Multifunctional Nanotechnology-Enabled Sensors for Rapid Capture and Detection of Pathogens
Abstract
:1. Introduction
2. Nanotechnology for Detection of Pathogens and Toxins: Opportunities and Challenges
- miniaturized portable instrumentation for field testing
- smart labels to indicate quality and safety
- delivery systems of active ingredients
- nano-barcodes or trackers for product traceability and authentication
3. Nanotechnology-Enabled Sensors and Sensing Systems for Detection of Pathogens
3.1. Aptamer-Based Nanosensing
3.2. Immuno-Based Nanosensor Strategies
3.3. Phage-Based Recognition
3.4. Molecularly Imprinted Polymers (MIP)
3.5. Antimicrobial Peptides
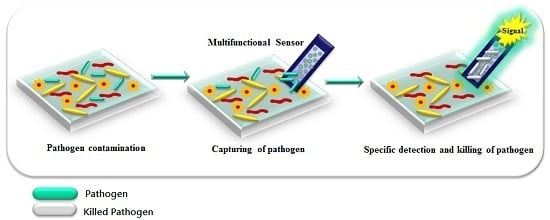
3.6. Multifunctional Platforms for Inactivation and Detection of Pathogens
4. Conclusions and Future Perspectives
Acknowledgments
Conflicts of Interest
References
- CDC. Foodborne Illness: Frequently Asked Questions; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2016. [Google Scholar]
- WHO. Global Burden of Foodborne Diseases; WHO Press: Geneva, Switzerland, 2015. [Google Scholar]
- Centers for Disease Control and Prevention. Surveillance for foodborne disease outbreaks—United States, 2009–2010. MMWR Morb. Mortal. Wkly. Rep. 2013, 62, 41. [Google Scholar]
- Centers for Disease Control and Prevention. Surveillance for Foodborne Disease Outbreaks, United States, 2011; Annual Report; US Department of Health and Human Services: Atlanta, GA, USA, 2014. [Google Scholar]
- Centers for Disease Control and Prevention. Surveillance for Foodborne Disease Outbreaks, United States, 2012; Annual Report; US Department of Health and Human Services: Atlanta, GA, USA, 2014. [Google Scholar]
- Centers for Disease Control and Prevention. Surveillance for Foodborne Disease Outbreaks, United States, 2013; Annual Report; US Department of Health and Human Services: Atlanta, GA, USA, 2015. [Google Scholar]
- De Boer, E.; Beumer, R.R. Methodology for detection and typing of foodborne microorganisms. Int. J. Food Microbiol. 1999, 50, 119–130. [Google Scholar] [CrossRef]
- Bülbül, G.; Hayat, A.; Andreescu, S. Portable Nanoparticle-Based Sensors for Food Safety Assessment. Sensors 2015, 15, 29826. [Google Scholar] [CrossRef] [PubMed]
- Sanvicens, N.; Pastells, C.; Pascual, N.; Marco, M.P. Nanoparticle-based biosensors for detection of pathogenic bacteria. TrAC Trends Anal. Chem. 2009, 28, 1243–1252. [Google Scholar] [CrossRef]
- Ray, P.C.; Khan, S.A.; Singh, A.K.; Senapati, D.; Fan, Z. Nanomaterials for targeted detection and photothermal killing of bacteria. Chem. Soc. Rev. 2012, 41, 3193–3209. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Andler, S.M.; Goddard, J.M.; Nugen, S.R.; Rotello, V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017, 46, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Mocan, T.; Matea, C.T.; Pop, T.; Mosteanu, O.; Buzoianu, A.D.; Puia, C.; Iancu, C.; Mocan, L. Development of nanoparticle-based optical sensors for pathogenic bacterial detection. J. Nanobiotechnol. 2017, 15, 25. [Google Scholar] [CrossRef] [PubMed]
- Pyrgiotakis, G.; Vedantam, P.; Cirenza, C.; McDevitt, J.; Eleftheriadou, M.; Leonard, S.S.; Demokritou, P. Optimization of a nanotechnology based antimicrobial platform for food safety applications using Engineered Water Nanostructures (EWNS). Sci. Rep. 2016, 6, 21073. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.Y.; Cang, J.; Roy, P.; Chang, H.T.; Huang, Y.C.; Huang, C.C. Synthesis and Antimicrobial Activity of Gold/Silver-Tellurium Nanostructures. ACS Appl. Mater. Interfaces 2014, 6, 8305–8312. [Google Scholar] [CrossRef] [PubMed]
- Ramani, M.; Ponnusamy, S.; Muthamizhchelvan, C.; Marsili, E. Amino acid-mediated synthesis of zinc oxide nanostructures and evaluation of their facet-dependent antimicrobial activity. Coll. Surf. B Biointerfaces 2014, 117, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Mageshwari, K.; Sathyamoorthy, R. Flower-shaped CuO Nanostructures: Synthesis, Characterization and Antimicrobial Activity. J. Mater. Sci. Technol. 2013, 29, 909–914. [Google Scholar] [CrossRef]
- De Azeredo, H.M.C. Antimicrobial nanostructures in food packaging. Trends Food Sci. Technol. 2013, 30, 56–69. [Google Scholar] [CrossRef]
- Wang, X.Y.; Cui, Q.; Yao, C.; Li, S.; Zhang, P.; Sun, H.; Lv, F.; Liu, L.; Li, L.; Wang, S. Conjugated Polyelectrolyte-Silver Nanostructure Pair for Detection and Killing of Bacteria. Adv. Mater. Technol. 2017, 2. [Google Scholar] [CrossRef]
- Duan, N.; Wu, S.; Dai, S.; Miao, T.; Chen, J.; Wang, Z. Simultaneous detection of pathogenic bacteria using an aptamer based biosensor and dual fluorescence resonance energy transfer from quantum dots to carbon nanoparticles. Microchim. Acta 2015, 182, 917–923. [Google Scholar] [CrossRef]
- Vargas, C.A.; Wilhelm, A.A.; Williams, J.; Lucas, P.; Reynolds, K.A.; Riley, M.R. Integrated Capture and Spectroscopic Detection of Viruses. Appl. Environ. Microbiol. 2009, 75, 6431–6440. [Google Scholar] [CrossRef] [PubMed]
- Othman, A.; Karimi, A.; Andreescu, S. Functional nanostructures for enzyme based biosensors: Properties, fabrication and applications. J. Mater. Chem. B 2016, 4, 7178–7203. [Google Scholar] [CrossRef]
- Armentano, I.; Rescignano, N.; Fortunati, E.; Mattioli, S.; Morena, F.; Martino, S.; Torre, L.; Kenny, J.M. Chapter 5—Multifunctional nanostructured biopolymeric materials for therapeutic applications A2. In Nanostructures for Novel Therapy; Ficai, D., Grumezescu, A.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 107–135. [Google Scholar]
- Baccar, H.; Mejri, M.B.; Hafaiedh, I.; Ktari, T.; Aouni, M.; Abdelghani, A. Surface plasmon resonance immunosensor for bacteria detection. Talanta 2010, 82, 810–814. [Google Scholar] [CrossRef] [PubMed]
- Celardo, I.; De Nicola, M.; Mandoli, C.; Pedersen, J.Z.; Traversa, E.; Ghibelli, L. Ce3+ Ions Determine Redox-Dependent Anti-apoptotic Effect of Cerium Oxide Nanoparticles. ACS Nano 2011, 5, 4537–4549. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Cao, C.; Han, S.J.; Sim, S.J. Detection of pathogen based on the catalytic growth of gold nanocrystals. Water Res. 2009, 43, 1425–1431. [Google Scholar] [CrossRef] [PubMed]
- Setterington, E.B.; Alocilja, E.C. Electrochemical Biosensor for Rapid and Sensitive Detection of Magnetically Extracted Bacterial Pathogens. Biosensors 2012, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Senapati, D.; Wang, S.; Griffin, J.; Neely, A.; Naylor, K.M.; Varisli, B.; Kalluri, J.R.; Ray, P.C. Gold Nanorod Based Selective Identification of Escherichia coli Bacteria Using Two-Photon Rayleigh Scattering Spectroscopy. ACS Nano 2009, 3, 1906–1912. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Li, J.; Pan, D.; Li, J.; Song, S.; Rong, M.; Li, Z.; Gao, J.; Lu, J. Gold Nanoparticle-Based Enzyme-Linked Antibody-Aptamer Sandwich Assay for Detection of Salmonella Typhimurium. ACS Appl. Mater. Interfaces 2014, 6, 16974–16981. [Google Scholar] [CrossRef] [PubMed]
- Ispas, C.R.; Crivat, G.; Andreescu, S. Review: Recent Developments in Enzyme-Based Biosensors for Biomedical Analysis. Anal. Lett. 2012, 45, 168–186. [Google Scholar] [CrossRef]
- Chua, A.; Yean, C.Y.; Ravichandran, M.; Lim, B.; Lalitha, P. A rapid DNA biosensor for the molecular diagnosis of infectious disease. Biosens. Bioelectron. 2011, 26, 3825–3831. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cheng, J.; Zhang, Y. Upconversion nanoparticle based LRET system for sensitive detection of MRSA DNA sequence. Biosens. Bioelectron. 2013, 43, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Thiruppathiraja, C.; Kamatchiammal, S.; Adaikkappan, P.; Santhosh, D.J.; Alagar, M. Specific detection of Mycobacterium sp. genomic DNA using dual labeled gold nanoparticle based electrochemical biosensor. Anal. Biochem. 2011, 417, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; Wu, J.; Flechsig, G.U.; Haake, D.A.; Wang, J. Redox cycling amplified electrochemical detection of DNA hybridization: Application to pathogen E. coli bacterial RNA. Anal. Chim. Acta 2011, 689, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, J. Bacteriophage-based nanoprobes for rapid bacteria separation. Nanoscale 2015, 7, 16230. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Alcaine, S.D.; Jiang, Z.; Rotello, V.M.; Nugen, S.R. Detection of Escherichia coli in Drinking Water Using T7 Bacteriophage-Conjugated Magnetic Probe. Anal. Chem. 2015, 87, 8977–8984. [Google Scholar] [CrossRef] [PubMed]
- Derda, R.; Lockett, M.R.; Tang, S.K.; Fuller, R.C.; Maxwell, E.J.; Breiten, B.; Cuddemi, C.A.; Ozdogan, A.; Whitesides, G.M. Filter-Based Assay for Escherichia coli in Aqueous Samples Using Bacteriophage-Based Amplification. Anal. Chem. 2013, 85, 7213–7220. [Google Scholar] [CrossRef] [PubMed]
- Franche, N.; Vinay, M.; Ansaldi, M. Substrate-independent luminescent phage-based biosensor to specifically detect enteric bacteria such as E. coli. Environ. Sci. Pollut. Res. 2017, 24, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Guntupalli, R.; Sorokulova, I.; Olsen, E.; Globa, L.; Pustovyy, O.; Vodyanoy, V. Biosensor for Detection of Antibiotic Resistant Staphylococcus Bacteria. J. Vis. Exp. 2013, 75, e50474. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, T.; Schwartz-Mittelmann, A.; Biran, D.; Ron, E.Z.; Rishpon, J. Combined Phage Typing and Amperometric Detection of Released Enzymatic Activity for the Specific Identification and Quantification of Bacteria. Anal. Chem. 2003, 75, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Olsen, E.V.; Sorokulova, I.B.; Petrenko, V.A.; Chen, I.H.; Barbaree, J.M.; Vodyanoy, V.J. Affinity-selected filamentous bacteriophage as a probe for acoustic wave biodetectors of Salmonella typhimurium. Biosens. Bioelectron. 2006, 21, 1434–1442. [Google Scholar] [CrossRef] [PubMed]
- Shabani, A.; Marquette, C.A.; Mandeville, R.; Lawrence, M.F. Magnetically-assisted impedimetric detection of bacteria using phage-modified carbon microarrays. Talanta 2013, 116, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Vinay, M.; Franche, N.; Grégori, G.; Fantino, J.R.; Pouillot, F.; Ansaldi, M. Phage-Based Fluorescent Biosensor Prototypes to Specifically Detect Enteric Bacteria Such as E. coli and Salmonella enterica Typhimurium. PLoS ONE 2015, 10, e0131466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbaspour, A.; Norouz-Sarvestani, F.; Noori, A.; Soltani, N. Aptamer-conjugated silver nanoparticles for electrochemical dual-aptamer-based sandwich detection of staphylococcus aureus. Biosens. Bioelectron. 2015, 68, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Wu, S.; Yu, Y.; Ma, X.; Xia, Y.; Chen, X.; Huang, Y.; Wang, Z. A dual-color flow cytometry protocol for the simultaneous detection of Vibrio parahaemolyticus and Salmonella typhimurium using aptamer conjugated quantum dots as labels. Anal. Chim. Acta 2013, 804, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Duan, N.; Wu, S.; Ma, X.; Xia, Y.; Wang, Z.; Wei, X. Impedimetric aptasensor for Staphylococcus aureus based on nanocomposite prepared from reduced graphene oxide and gold nanoparticles. Microchim. Acta 2014, 181, 967–974. [Google Scholar] [CrossRef]
- Lian, Y.; He, F.; Wang, H.; Tong, F. A new aptamer/graphene interdigitated gold electrode piezoelectric sensor for rapid and specific detection of Staphylococcus aureus. Biosens. Bioelectron. 2015, 65, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Duan, N.; Shi, Z.; Fang, C.; Wang, Z. Simultaneous Aptasensor for Multiplex Pathogenic Bacteria Detection Based on Multicolor Upconversion Nanoparticles Labels. Anal. Chem. 2014, 86, 3100–3107. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, J.; Zheng, M.; Zhong, Y.; Yang, J.; Zhao, Y.; Wu, W.; Ye, W.; Wen, J.; Wang, Q.; Lu, J. Correction: An Aptamer-Based Biosensor for Colorimetric Detection of Escherichia coli O157:H7. PLoS ONE 2013, 8, e48999. [Google Scholar] [CrossRef]
- Wu, W.-H.; Li, M.; Wang, Y.; Ouyang, H.X.; Wang, L.; Li, C.X.; Cao, Y.C.; Meng, Q.H.; Lu, J.X. Aptasensors for rapid detection of Escherichia coli O157:H7 and Salmonella typhimurium. Nanoscale Res. Lett. 2012, 7, 658. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.M.; Kim, D.-K.; Lee, S.Y. Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta 2015, 132, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Zelada-Guillén, G.A.; Bhosale, S.V.; Riu, J.; Rius, F.X. Real-Time Potentiometric Detection of Bacteria in Complex Samples. Anal. Chem. 2010, 82, 9254–9260. [Google Scholar] [CrossRef] [PubMed]
- Zelada-Guillén, G.A.; Riu, J.; Düzgün, A.; Rius, F.X. Immediate Detection of Living Bacteria at Ultralow Concentrations Using a Carbon Nanotube Based Potentiometric Aptasensor. Angew. Chem. Int. Ed. 2009, 48, 7334–7337. [Google Scholar] [CrossRef] [PubMed]
- Zuo, P. A PDMS/paper/glass hybrid microfluidic biochip integrated with aptamer-functionalized graphene oxide nano-biosensors for one-step multiplexed pathogen detection. Lab Chip 2013, 13, 3921. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Ragavan, K.V.; Thakur, M.S.; Raghavarao, K.S. Recent advances in nanoparticle based aptasensors for food contaminants. Biosens. Bioelectron. 2015, 74, 612–627. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Marrakchi, M.; Xu, D.; Dong, H.; Andreescu, S. Biosensors based on modularly designed synthetic peptides for recognition, detection and live/dead differentiation of pathogenic bacteria. Biosens. Bioelectron. 2016, 80, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Emanuel, P.A.; Dang, J.; Gebhardt, J.S.; Aldrich, J.; Garber, E.A.; Kulaga, H.; Stopa, P.; Valdes, J.J.; Dion-Schultz, A. Recombinant antibodies: A new reagent for biological agent detection. Biosens. Bioelectron. 2000, 14, 751–759. [Google Scholar] [CrossRef]
- Jiang, H.; Jiang, D.; Shao, J.; Sun, X. Magnetic molecularly imprinted polymer nanoparticles based electrochemical sensor for the measurement of Gram-negative bacterial quorum signaling molecules (N-acyl-homoserine-lactones). Biosens. Bioelectron. 2016, 75, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Tokonami, S.; Nakadoi, Y.; Takahashi, M.; Ikemizu, M.; Kadoma, T.; Saimatsu, K.; Dung, L.Q.; Shiigi, H.; Nagaoka, T. Label-Free and Selective Bacteria Detection Using a Film with Transferred Bacterial Configuration. Anal. Chem. 2013, 85, 4925–4929. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, E.; Majidi, D.; Ozgur, E.; Denizli, A. Whole cell imprinting based Escherichia coli sensors: A study for SPR and QCM. Sens. Actuators B Chem. 2015, 209, 714–721. [Google Scholar] [CrossRef]
- Jia, F.; Duan, N.; Wu, S.; Dai, R.; Wang, Z.; Li, X. Impedimetric Salmonella aptasensor using a glassy carbon electrode modified with an electrodeposited composite consisting of reduced graphene oxide and carbon nanotubes. Microchim. Acta 2016, 183, 337–344. [Google Scholar] [CrossRef]
- Kang, X.; Pang, G.; Chen, Q.; Liang, X. Fabrication of Bacillus cereus electrochemical immunosensor based on double-layer gold nanoparticles and chitosan. Sens. Actuators B Chem. 2013, 177, 1010–1016. [Google Scholar] [CrossRef]
- Chau, C.F.; Wu, S.H.; Yen, G.C. The development of regulations for food nanotechnology. Trends Food Sci. Technol. 2007, 18, 269–280. [Google Scholar] [CrossRef]
- Ariyarathna, I.R.; Rajakaruna, R.M.P.I.; Karunaratne, D.N. The rise of inorganic nanomaterial implementation in food applications. Food Control 2017, 77, 251–259. [Google Scholar] [CrossRef]
- Cao, Y.; Li, J.; Liu, F.; Li, X.; Jiang, Q.; Cheng, S.; Gu, Y. Consideration of interaction between nanoparticles and food components for the safety assessment of nanoparticles following oral exposure: A review. Environ. Toxicol. Pharmacol. 2016, 46, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.; Takhistov, P.; McClements, D.J. Functional materials in food nanotechnology. J. Food Sci. 2006, 71, R107–R116. [Google Scholar] [CrossRef]
- Fonseca, L.; Cane, C.; Mazzolai, B. Application of micro and nanotechnologies to food safety and quality monitoring. Meas. Control 2007, 40, 116–119. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, Q. The role of nanomaterials in electroanalytical biosensors: A mini review. J. Electroanal. Chem. 2016, 781, 401–409. [Google Scholar] [CrossRef]
- Irkin, R.; Esmer, O.K. Novel food packaging systems with natural antimicrobial agents. J. Food Sci. Technol. Mysore 2015, 52, 6095–6111. [Google Scholar] [CrossRef] [PubMed]
- Hoseinnejad, M.; Jafari, S.M.; Katouzian, I. Inorganic and metal nanoparticles and their antimicrobial activity in food packaging applications. Crit. Rev. Microbiol. 2017, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Bibi, F.; Guillaume, C.; Gontard, N.; Sorli, B. A review: RFID technology having sensing aptitudes for food industry and their contribution to tracking and monitoring of food products. Trends Food Sci. Technol. 2017, 62, 91–103. [Google Scholar] [CrossRef]
- Karuppuswami, S.; Kaur, A.; Arangali, H.; Chahal, P.P. A Hybrid Magnetoelastic Wireless Sensor for Detection of Food Adulteration. IEEE Sens. J. 2017, 17, 1706–1714. [Google Scholar] [CrossRef]
- Mills, A.; Hawthorne, D.; Burns, L.; Hazafy, D. Novel temperature-activated humidity-sensitive optical sensor. Sens. Actuators B Chem. 2017, 240, 1009–1015. [Google Scholar] [CrossRef]
- Maloney, N.; Lukacs, G.; Jensen, J.; Hegner, M. Nanomechanical sensors for single microbial cell growth monitoring. Nanoscale 2014, 6, 8242–8249. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Duncan, T.V. Nanoscale sensors for assuring the safety of food products. Curr. Opin. Biotechnol. 2017, 44, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An Emerging Class of Molecules That Rival Antibodies in Diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Zayats, M.; Huang, Y.; Gill, R.; Ma, C.A.; Willner, I. Label-Free and Reagentless Aptamer-Based Sensors for Small Molecules. J. Am. Chem. Soc. 2006, 128, 13666–13667. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, C.K. Aptasensors—The future of biosensing? Anal. Bioanal. Chem. 2002, 372, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.R.; Johnson, A.T.C.; Klein, M.L. Probing the Structure of DNA–Carbon Nanotube Hybrids with Molecular Dynamics. Nano Lett. 2008, 8, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Brown, D.G. Electrostatic Behavior of the Charge-Regulated Bacterial Cell Surface. Langmuir 2008, 24, 5003–5009. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.S. Branching and size of CTAB-coated gold nanostars control the colorimetric detection of bacteria. RSC Adv. 2014, 4, 10660. [Google Scholar] [CrossRef]
- Chen, J.; Jackson, A.A.; Rotello, V.M.; Nugen, S.R. Colorimetric Detection of Escherichia coli Based on the Enzyme-Induced Metallization of Gold Nanorods. Small 2016, 12, 2469–2475. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.J. Discrimination between bacterial phenotypes using glyco-nanoparticles and the impact of polymer coating on detection readouts. J. Mater. Chem. 2014, 2, 1490. [Google Scholar] [CrossRef]
- Li, B.M.; Yu, Q.L.; Duan, Y.X. Fluorescent labels in biosensors for pathogen detection. Crit. Rev. Biotechnol. 2015, 35, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.H.; Zhou, X.M.; Xing, D. Quantum Dots and Graphene Oxide Fluorescent Switch Based Multivariate Testing Strategy for Reliable Detection of Listeria monocytogenes. ACS Appl. Mater. Interfaces 2014, 6, 9988–9996. [Google Scholar] [CrossRef] [PubMed]
- Zuo, P.; Lu, X.; Sun, Z.; Guo, Y.; He, H. A review on syntheses, properties, characterization and bioanalytical applications of fluorescent carbon dots. Microchim. Acta 2016, 183, 519–542. [Google Scholar] [CrossRef]
- Liu, J.; Wagan, S.; Dávila Morris, M.; Taylor, J.; White, R.J. Achieving Reproducible Performance of Electrochemical, Folding Aptamer-Based Sensors on Microelectrodes: Challenges and Prospects. Anal. Chem. 2014, 86, 11417–11424. [Google Scholar] [CrossRef] [PubMed]
- Swanink, C.M.; Meis, J.F.; Rijs, A.J.; Donnelly, J.P.; Verweij, P.E. Specificity of a sandwich enzyme-linked immunosorbent assay for detecting Aspergillus galactomannan. J. Clin. Microbiol. 1997, 35, 257–260. [Google Scholar] [PubMed]
- Guner, A.; Çevik, E.; Şenel, M.; Alpsoy, L. An electrochemical immunosensor for sensitive detection of Escherichia coli O157:H7 by using chitosan, MWCNT, polypyrrole with gold nanoparticles hybrid sensing platform. Food Chem. 2017, 229, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Guo, J.; Bao, X.; Chen, T.; Weng, W.; Chen, S.; Yang, M. Rapid and Sensitive Detection of Bacteria Response to Antibiotics Using Nanoporous Membrane and Graphene Quantum Dot (GQDs)-Based Electrochemical Biosensors. Materials 2017, 10, 603. [Google Scholar] [CrossRef] [PubMed]
- Inbaraj, B.S.; Chen, B.H. Nanomaterial-based sensors for detection of foodborne bacterial pathogens and toxins as well as pork adulteration in meat products. J. Food Drug Anal. 2016, 24, 15–28. [Google Scholar] [CrossRef]
- Li, Y.S.; Church, J.S. Raman spectroscopy in the analysis of food and pharmaceutical nanomaterials. J. Food Drug Anal. 2014, 22, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Deisingh, A.K.; Thompson, M. Biosensors for the detection of bacteria. Can. J. Microbiol. 2004, 50, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Mathelié-Guinlet, M.; Gammoudi, I.; Beven, L.; Moroté, F.; Delville, M.H.; Grauby-Heywang, C.; Cohen-Bouhacina, T. Silica Nanoparticles Assisted Electrochemical Biosensor for the Detection and Degradation of Escherichia Coli Bacteria. Procedia Eng. 2016, 168, 1048–1051. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, J.; Gui, R.; Jin, H.; Xia, Y. Carbon nanomaterials-based electrochemical aptasensors. Biosens. Bioelectron. 2016, 79, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Velu, R.; Frost, N.; DeRosa, M.C. Linkage inversion assembled nano-aptasensors (LIANAs) for turn-on fluorescence detection. Chem. Commun. 2015, 51, 14346–14349. [Google Scholar] [CrossRef] [PubMed]
- Rowland, C.E.; Brown, C.W.; Delehanty, J.B.; Medintz, I.L. Nanomaterial-based sensors for the detection of biological threat agents. Mater.Today 2016, 19, 464–477. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, J. Oriented immobilization of proteins on solid supports for use in biosensors and biochips: A review. Microchim. Acta 2016, 183, 1–19. [Google Scholar] [CrossRef]
- Wang, H.; Yang, R.; Yang, L.; Tan, W. Nucleic Acid Conjugated Nanomaterials for Enhanced Molecular Recognition. ACS Nano 2009, 3, 2451–2460. [Google Scholar] [CrossRef] [PubMed]
- Korayem, M.H.; Estaji, M.; Homayooni, A. Noncalssical multiscale modeling of ssDNA manipulation using a CNT-nanocarrier based on AFM. Coll. Surf. B Biointerfaces 2017, 158, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Jeddi, I.; Saiz, L. Three-dimensional modeling of single stranded DNA hairpins for aptamer-based biosensors. Sci. Rep. 2017, 7, 1178. [Google Scholar] [CrossRef] [PubMed]
- Nasiri Khonsari, Y.; Sun, S. Recent trends in electrochemiluminescence aptasensors and their applications. Chem. Commun. 2017, 53, 9042–9054. [Google Scholar] [CrossRef] [PubMed]
- Guttman, B.; Raya, R.; Kutter, E. Basic phage biology. Bacteriophages Biol. Appl. 2005, 4. [Google Scholar] [CrossRef]
- Tawil, N. Bacteriophages: Biosensing tools for multi-drug resistant pathogens. Analyst 2014, 139, 1224. [Google Scholar] [CrossRef] [PubMed]
- Baggesen, D.L.; Wegener, H.C. Phage types of Salmonella enterica ssp. enterica serovar typhimurium isolated from production animals and humans in Denmark. Acta Vet. Scand. 1993, 35, 349–354. [Google Scholar]
- Yanez-Sedeno, P.; Campuzano, S.; Pingarron, J.M. Electrochemical sensors based on magnetic molecularly imprinted polymers: A review. Anal. Chim. Acta 2017, 960, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Gama, M.R.; Bottoli, C.B. Molecularly imprinted polymers for bioanalytical sample preparation. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1043, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Tokonami, S.; Shiigi, H.; Nagaoka, T. Review: Micro- and nanosized molecularly imprinted polymers for high-throughput analytical applications. Anal. Chim. Acta 2009, 641, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Idil, N.; Mattiasson, B. Imprinting of Microorganisms for Biosensor Applications. Sensors 2017, 17, 708. [Google Scholar] [CrossRef] [PubMed]
- Haupt, K.; Mosbach, K. Molecularly Imprinted Polymers and Their Use in Biomimetic Sensors. Chem. Rev. 2000, 100, 2495–2504. [Google Scholar] [CrossRef] [PubMed]
- Mayes, A.G.; Mosbach, K. Molecularly imprinted polymers: Useful materials for analytical chemistry? TrAC Trends Anal. Chem. 1997, 16, 321–332. [Google Scholar] [CrossRef]
- Cai, D.; Ren, L.; Zhao, H.; Xu, C.; Zhang, L.; Yu, Y.; Wang, H.; Lan, Y.; Roberts, M.F.; Chuang, J.H.; et al. A molecular-imprint nanosensor for ultrasensitive detection of proteins. Nat. Nanotechnol. 2010, 5, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Whitcombe, M.J. The rational development of molecularly imprinted polymer-based sensors for protein detection. Chem. Soc. Rev. 2011, 40, 1547. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.V.R.; Yedery, R.D.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Lupetti, A.; Welling, M.M.; Pauwels, E.K.; Nibbering, P.H. Radiolabelled antimicrobial peptides for infection detection. Lancet Infect. Dis. 2003, 3, 223–229. [Google Scholar] [CrossRef]
- Welling, M.M.; Lupetti, A.; Balter, H.S.; Lanzzeri, S.; Souto, B.; Rey, A.M.; Savio, E.O.; Paulusma-Annema, A.; Pauwels, E.K.; Nibbering, P.H. 99mTc-Labeled Antimicrobial Peptides for Detection of Bacterial and Candida albicans Infections. J. Nucl. Med. 2001, 42, 788–794. [Google Scholar] [PubMed]
- Kulagina, N.V.; Lassman, M.E.; Ligler, F.S.; Taitt, C.R. Antimicrobial Peptides for Detection of Bacteria in Biosensor Assays. Anal. Chem. 2005, 77, 6504–6508. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.H.; Woo, D.H.; Chang, M.S.; Chun, M.S. Microfluidic based biosensing for Escherichia coli detection by embedding antimicrobial peptide-labeled beads. Sens. Actuators B Chem. 2014, 191, 211–218. [Google Scholar] [CrossRef]
- Kulagina, N.V.; Shaffer, K.M.; Anderson, G.P.; Ligler, F.S.; Taitt, C.R. Antimicrobial peptide-based array for Escherichia coli and Salmonella screening. Anal. Chim. Acta 2006, 575, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Mannoor, M.S.; Zhang, S.; Link, A.J.; McAlpine, M.C. Electrical detection of pathogenic bacteria via immobilized antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2010, 107, 19207–19212. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, C.; Scott, M.G.; Karunaratne, N.; Yan, H.; Hancock, R.E. Salt-Resistant Alpha-Helical Cationic Antimicrobial Peptides. Antimicrob. Agents Chemother. 1999, 43, 1542–1548. [Google Scholar] [PubMed]
- Rydlo, T.; Rotem, S.; Mor, A. Antibacterial Properties of Dermaseptin S4 Derivatives under Extreme Incubation Conditions. Antimicrob. Agents Chemother. 2006, 50, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Xu, D.; Jiang, L.; Zhang, L.; Dustin, D.; Lund, R.; Liu, L.; Dong, H. Filamentous supramolecular peptide-drug conjugates as highly efficient drug delivery vehicles. Chem. Commun. 2014, 50, 4827–4830. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, L.; Singh, A.; Dustin, D.; Yang, M.; Liu, L.; Lund, R.; Sellati, T.J.; Dong, H. Designed supramolecular filamentous peptides: Balance of nanostructure, cytotoxicity and antimicrobial activity. Chem. Commun. 2015, 51, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-F.; Cheng, S.; Ge, S.L.; Wang, H.; Wang, Q.J.; He, P.G.; Fang, Y.Z. Ultrasensitive Detection of Bacteria by Microchip Electrophoresis Based on Multiple-Concentration Approaches Combining Chitosan Sweeping, Field-Amplified Sample Stacking, and Reversed-Field Stacking. Anal. Chem. 2012, 84, 1687–1694. [Google Scholar] [CrossRef] [PubMed]
- Alhogail, S.; Suaifan, G.A.R.Y.; Zourob, M. Rapid colorimetric sensing platform for the detection of Listeria monocytogenes foodborne pathogen. Biosens. Bioelectron. 2016, 86, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Jokerst, J.C.; Adkins, J.A.; Bisha, B.; Mentele, M.M.; Goodridge, L.D.; Henry, C.S. Development of a Paper-Based Analytical Device for Colorimetric Detection of Select Foodborne Pathogens. Anal. Chem. 2012, 84, 2900–2907. [Google Scholar] [CrossRef] [PubMed]
- Adkins, J.A.; Boehle, K.; Friend, C.; Chamberlain, B.; Bisha, B.; Henry, C.S. Colorimetric and Electrochemical Bacteria Detection Using Printed Paper- and Transparency-Based Analytic Devices. Anal. Chem. 2017, 89, 3613–3621. [Google Scholar] [CrossRef] [PubMed]
- Tsai, T.T.; Huang, T.H.; Chang, C.J.; Ho, N.Y.; Tseng, Y.T.; Chen, C.F. Antibacterial cellulose paper made with silver-coated gold nanoparticles. Sci. Rep. 2017, 7, 3155. [Google Scholar] [CrossRef] [PubMed]
- Arfat, Y.A.; Ejaz, M.; Jacob, H.; Ahmed, J. Deciphering the potential of guar gum/Ag-Cu nanocomposite films as an active food packaging material. Carbohydr. Polym. 2017, 157, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Tiimob, B.J.; Mwinyelle, G.; Abdela, W.; Samuel, T.; Jeelani, S.; Rangari, V.K. Nanoengineered Eggshell-Silver Tailored Copolyester Polymer Blend Film with Antimicrobial Properties. J. Agric. Food Chem. 2017, 65, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.B.; Peng, C.; Luo, W.; Lv, M.; Li, X.; Li, D.; Huang, Q.; Fan, C. Graphene-Based Antibacterial Paper. ACS Nano 2010, 4, 4317–4323. [Google Scholar] [CrossRef] [PubMed]
- Lichter, J.A.; Van Vliet, K.J.; Rubner, M.F. Design of Antibacterial Surfaces and Interfaces: Polyelectrolyte Multilayers as a Multifunctional Platform. Macromolecules 2009, 42, 8573–8586. [Google Scholar] [CrossRef]
- Emamifar, A. Applications of Antimicrobial Polymer Nanocomposites in Food Packaging. In Advances in Nanocomposite Technology; Hashim, A., Ed.; InTech; Available online: https://www.intechopen.com/books/advances-in-nanocomposite-technology/applications-of-antimicrobial-polymer-nanocomposites-in-food-packaging (accessed on 15 September 2017).
- Wang, H.Y.; Zhou, Y.; Jiang, X.; Sun, B.; Zhu, Y.; Wang, H.; Su, Y.; He, Y. Simultaneous Capture, Detection, and Inactivation of Bacteria as Enabled by a Surface-Enhanced Raman Scattering Multifunctional Chip. Angew. Chem.-Int. Ed. 2015, 54, 5132–5136. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, Y.; Zhang, D. A novel multifunctional electrochemical platform for simultaneous detection, elimination, and inactivation of pathogenic bacteria based on the Vancomycin-functionalised AgNPs/3D-ZnO nanorod arrays. Biosens. Bioelectron. 2017, 98, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Tam, L.T.; Dinh, N.X.; Van Cuong, N.; Mølhave, K.; Le, A. Graphene Oxide/Silver Nanohybrid as Multi-functional Material for Highly Efficient Bacterial Disinfection and Detectionof Organic Dye. J. Electron. Mater. 2016, 45, 5321–5333. [Google Scholar] [CrossRef]
- Chen, L.; Razavi, F.S.; Mumin, A.; Guo, X.; Sham, T.K.; Zhang, J. Multifunctional nanoparticles for rapid bacterial capture, detection, and decontamination. RSC Adv. 2013, 3, 2390–2397. [Google Scholar] [CrossRef]
- Kuswandi, B. Environmental friendly food nano-packaging. Environ. Chem. Lett. 2017, 15, 205–221. [Google Scholar] [CrossRef]


















| Ref. | Target Bacteria | NPs Used in the Sensor | NPs Function | LOD | Real Sample | Time | Detection Method | Range |
|---|---|---|---|---|---|---|---|---|
| [53] | Salmonella Typhibacteria | SWCNT | Conductive support for aptamer where change in conformation occurs in presence of target bacteria | 1 CFU/mL | - | Few seconds | Potentiometric | 0.2–103 CFU/mL |
| [52] | E. coli CECT 675 as a nonpathogenic surrogate for pathogenic E. coli O157:H7 | SWCNT | Conductive support for aptamer where change in conformation occurs in presence of target bacteria | LOD 12 CFU in 2 mL of milk and 26 CFU/mL in apple juice | Milk and apple juice | Couple of minutes | Potentiometric | linear response of up to 104 CFU/mL |
| [50] | E. coli O157:H7 and Salmonella typhimurium | AuNPs | Color change due to target induced aggregation | 105 CFU/mL | 20 min or less | Optical/Colorimetric UV-Vis | ||
| [45] | Vibrio parahaemolyticus and Salmonella typhimurium | CDs | Fluorescent label | 5 × 103 CFU/mL | Shrimp | Optical/Fluorescence | 3.8 × 104–3.8 × 107 CFU/mL | |
| [54] | Lactobacillus acidophilus, Staphylococcus aureus and Salmonella enterica | Gaphene oxide (GO) nanomaterial | Fluorescent signal adsorbent | 11.0 CFU/mL for Lactobacillus acidophilus 61.0 CFU/mL for S. enterica and 800.0 CFU/mL and S. aureus | 10 min | Optical/Fluorescence | 9.4–150.0 CFU/mL for Lactobacillus acidophilus 42.2–675.0 CFU/mL for S. enterica and 104–106 CFU/mL for S. aureus | |
| [46] | Staphylococcus aureus | AuNPs-reduced graphene oxide nanocomposite | Signal-amplification and support for aptamer | 10 CFU/mL | water and fish | 60 min | Electrochemical/impedance | 10–106 CFU/mL |
| [49] | E. coli O157:H7 | nanoscale polydiacetylene polymer (PDA ) | Generates color change | 104 CFU/mL | Clinical fecal specimens | 2 h | Optical/colorimetric UV-Vis | 104–108 CFU/mL |
| [51] | Lactobacillus acidophilus Salmonella typhimurium Pseudomonas aeruginosa | Au layer | The combination of gold and silicon NPs (MG-NP) forms a dilectric layer; attachment of biomolecule changes the peak extinction intensity | 30 CFU per assay | - | - | Optical/localized surface plasmon resonance LSPR | 109–104 CFU/mL |
| [48] | Staphylococcus aureus, Vibrio parahemolyticus, and Salmonella typhimurium | 1-Rare earth upconversion nanoparticles (UCNPs) (NaYF4: Yb, Tm NaYF4: Yb, Ho NaYF4: Yb, Er/Mn), 2-magnetic nanoparticles Fe2O3 | 1-luminescence labels for aptamers 2-separation and concentration | 25, 10, and 15 CFU/mL for S. aureus, V. parahemolyticus, and S. typhimurium, respectively | Milk and shrimp | - | Optical/luminescence | 50–106 CFU/mL |
| [19] | Vibrio parahaemolyticus and Salmonella typhimurium | 1-QDs 2-novel amorphous carbon nanoparticles (CNPs) | 1-Fluorescence emitter 2-Fluoresence acceptor | 25 CFU/mL for V. parahaemolyticus, and 35CFU/mL for S. typhimurium | Chicken and shrimps | - | Optical/dual fluorescence resonance energy transfer (FRET) | 50–106 CFU/mL |
| [47] | Staphylococcus aureus | graphene to interdigital gold electrodes connected to a series electrode piezoelectric quartz crystal | - | 41 CFU/mL | Milk | 60 min | Mechanical/series electrode piezoelectric quartz crystal SPQC | 4.1 × 101–4.1 × 105 CFU/mL |
| [44] | Staphylococcus aureus (S. aureus) | AgNPs | Origin of electrochemical signal | 1.0 CFU/mL | Real water | - | Electrochemical/stripping voltammetry | 10–1 × 106 CFU/mL |
| [28] | Salmonella enterica serovar Typhimurium | antibodies -horseradish peroxidase-gold nanoparticles | Amplification of color | 1 × 103 CFU/mL | milk | <3 h | Optical | 1 × 103–1 × 108 CFU/mL |
| [61] | Salmonella | multi-walled carbon nanotubes (MWCNTs) | Signal-amplification and a support material for the bioreceptor (aptamer) | 25 CFU/mL | chicken | 60 min | Amperometric: Cyclic voltammetry and impedimetric | 75–7.5 × 105 CFU⋅mL−1 |
| Ref. | Target Bacteria | NPs | NPs Function | LOD | Real Sample | Time | Detection Method | Range |
|---|---|---|---|---|---|---|---|---|
| [23] | E. coli K12 (gram negative) and Lactobacillus fermentium (gram positive) | AuNPs | amplifying the SPR signal | 104 CFU/mL and 103 CFU/mL in presence of Au NPs | - | 1 min | SPR | 105–107 CFU/mL |
| [24] | Pseudomonas aeruginosa and Staphylococcus aureus | AuNPs | Signaling- origin of color | - | Sputum | 5 min | Visually and Optical Density at 600 nm (OD600) | 500–5000 CFU/mL |
| [25] | Giardia lamblia cysts | AuNPs | Signaling- origin of color | 1.088 × 103 cells mL−1 | - | - | UV-Vis | 103–104 cells/mL |
| [26] | Bacillus and E. coli O157:H7 | magnetic/polyaniline core/shell nanoparticle (c/sNP) | Separation and electrical conductive based material | 40 CFU/mL and 6 CFU/mL | - | ~1 h | Amperometric: Cyclic voltammetry | 100–102 CFU/mL |
| [27] | E. coli O157:H7 | Au nanorods | Signaling- origin of color | 50 CFU/mL | - | 15 min | two-photon Rayleigh scattering (TPRS) | 50–2100 CFU/mL |
| [62] | Bacillus cereus | AuNPs | Increase sensitivity and stability | 10.0 CFU/mL | Milk | - | Amperometric: Cyclic voltammetry | 5.0 × 101–5.0 × 104 CFU/mL |
| Ref. | Phage | Target Bacteria | LOD | Sample | Time | Detection Method | Range |
|---|---|---|---|---|---|---|---|
| [35] | T7 | E. coli | 10 CFU/mL | Drinking water | 2.5 h | Optical/colorimetric | - |
| [36] | M13KE phage | E. coli K12 | 5 CFU/L | Water | overnight | Colorimetric-culture based assay | - |
| 50 CFU/L water (or 5 CFU/mL orange juice and skim milk) | Water, orange juice and skim milk | <4 h | Colorimetric-solution based assay | - | |||
| [34] | T7 | E. coli K12 | - | - | - | Bacteria culture | 102–107 CFU/mL |
| [43] | Engineered HK620 | E. coli TD2158 and Salmonella | 10 bacteria/mL | Sea water | 1 h | Optical/Fluorescence | |
| [37] | Engineered HK620 and HK97 | E. coli | 104 bacteria/mL | - | 1.5 h | luminescence | - |
| [39] | virulent phage-typing (λ vir) | E. coli (K-12, MG1655) | 1 CFU/100 mL | - | 6–8 h | Electrochemical/amperometric | 102–105 with extended incubation time and 105–109 without time extension |
| [41] | T4 | E. coli K12 | 103 CFU/mL | Milk | - | Electrochemical/impedimetric | 103–108 CFU/mL |
| [40] | Filamentous phage (clone E2—displaying foreign peptide VTPPTQHQ | Salmonella typhimurium | 102 cells/mL | - | <180 s | Mechanical/QCM | 101–107 cells/mL |
| [38] | S. aureus bacteriophage | Staphylococcal and methicillin resistant (MRSA) and sensitive (MSSA) S. aureus species | 104 CFU/mL surface plasmon resonance | - | 16 min | Mechanical/QCM | - |
| [42] | T4 and BP14 phage was used to detect MRSA | E. coli O157:H7 and methicillin-resistant Staphylococcus aureus (MRSA) | 103 CFU/mL | 20 min | Optical/SPR | - |
| Ref. | MIP | Target Bacteria | NPs Used in the Sensor | NPs Function | LOD | Real Sample | Time | Detection Method | Range |
|---|---|---|---|---|---|---|---|---|---|
| [59] | Polypyrrole (PPy) | Pseudomonas aeruginosa | - | - | 103 CFU/mL | Apple juice | 3 min | Mechanical/QCM | 103 to 109 CFU/mL |
| [60] | - | E. coli | - | - | 1.54 × 106 CFU/mL, 3.72 × 105 CFU/mL with SPR and QCM | Apple juice | 113 s for SPR 56 s for QCM respectively, while respectively. | 1-Optical/SPR 2-Mechanical/QCM | 5.13 × 106 CFU/mL, 1.24 × 106 CFU/mL with SPR and QCM |
| [58] | 2,5-dimethyl-4-hydroxy-3(2H)-furanone (DMHF) | Aeromonas hydrophila and Pseudomonas aeruginosa | Magnetic Fe3O4@SiO2–NH2 (MNPs) | Faciliate separation | AHL LOD 8 × 10−10 mol L−1 | Bacteria supernatant spiked samples | - | Electrochemical/ Differential Pulse Voltammetry (DPV) | 2.5 × 10−9–1.0 × 10−7 mol/L |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mustafa, F.; Hassan, R.Y.A.; Andreescu, S. Multifunctional Nanotechnology-Enabled Sensors for Rapid Capture and Detection of Pathogens. Sensors 2017, 17, 2121. https://doi.org/10.3390/s17092121
Mustafa F, Hassan RYA, Andreescu S. Multifunctional Nanotechnology-Enabled Sensors for Rapid Capture and Detection of Pathogens. Sensors. 2017; 17(9):2121. https://doi.org/10.3390/s17092121
Chicago/Turabian StyleMustafa, Fatima, Rabeay Y. A. Hassan, and Silvana Andreescu. 2017. "Multifunctional Nanotechnology-Enabled Sensors for Rapid Capture and Detection of Pathogens" Sensors 17, no. 9: 2121. https://doi.org/10.3390/s17092121
APA StyleMustafa, F., Hassan, R. Y. A., & Andreescu, S. (2017). Multifunctional Nanotechnology-Enabled Sensors for Rapid Capture and Detection of Pathogens. Sensors, 17(9), 2121. https://doi.org/10.3390/s17092121