Meat and Fish Freshness Assessment by a Portable and Simplified Electronic Nose System (Mastersense)
Abstract
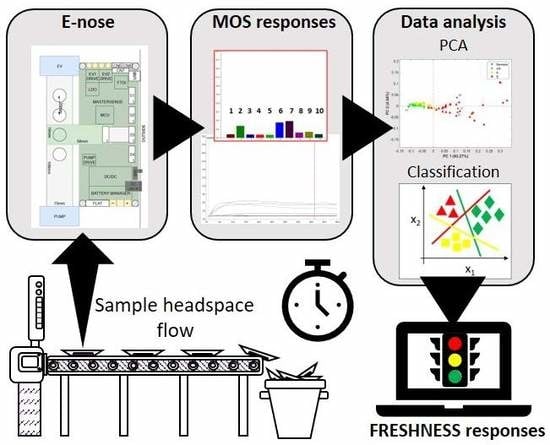
:1. Introduction
2. Materials and Methods
2.1. Electronic Nose System Development
2.1.1. Measuring Chamber
2.1.2. Acquisition System
- Off-line mode: the control software and the pattern recognition system are installed on a PC sending commands to the e-nose and receiving data; the connection could be wireless (WIFI) or wired (Ethernet).
- In-line: the control software and the pattern recognition system are included directly in the e-nose firmware. The e-nose sends the result of pattern recognition to PC to control the production line.
- On-line mode: different e-noses could be wireless connected to a cloud platform and send the collected data to a server via an internet connection. The control software and the pattern recognition system are in the cloud. The users will be able to access the data by a smart client application for computer, or by mobile applications for tablet and smartphone.
2.2. System Testing, Data Acquisition, and Laboration
2.2.1. Samples and Sample Preparation
2.2.2. E-nose Analysis
2.2.3. Microbial Analysis
2.2.4. Data Analysis and Predictive Model Development
3. Results and Discussion
3.1. Sensor Selection
3.2. Microbiological Analysis and Class Identification
- Unspoiled samples: ANOVA group “a”; microbial count < 106
- Acceptable samples: ANOVA group “b”; 106 < microbial count < 107
- Spoiled Samples: ANOVA “c; d; e”; microbial count > 107
3.3. PCA of Electronic Nose Data
3.4. Classification Model Development (KNN and PLS-DA)
3.4.1. K-NN Models
3.4.2. PLS-DA Models
3.4.3. Classification Results Comparison
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sen, D.P. Advaced in Fish Processing Technology; Allied Pub.: New Delhi, India, 2005; p. 848. [Google Scholar]
- Luning, P.A.; Jacxsens, L.; Rovira, J.; Osés, S.M.; Uyttendaele, M.; Marcelis, W.J. A concurrent diagnosis of microbiological food safety output and food safety management system performance: Cases from meat processing industries. Food Control 2011, 22, 555–565. [Google Scholar] [CrossRef]
- Commission Regulation (EC) N. On microbiological criteria for foodstuffs. Available online: https://www.fsai.ie/uploadedFiles/Reg2073_2005(1).pdf (accessed on 13 June 2019).
- Linee Guida per l’analisi del rischio nel campo della microbiologia degli alimenti. Available online: https://www.ceirsa.org/docum/allegato_punto4.pdf (accessed on 13 June 2019).
- Martinsdottir, E.; Luten, J.B.; Schelvis, A.A.M.; Hydig, G. Development of QIM-past and future. In Quality of Fish from Catch to Consumers; Luten, J.B., Oehlenschlager, J., Olafsdottir, G., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2003; pp. 265–272. [Google Scholar]
- Stutz, H.K.; Silverman, G.J.; Angelini, P.; Levin, R.E. Bacteria and Volatile Compounds Associated with Ground Beef Spoilage. J. Food Sci. 1991, 56, 1147–1153. [Google Scholar] [CrossRef]
- Wu, L.; Pu, H.; Sun, D.W. Novel techniques for evaluating freshness quality attributes of fish: A review of recent developments. Trends Food Sci. Technol. 2019, 83, 259–273. [Google Scholar] [CrossRef]
- Johnson, J.; Atkin, D.; Lee, K.; Sell, M.; Chandra, S. Determining meat freshness using electrochemistry: Are we ready for the fast and furious? Meat Sci. 2019, 150, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Applications of Electronic-Nose Technologies for Noninvasive Early Detection of Plant, Animal and Human Diseases. Chemosensors 2018, 6, 45–51. [Google Scholar] [CrossRef]
- Loutfi, A.; Coradeschi, S.; Mani, G.K.; Shankar, P.; Rayappan, J.B.B. Electronic noses for food quality: A review. J. Food Eng. 2015, 144, 103–111. [Google Scholar] [CrossRef]
- Deisingh, A.K.; Stone, D.C.; Thompson, M. Applications of electronic noses and tongues in food analysis. Int. J. Food Sci. Technol. 2004, 39, 587–604. [Google Scholar] [CrossRef]
- Gliszczyńska-Świgło, A.; Chmielewski, J. Electronic Nose as a Tool for Monitoring the Authenticity of Food. A Review. Food Anal. Methods 2017, 10, 1800–1816. [Google Scholar] [CrossRef]
- El Barbri, N.; Llobet, E.; El Bari, N.; Correig, X.; Bouchikhi, B. Electronic nose based on metal oxide semiconductor sensors as an alternative technique for the spoilage classification of red meat. Sensors 2008, 8, 142–156. [Google Scholar] [CrossRef]
- Boothe, D.D.H.; Arnold, J.W. Electronic nose analysis of volatile compounds from poultry meat samples, fresh and after refrigerated storage. J. Sci. Food Agric. 2002, 82, 315–322. [Google Scholar] [CrossRef]
- O’Connell, M.; Valdora, G.; Peltzer, G.; Martín Negri, R. A practical approach for fish freshness determinations using a portable electronic nose. Sens. Actuators B Chem. 2001, 80, 149–154. [Google Scholar] [CrossRef]
- Berna, A. Metal oxide sensors for electronic noses and their application to food analysis. Sensors 2010, 10, 3882–3910. [Google Scholar] [CrossRef] [PubMed]
- Ankara, Z.; Kammerer, T.; Gramm, A.; Schütze, A. Low power virtual sensor array based on a micromachined gas sensor for fast discrimination between H2, CO and relative humidity. Sens. Actuators B Chem. 2004, 100, 240–245. [Google Scholar] [CrossRef]
- Ramírez, H.L.; Soriano, A.; Gómez, S.; Iranzo, J.U.; Briones, A.I. Evaluation of the Food Sniffer electronic nose for assessing the shelf life of fresh pork meat compared to physicochemical measurements of meat quality. Eur. Food Res. Technol. 2018, 244, 1047–1055. [Google Scholar] [CrossRef]
- Ni, Y.; Kokot, S. Does chemometrics enhance the performance of electroanalysis? Anal. Chim. Acta 2008, 626, 130–146. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.H.; Sun, D.W.; Zeng, X.A.; Liu, D. Recent Advances in Methods and Techniques for Freshness Quality Determination and Evaluation of Fish and Fish Fillets: A Review. Crit. Rev. Food Sci. Nutr. 2015, 55, 1012–1225. [Google Scholar] [CrossRef] [PubMed]
- Najam ul, Hasan; Ejaz, N.; Ejaz, W.; Kim, H.S. Meat and fish freshness inspection system based on odor sensing. Sensors 2012, 12, 15542–15557. [Google Scholar]
- Natale, C.D.; Olafsdottir, G.; Einarsson, S.; Martinelli, E.; Paolesse, R.; D’Amico, A. Comparison and integration of different electronic noses for freshness evaluation of cod-fish fillets. Sens. Actuators B Chem. 2001, 77, 572–578. [Google Scholar] [CrossRef]
- UST Umweltsensortechnik GmbH sensors. Available online: http://www.umweltsensortechnik.de/en/gas-sensors/mox-gas-sensors-overview.html (accessed on 13 June 2019).
- Brooks, J.C.; Alvarado, M.; Stephens, T.P.; Kellermeier, J.D.; Tittor, A.W.; Miller, M.F.; Brashears, M.M. Spoilage and Safety Characteristics of Ground Beef Packaged in Traditional and Modified AtmospherePackages. J. Food Prot. 2016, 71, 293–301. [Google Scholar] [CrossRef]
- ISO 4833-2: Microbiology of the Food Chain-Horizontal Method for the Enumeration of Microorganisms: Part 1: Colony Count at 30 °C by the Pour Plate Technique. 2013. Available online: https://www.iso.org/standard/53728.html (accessed on 13 June 2019).
- Wold, S.; Esbensen, K.; Geladi, P. Priciple component analysis. Chemom Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Wu, Y.; Ianakiev, K.; Govindaraju, V. Improved k-nearest neighbor classification. Pattern Recognit. 2002, 35, 2311–2318. [Google Scholar] [CrossRef]
- Ballabio, D.; Consonni, V. Classification tools in chemistry. Part 1: Linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer Aided Design of Experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Roggo, Y.; Duponchel, L.; Huvenne, J.P. Comparison of supervised pattern recognition methods with McNemar’s statistical test: Application to qualitative analysis of sugar beet by near-infrared spectroscopy. Anal. Chim. Acta 2003, 477, 187–200. [Google Scholar] [CrossRef]
- Grassi, S.; Casiraghi, E.; Alamprese, C. Handheld NIR device: A non-targeted approach to assess authenticity of fish fillets and patties. Food Chem. 2018, 243, 382–388. [Google Scholar] [CrossRef]
- Panigrahi, S.; Balasubramanian, S.; Gu, H.; Logue, C.M.; Marchello, M. Design and development of a metal oxide based electronic nose for spoilage classification of beef. Sens. Actuators B Chem. 2006, 119, 2–14. [Google Scholar] [CrossRef]
- Panigrahi, S.; Balasubramanian, S.; Gu, H.; Logue, C.; Marchello, M. Neural-network-integrated electronic nose system for identification of spoiled beef. LWT-Food Sci. Technol. 2006, 3, 135–145. [Google Scholar] [CrossRef]
- Olafsdottir, G.; Lauzon, H.L.; Martinsdóttir, E.; Oehlenschläger, J.; Kristbergsson, K. Evaluation of shelf life of superchilled cod (Gadus morhua) fillets and the influence of temperature fluctuations during storage on microbial and chemical quality indicators. J. Food Sci. 2006, 71, 97–109. [Google Scholar] [CrossRef]




| Sensor Name | Sensor Type | Sensor Sensitivity |
|---|---|---|
| S1 | GGS 8530 | Sensor for the detection of C2H5OH, with low cross-sensitivity to CH4, CO and H2 |
| S2 | GGS 5430 | Sensor especially sensitive to NO2 (nitrogen dioxide) and O3 (ozone) |
| S3 | GGS 7330 | Sensor for the detection of NOX |
| S4 | GGS 6530 | Sensor for the detection of H2, with low cross-sensitivity to CH4, CO and alcohol |
| S5 | GGS 3530 | Sensor for the detection of hydrocarbonates, optimal for C1 C8-hydrocarbonate |
| S6 | GGS 2530 | Sensor with a high sensitivity to CO, H2 and C2H5OH and a low cross-sensitivity to CH4 |
| S7 | GGS10530 | Sensor for the detection of selected VOCs in the trace range |
| S8 | GGS 1530 | Universal sensor with many applications |
| S9 | GGS 4430 | Sensor for NH3 (ammonia), with low cross-sensitivity to CH4, CO and H2 |
| S10 | GGS 1430 | Universal sensor with many applications |
| BEEF | POULTRY | SALMON | EUROPEAN PLAICE | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Series 1 | Series 2 | Series 3 | Series 1 | Series 2 | Series 3 | Series 1 | Series 2 | Series 1 | Series 2 | |
| Times (days) from the day of packaging | 0 | 0 | 0 | - | - | 0 | - | - | - | - |
| 1 | - | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 2 | - | - | 2 | - | 2 | 2 | 2 | 2 | 2 | |
| 3 | 3 | 3 | 3 | - | 3 | 3 | 3 | 3 * | 3 * | |
| 4 | 4 | 4 | 4 | 4 | - | 4 * | 4 * | 4 | 4 | |
| - | 5 | 5 | - | 5 | - | 5 | 5 | 5 | 5 | |
| - | 6 | - | - | 6 | - | - | - | - | - | |
| 7 * | 7 * | - | 7 * | 7 * | - | - | - | - | - | |
| 9 | - | - | 8 | 8 | - | 8 | 8 | - | - | |
| 8 | - | - | 9 | - | - | - | - | - | - | |
| 10 | 10 | - | 10 | - | - | - | - | - | - | |
| - | - | - | - | 11 | - | - | - | - | - | |
| A-Priori | ||||
|---|---|---|---|---|
| Class 1 | Class 2 | |||
| Predicted | Class 1 | TP | FP | |
| Class 2 | FN | VN | ||
| BEEF | POULTRY | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Series | Time (Days) | CFU/g | ANOVA LSD * | Group | Series | Time (Days) | CFU/g | ANOVA LSD * | Group |
| 1 | 0 | 1.60 × 104 | a | Unspoiled | 1 | 1 | 3.40 × 104 | ab | Unspoiled |
| 1 | 1 | 1.76 × 105 | a | Unspoiled | 1 | 2 | 4.03 × 105 | b | Unspoiled |
| 1 | 2 | 6.50 × 103 | a | Unspoiled | 1 | 3 | 1.24 × 106 | bc | Acceptable |
| 1 | 3 | 4.70 × 104 | a | Unspoiled | 1 | 4 | 1.25 × 107 | bc | Acceptable |
| 1 | 4 | 2.25 × 105 | a | Unspoiled | 1 | 7 | 2.62 × 108 | d | Spoiled |
| 1 | 7 | 8.36 × 106 | b | Acceptable | 1 | 8 | 2.51 × 109 | e | Spoiled |
| 1 | 8 | 1.54 × 108 | d | Spoiled | 1 | 9 | 5.71 × 109 | f | Spoiled |
| 1 | 9 | 2.37 × 109 | e | Spoiled | 1 | 10 | 1.66 × 1010 | f | Spoiled |
| 1 | 10 | 9.20 × 109 | e | Spoiled | 2 | 1 | 4.00 × 105 | b | Unspoiled |
| 2 | 0 | 1.50 × 104 | a | Unspoiled | 2 | 4 | 3.60 × 106 | bc | Acceptable |
| 2 | 3 | 2.52 × 105 | a | Unspoiled | 2 | 5 | 7.00 × 107 | c | Spoiled |
| 2 | 4 | 5.54 × 105 | a | Unspoiled | 2 | 6 | 2.83 × 108 | d | Spoiled |
| 2 | 5 | 5.27 × 106 | b | Acceptable | 2 | 7 | 1.94 × 109 | e | Spoiled |
| 2 | 6 | 2.08 × 107 | c | Spoiled | 2 | 8 | 2.60 × 109 | e | Spoiled |
| 2 | 7 | 1.56 × 108 | d | Spoiled | 2 | 11 | 7.57 × 108 | de | Spoiled |
| 2 | 10 | 5.15 × 109 | e | Spoiled | 3 | 0 | 3.35 × 104 | ab | Unspoiled |
| 3 | 0 | 2.00 × 105 | a | Unspoiled | 3 | 0 | 4.67 × 103 | a | Unspoiled |
| 3 | 3 | 4.22 × 106 | b | Acceptable | 3 | 1 | 1.25 × 104 | ab | Unspoiled |
| 3 | 4 | 3.30 × 106 | b | Acceptable | 3 | 1 | 2.53 × 104 | ab | Unspoiled |
| 3 | 5 | 2.04 × 108 | d | Spoiled | 3 | 2 | 2.03 × 104 | ab | Unspoiled |
| * Different letters in each column indicate significant difference at 95% confidence levels as obtained by LSD test. | 3 | 2 | 2.43 × 104 | ab | Unspoiled | ||||
| 3 | 3 | 2.25 × 104 | ab | Unspoiled | |||||
| EUROPEAN PLAICE | SALMON | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Series | Time (days) | CFU/g | ANOVA LSD * | Group | Series | Time (days) | CFU/g | ANOVA LSD * | Group |
| 1 | 1 | 7.20 × 106 | b | Acceptable | 1 | 1 | 6.09 × 104 | a | Unspoiled |
| 1 | 1 | 1.13 × 105 | a | Unspoiled | 1 | 1 | 8.05 × 104 | a | Unspoiled |
| 1 | 2 | 1.19 × 107 | b | Acceptable | 1 | 2 | 1.46 × 105 | a | Unspoiled |
| 1 | 2 | 1.75 × 107 | b | Acceptable | 1 | 2 | 1.10 × 105 | a | Unspoiled |
| 1 | 3 | 2.07 × 108 | e | Spoiled | 1 | 3 | 1.36 × 107 | b | Acceptable |
| 1 | 3 | 1.87 × 108 | e | Spoiled | 1 | 3 | 1.10 × 107 | b | Acceptable |
| 1 | 4 | 1.53 × 108 | d | Spoiled | 1 | 4 | 1.49 × 106 | ab | Unspoiled |
| 1 | 4 | 1.96 × 108 | e | Spoiled | 1 | 4 | 1.30 × 107 | bc | Acceptable |
| 1 | 5 | 9.67 × 107 | c | Spoiled | 1 | 5 | 1.85 × 107 | bc | Acceptable |
| 1 | 5 | 1.24 × 108 | c | Spoiled | 1 | 5 | 2.90 × 107 | bc | Acceptable |
| 2 | 1 | 3.23 × 106 | ab | Unspoiled | 1 | 8 | 4.15 × 108 | d | Spoiled |
| 2 | 1 | 2.14 × 106 | ab | Unspoiled | 1 | 8 | 3.41 × 108 | d | Spoiled |
| 2 | 1 | 1.77 × 106 | ab | Unspoiled | 2 | 1 | 1.56 × 105 | a | Unspoiled |
| 2 | 2 | 1.18 × 107 | b | Acceptable | 2 | 1 | 1.04 × 105 | a | Unspoiled |
| 2 | 2 | 1.68 × 107 | b | Acceptable | 2 | 2 | 6.10 × 105 | ab | Unspoiled |
| 2 | 3 | 2.31 × 107 | b | Acceptable | 2 | 2 | 5.10 × 104 | a | Unspoiled |
| 2 | 3 | 1.52 × 107 | b | Acceptable | 2 | 3 | 8.52 × 105 | ab | Unspoiled |
| 2 | 4 | 2.14 × 108 | e | Spoiled | 2 | 3 | 1.17 × 106 | ab | Unspoiled |
| 2 | 4 | 2.91 × 108 | f | Spoiled | 2 | 4 | 3.40 × 107 | c | Acceptable |
| 2 | 5 | 1.51 × 109 | h | Spoiled | 2 | 4 | 2.17 × 107 | c | Acceptable |
| 2 | 5 | 5.25 × 108 | g | Spoiled | 2 | 5 | 1.01 × 108 | d | Spoiled |
| * Different letters in each column indicate significant difference at 95% confidence levels as obtained by LSD test. | 2 | 5 | 1.23 × 108 | d | Spoiled | ||||
| 2 | 8 | 2.69 × 109 | e | Spoiled | |||||
| 2 | 8 | 8.57 × 108 | de | Spoiled | |||||
| BEEF | POULTRY | EUROPEAN PLAICE | SALMON | |
|---|---|---|---|---|
| (CFU/g) | (CFU/g) | (CFU/g) | (CFU/g) | |
| Green | ≤106 | ≤106 | ≤3 × 106 | ≤1.5 × 106 |
| Yellow | 106 < x ≤ 107 | 106 < x ≤ 1.2 × 107 | 3 × 106 < x ≤ 5 × 107 | 1.5 × 106 < x ≤ 5 × 107 |
| Red | >107 | >1.2 × 107 | >5 × 107 | >5 × 107 |
| CALIBRATION | CROSS-VALIDATION | PREDICTION | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Class | US | A | S | US | A | S | US | A | S |
| BEEF | |||||||||
| Samples | 34 | 13 | 8 | 34 | 13 | 8 | 2 | 3 | 20 |
| Sensitivity | 0.91 | 0.77 | 0.75 | 0.94 | 0.77 | 0.75 | 0.100 | 0.67 | 0.95 |
| Specificity | 0.86 | 0.88 | 1.00 | 0.86 | 0.90 | 1.00 | 0.95 | 0.95 | 1.00 |
| p-values | 0.91 | 0.81 | 0.86 | 0.91 | 0.81 | 0.86 | 1.00 | 0.80 | 1.00 |
| POULTRY | |||||||||
| Samples | 37 | 10 | 11 | 37 | 10 | 11 | 3 | 2 | 25 |
| Sensitivity | 0.89 | 0.70 | 0.91 | 0.92 | 0.70 | 0.91 | 0.66 | 1.00 | 0.84 |
| Specificity | 0.86 | 0.89 | 1.00 | 0.86 | 0.92 | 1.00 | 1.00 | 0.82 | 1.00 |
| p-values | 0.91 | 0.78 | 1.00 | 0.91 | 0.78 | 1.00 | 0.91 | 0.90 | 1.00 |
| EUROPEAN PLAICE | |||||||||
| Samples | 11 | 14 | 18 | 11 | 14 | 18 | 1 | 7 | 12 |
| Sensitivity | 1.00 | 0.79 | 0.94 | 1.00 | 0.71 | 0.89 | 1.00 | 0.75 | 1.00 |
| Specificity | 0.97 | 0.97 | 0.92 | 0.97 | 0.93 | 0.88 | 1.00 | 1.00 | 0.78 |
| p-values | 0.92 | 0.92 | 0.90 | 0.92 | 0.91 | 0.89 | 1.00 | 1.00 | 0.95 |
| SALMON | |||||||||
| Samples | 29 | 13 | 5 | 29 | 13 | 5 | 4 | 8 | 13 |
| Sensitivity | 0.97 | 0.85 | 0.80 | 0.93 | 0.85 | 0.80 | 0.75 | 1.00 | 0.92 |
| Specificity | 0.89 | 0.94 | 1.00 | 0.89 | 0.91 | 1.00 | 1.00 | 0.88 | 1.00 |
| p-values | 0.93 | 0.84 | 1.00 | 0.93 | 0.83 | 1.00 | 0.96 | 0.96 | 1.00 |
| CALIBRATION | CROSS-VALIDATION | PREDICTION | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Class | US | A | S | US | A | S | US | A | S |
| BEEF | |||||||||
| Samples | 34 | 13 | 8 | 34 | 13 | 8 | 2 | 3 | 20 |
| Sensitivity | 0.82 | 0.69 | 0.88 | 0.82 | 0.54 | 0.88 | 1.00 | 0.67 | 1.00 |
| Specificity | 0.91 | 0.86 | 0.94 | 0.81 | 0.86 | 0.94 | 1.00 | 1.00 | 0.80 |
| p-values | 0.93 | 0.80 | 0.80 | 0.90 | 0.80 | 0.80 | 1.00 | 1.00 | 0.95 |
| POULTRY | |||||||||
| Samples | 37 | 10 | 11 | 37 | 10 | 11 | 3 | 2 | 25 |
| Sensitivity | 0.81 | 0.80 | 0.91 | 0.84 | 0.70 | 0.91 | 0.67 | 1.00 | 0.92 |
| Specificity | 1.00 | 0.83 | 0.96 | 1.00 | 0.85 | 0.94 | 1.00 | 0.89 | 1.00 |
| p-values | 1.00 | 0.80 | 0.76 | 1.00 | 0.80 | 0.76 | 1.00 | 0.80 | 1.00 |
| EUROPEAN PLAICE | |||||||||
| Samples | 11 | 14 | 18 | 11 | 14 | 18 | 1 | 7 | 12 |
| Sensitivity | 1.00 | 0.93 | 0.83 | 1.00 | 0.79 | 0.78 | 1.00 | 1.00 | 0.92 |
| Specificity | 1.00 | 0.90 | 0.96 | 1.00 | 0.86 | 0.88 | 1.00 | 0.92 | 1.00 |
| p-values | 1.00 | 0.81 | 0.93 | 1.00 | 0.73 | 0.82 | 1.00 | 0.88 | 1.00 |
| SALMON | |||||||||
| Samples | 29 | 13 | 5 | 29 | 13 | 5 | 4 | 8 | 13 |
| Sensitivity | 0.97 | 0.92 | 1.00 | 0.97 | 0.85 | 0.40 | 1.00 | 0.75 | 100 |
| Specificity | 1.00 | 0.97 | 0.98 | 1.00 | 0.88 | 0.95 | 1.00 | 1.00 | 0.83 |
| p-values | 1.00 | 0.92 | 0.83 | 1.00 | 0.90 | 0.80 | 1.00 | 1.00 | 0.86 |
| Model | Beef | Poultry | European Plaice | Salmon | |
|---|---|---|---|---|---|
| Sensitivity | KNN | 0.92 | 0.83 | 0.91 | 0.92 |
| PLS-DA | 0.96 | 0.90 | 0.95 | 0.92 | |
| Specificity | KNN | 0.97 | 0.93 | 0.92 | 0.96 |
| PLS-DA | 0.84 | 0.99 | 0.97 | 0.91 | |
| E | KNN | 0.12 | 0.17 | 0.10 | 0.08 |
| PLS-DA | 0.04 | 0.10 | 0.05 | 0.08 | |
| p-values | 0.25 | 0.25 | 0.625 | 1 | |
| H0: KNN = PLS-DA | Equal predictive accuracies | Equal predictive accuracies | Equal predictive accuracies | Equal predictive accuracies | |
| E, classification loss that summarises the accuracy of the classes predicted by k-NN or PLS-DA, p-value, p-value of the test, H0, Hypothesis test. | |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grassi, S.; Benedetti, S.; Opizzio, M.; di Nardo, E.; Buratti, S. Meat and Fish Freshness Assessment by a Portable and Simplified Electronic Nose System (Mastersense). Sensors 2019, 19, 3225. https://doi.org/10.3390/s19143225
Grassi S, Benedetti S, Opizzio M, di Nardo E, Buratti S. Meat and Fish Freshness Assessment by a Portable and Simplified Electronic Nose System (Mastersense). Sensors. 2019; 19(14):3225. https://doi.org/10.3390/s19143225
Chicago/Turabian StyleGrassi, Silvia, Simona Benedetti, Matteo Opizzio, Elia di Nardo, and Susanna Buratti. 2019. "Meat and Fish Freshness Assessment by a Portable and Simplified Electronic Nose System (Mastersense)" Sensors 19, no. 14: 3225. https://doi.org/10.3390/s19143225
APA StyleGrassi, S., Benedetti, S., Opizzio, M., di Nardo, E., & Buratti, S. (2019). Meat and Fish Freshness Assessment by a Portable and Simplified Electronic Nose System (Mastersense). Sensors, 19(14), 3225. https://doi.org/10.3390/s19143225