soligo, D.; Bossolasco, P.; Servida, F.; Lumini, C.; Deliliers, G.L. Isolation of bone marrow mesenchymal stem cells by anti-nerve growth factor receptor antibodies. Exp. Hematol. 2002, 30, 783–791. [Google Scholar] [CrossRef]
- Tomlinson, M.J.; Tomlinson, S.; Yang, X.B.; Kirkham, J. Cell separation: Terminology and practical considerations. J. Tissue Eng. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xue, C.; Zhao, Y.; Chen, D.; Wu, M.H.; Wang, J. Microfluidic impedance flow cytometry enabling high-throughput single-cell electrical property characterization. Int. J. Mol. Sci. 2015, 16, 9804–9830. [Google Scholar] [CrossRef] [PubMed]
- Inglis, D.W.; Riehn, R.; Sturm, J.C.; Austin, R.H. Microfluidic high gradient magnetic cell separation. J. Appl. Phys. 2006, 99, 08K101. [Google Scholar] [CrossRef]
- Kiermer, V. FACS-on-a-chip. Nat. Methods 2005, 2, 91. [Google Scholar] [CrossRef]
- Rembaum, A.; Yen, R.C.K.; Kempner, D.H.; Ugelstad, J. Cell labeling and magnetic separation by means of immunoreagents based on polyacrolein microspheres. J. Immunol. Methods 1982, 52, 341–351. [Google Scholar] [CrossRef]
- Lundahl, J.; Hallden, G.; Hallgren, M.; Sköld, C.M.; Hed, J. Altered expression of CD11b/CD18 and CD62L on human monocytes after cell preparation procedures. J. Immunol. Methods 1995, 180, 93–100. [Google Scholar] [CrossRef]
- Toner, M.; Irimia, D. Blood-on-a-chip. Annu. Rev. Biomed. Eng. 2005, 7, 77–103. [Google Scholar] [CrossRef] [PubMed]
- Pohl, H.A.; Hawk, I. Separation of living and dead cells by dielectrophoresis. Science 1966, 152, 647–649. [Google Scholar] [CrossRef] [PubMed]
- Elitas, M.; Martinez-Duarte, R.; Dhar, N.; McKinney, J.D.; Renaud, P. Dielectrophoresis-based purification of antibiotic-treated bacterial subpopulations. Lab Chip 2014, 14, 1850–1857. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Huang, Y.; Wang, X.; Wang, X.B.; Becker, F.F.; Gascoyne, P.R.C. Dielectric properties of human leukocyte subpopulations determined by electrorotation as a cell separation criterion. Biophys. J. 1999, 76, 3307–3314. [Google Scholar] [CrossRef]
- Pohl, H.A.; Crane, J.S. Dielectrophoresis of cells. Biophys. J. 1971, 11, 711–727. [Google Scholar] [CrossRef]
- Elitas, M.; Dhar, N.; Schneider, K.; Valero, A.; Braschler, T.; McKinney, J.D.; Renaud, P. Dielectrophoresis as a single cell characterization method for bacteria. Biomed. Phys. Eng. Express 2017, 3, 15005. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Shim, S. Isolation of circulating tumor cells by dielectrophoresis. Cancers 2014, 6, 545–579. [Google Scholar] [CrossRef] [PubMed]
- Pethig, R.; Menachery, A.; Pells, S.; De Sousa, P. Dielectrophoresis: A review of applications for stem cell research. BioMed Res. Int. 2010, 2010, 182581. [Google Scholar] [CrossRef] [PubMed]
- Gascoyne, P.R.C.; Vykoukal, J. Particle separation by dielectrophoresis. Electrophoresis 2002, 23, 1973. [Google Scholar] [CrossRef]
- Pethig, R.R. Dielectrophoresis: Theory, Methodology and Biological Applications; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Carpenter, E.L.; Rader, J.; Ruden, J.; Rappaport, E.F.; Hunter, K.N.; Hallberg, P.L.; Krytska, K.; O’Dwyer, P.J.; Mosse, Y.P. Dielectrophoretic Capture and Genetic Analysis of Single Neuroblastoma Tumor Cells. Front. Oncol. 2014, 4, 201. [Google Scholar] [CrossRef] [PubMed]
- Tada, S. Separation of live/dead cells by the use of three dimensional non-uniform AC electric field. In Proceedings of the World Automation Congress (WAC), Waikoloa, HI, USA, 3–7 August 2014; pp. 66–70. [Google Scholar]
- Lapizco-Encinas, B.H.; Simmons, B.A.; Cummings, E.B.; Fintschenko, Y. Dielectrophoretic concentration and separation of live and dead bacteria in an array of insulators. Anal. Chem. 2004, 76, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Bashir, R. Dielectrophoretic separation and manipulation of live and heat-treated cells of Listeria on microfabricated devices with interdigitated electrodes. Sens. Actuators B Chem. 2002, 86, 215–221. [Google Scholar] [CrossRef]
- Markx, G.H.; Talary, M.S.; Pethig, R. Separation of viable and non-viable yeast using dielectrophoresis. J. Biotechnol. 1994, 32, 29–37. [Google Scholar] [CrossRef]
- Qian, C.; Huang, H.; Chen, L.; Li, X.; Ge, Z.; Chen, T.; Yang, Z.; Sun, L. Dielectrophoresis for bioparticle manipulation. Int. J. Mol. Sci. 2014, 15, 18281–18309. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Showers, D.; Vedantam, P.; Tzeng, T.R.; Qian, S.; Xuan, X. Microfluidic separation of live and dead yeast cells using reservoir-based dielectrophoresis. Biomicrofluidics 2012, 6, 34102. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.T.; Lin, R.Z.; Chang, W.Y.; Chang, H.Y.; Liu, C.H. Rapid heterogeneous liver-cell on-chip patterning via the enhanced field-induced dielectrophoresis trap. Lab Chip 2006, 6, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Zahn, J.D. Methods in Bioengineering: Biomicrofabrication and Biomicrofluidics; Artech House: Boston, MA, USA, 2009. [Google Scholar]
- Shafiee, H.; Sano, M.B.; Henslee, E.A.; Caldwell, J.L.; Davalos, R.V. Selective isolation of live/dead cells using contactless dielectrophoresis (cDEP). Lab Chip 2010, 10, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Duarte, R.; Gorkin, R.A., III; Abi-Samra, K.; Madou, M.J. The integration of 3D carbon-electrode dielectrophoresis on a CD-like centrifugal microfluidic platform. Lab Chip 2010, 10, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.; Natu, R.; Larraga-Martinez, M.F.; Martinez-Duarte, R. Enrichment of diluted cell populations from large sample volumes using 3D carbon-electrode dielectrophoresis. Biomicrofluidics 2016, 10, 33107. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Duarte, R.; Renaud, P.; Madou, M.J. A novel approach to dielectrophoresis using carbon electrodes. Electrophoresis 2011, 32, 2385–2392. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.d.C.; Torrents, E.; Martínez-Duarte, R.; Madou, M.J.; Juárez, A. On-line separation of bacterial cells by carbon-electrode dielectrophoresis. Electrophoresis 2010, 31, 2921–2928. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Duarte, R.; Camacho-Alanis, F.; Renaud, P.; Ros, A. Dielectrophoresis of lambda-DNA using 3D carbon electrodes. Electrophoresis 2013, 34, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Van Furth, R.; Cohn, Z.A. The origin and kinetics of mononuclear phagocytes. J. Exp. Med. 1968, 128, 415–435. [Google Scholar] [CrossRef] [PubMed]
- Galkina, E.; Ley, K. Immune and Inflammatory Mechanisms of Atherosclerosis. Annu. Rev. Immunol. 2009, 27, 165–197. [Google Scholar] [CrossRef] [PubMed]
- Mee, A.S.; Berney, J.; Jewell, D.P. Monocytes in inflammatory bowel disease: Absolute monocyte counts. J. Clin. Pathol. 1980, 33, 917–920. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.K.; Obara, C.J.; Rivollier, A.; Pletnev, A.G.; Kelsall, B.L.; Murphy, P.M. Chemokine receptor Ccr2 is critical for monocyte accumulation and survival in West Nile virus encephalitis. J. Immunol. 2011, 186, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Dawson, T.C.; Beck, M.A.; Kuziel, W.A.; Henderson, F.; Maeda, N. Contrasting effects of CCR5 and CCR2 deficiency in the pulmonary inflammatory response to influenza A virus. Am. J. Pathol. 2000, 156, 1951–1959. [Google Scholar] [CrossRef]
- Aldridge, J.R.; Moseley, C.E.; Boltz, D.A.; Negovetich, N.J.; Reynolds, C.; Franks, J.; Brown, S.A.; Doherty, P.C.; Webster, R.G.; Thomas, P.G. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc. Natl. Acad. Sci. USA 2009, 106, 5306–5311. [Google Scholar] [CrossRef] [PubMed]
- Osterholzer, J.J.; Chen, G.H.; Olszewski, M.A.; Curtis, J.L.; Huffnagle, G.B.; Toews, G.B. Accumulation of CD11b+ lung dendritic cells in response to fungal infection results from the CCR2-mediated recruitment and differentiation of Ly-6Chigh monocytes. J. Immunol. 2009, 183, 8044–8053. [Google Scholar] [CrossRef] [PubMed]
- Ersland, K.; Wüthrich, M.; Klein, B.S. Dynamic interplay among monocyte-derived, dermal, and resident lymph node dendritic cells during the generation of vaccine immunity to fungi. Cell Host Microbe 2010, 7, 474–487. [Google Scholar] [CrossRef] [PubMed]
- Tsou, C.L.; Peters, W.; Si, Y.; Slaymaker, S.; Aslanian, A.M.; Weisberg, S.P.; Mack, M.; Charo, I.F. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J. Clin. Investig. 2007, 117, 902. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.; Serbina, N.V.; Brandl, K.; Zhong, M.X.; Leiner, I.M.; Charo, I.F.; Pamer, E.G. Additive roles for MCP-1 and MCP-3 in CCR2-mediated recruitment of inflammatory monocytes during Listeria monocytogenes infection. J. Immunol. 2008, 180, 6846–6853. [Google Scholar] [CrossRef] [PubMed]
- Kipnis, A.; Basaraba, R.J.; Orme, I.M.; Cooper, A.M. Role of chemokine ligand 2 in the protective response to early murine pulmonary tuberculosis. Immunology 2003, 109, 547–551. [Google Scholar] [CrossRef] [PubMed]
- La Manna, M.P.; Orlando, V.; Dieli, F.; Di Carlo, P.; Cascio, A.; Cuzzi, G.; Palmieri, F.; Goletti, D.; Caccamo, N. Quantitative and qualitative profiles of circulating monocytes may help identifying tuberculosis infection and disease stages. PLoS ONE 2017, 12, e0171358. [Google Scholar] [CrossRef] [PubMed]
- Bobdey, S.; Ganesh, B.; Mishra, P.; Jain, A. Role of Monocyte Count and Neutrophil-to-Lymphocyte Ratio in Survival of Oral Cancer Patients. Int. Arch. Otorhinolaryngol. 2017, 21, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, C.M.; Arico, M.; Basso, G.; Biondi, A.; Rajnoldi, A.C.; Creutzig, U.; Haas, O.; Harbott, J.; Hasle, H.; Kerndrup, G. Chronic myelomonocytic leukemia in childhood: A retrospective analysis of 110 cases. Blood 1997, 89, 3534–3543. [Google Scholar] [PubMed]
- Sprangers, S.; Vries, T.J.D.; Everts, V. Monocyte heterogeneity: Consequences for monocyte-derived immune cells. J. Immunol. Res. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Pethig, R. Dielectrophoresis: Status of the theory, technology, and applications. Biomicrofluidics 2010, 4, 22811. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.; Green, N.G. AC Electrokinetics: Colloids and Nanoparticles; Research Studies Press: Philadelphia, PA, USA, 2003. [Google Scholar]
- Valero, A.; Braschler, T.; Renaud, P. A unified approach to dielectric single cell analysis: Impedance and dielectrophoretic force spectroscopy. Lab Chip 2010, 10, 2216–2225. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.L.; Gascoyne, P.R.C.; Becker, F.F.; Pethig, R. Electrorotation of liposomes: Verification of dielectric multi-shell model for cells. Biochim. Biophys. Acta-Lipids Lipid Metab. 1997, 1349, 182–196. [Google Scholar] [CrossRef]
- Jönsson, M.; Welch, K.; Hamp, S.; Strømme, M. Bacteria counting with impedance spectroscopy in a micro probe station. J. Phys. Chem. B 2006, 110, 10165–10169. [Google Scholar] [CrossRef] [PubMed]
- Schwan, H.P. Electrical properties of tissue and cell suspensions. Adv. Biol. Med. Phys. 1957, 5, 147. [Google Scholar] [PubMed]
- Erdem, N.; Yildizhan, Y.; Elitasş, M. A numerical approach for dielectrophoretic characterization and separation of human hematopoietic cells. Int. J. Eng. Res. Technol. 2017, 6, 1079–1082. [Google Scholar]
- Mernier, G.; Martinez-Duarte, R.; Lehal, R.; Radtke, F.; Renaud, P. Very high throughput electrical cell lysis and extraction of intracellular compounds using 3D carbon electrodes in lab-on-a-chip devices. Micromachines 2012, 3, 574–581. [Google Scholar] [CrossRef]
- Prodan, E.; Prodan, C.; Miller, J.H. The dielectric response of spherical live cells in suspension: An analytic solution. Biophys. J. 2008, 95, 4174–4182. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.B.; Liu, S.L.; Li, J.T.; Wu, M.H. Label-free Live and Dead Cell Separation Method Using a High-Efficiency Optically-Induced Dielectrophoretic (ODEP) Force-based Microfluidic Platform. Int. J. Autom. Smart Technol. 2014, 4, 83–91. [Google Scholar]
- Martinez-Duarte, R. Microfabrication technologies in dielectrophoresis applications—A review. Electrophoresis 2012, 33, 3110–3132. [Google Scholar] [CrossRef] [PubMed]





| Cell Type | Method | Separation/Removal Efficiency | Advantages | Disadvantages |
|---|---|---|---|---|
| Human Leukemia (THP-1) | Contactless dielectrophoresis [38] | 95% Removal Efficiency | High efficiency, inexpensive fabrication process | Lysing due to electrical field |
| Yeast | Reservoir-based dielectrophoresis [35] | N/A | No mechanical or electrical parts inside the microchannel | Clogging due to trapped cells |
| Chondrocytes | Optically-induced dielectrophoresis [68] | 96.4 ± 2.2% Purity of the isolated live cells | No mechanical or electrical parts inside the microchannel | N/A |
| Listeria innocua | Interdigitated microelectrodes [32] | 90% Separation Efficiency | Cells were viable for downstream analysis | N/A |
| Escheria coli | Insulator-based dielectrophoresis [31] | N/A | Differential DEP trapping was achieved | N/A |
| U937 Monocyte | 3D carbon-dielectrophoresis | >90% | High efficiency, cells were viable for downstream analysis, no electrolysis wider electrochemical stability | N/A |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yildizhan, Y.; Erdem, N.; Islam, M.; Martinez-Duarte, R.; Elitas, M. Dielectrophoretic Separation of Live and Dead Monocytes Using 3D Carbon-Electrodes. Sensors 2017, 17, 2691. https://doi.org/10.3390/s17112691
Yildizhan Y, Erdem N, Islam M, Martinez-Duarte R, Elitas M. Dielectrophoretic Separation of Live and Dead Monocytes Using 3D Carbon-Electrodes. Sensors. 2017; 17(11):2691. https://doi.org/10.3390/s17112691
Chicago/Turabian StyleYildizhan, Yagmur, Nurdan Erdem, Monsur Islam, Rodrigo Martinez-Duarte, and Meltem Elitas. 2017. "Dielectrophoretic Separation of Live and Dead Monocytes Using 3D Carbon-Electrodes" Sensors 17, no. 11: 2691. https://doi.org/10.3390/s17112691
APA StyleYildizhan, Y., Erdem, N., Islam, M., Martinez-Duarte, R., & Elitas, M. (2017). Dielectrophoretic Separation of Live and Dead Monocytes Using 3D Carbon-Electrodes. Sensors, 17(11), 2691. https://doi.org/10.3390/s17112691