Zinc Binding to NAP-Type Neuroprotective Peptides: Nuclear Magnetic Resonance Studies and Molecular Modeling
Abstract
:1. Introduction
2. Results
2.1. Mass Spectrometric Studies
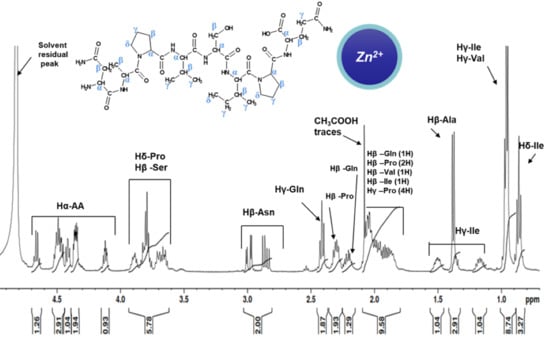
2.2. NMR Study
2.3. Molecular Modeling
3. Discussion
4. Materials and Methods
4.1. Peptide Synthesis
4.2. Instruments
4.2.1. Mass Spectrometry
4.2.2. NMR Spectroscopy
4.2.3. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jamerlan, A.; An, S.S.A. The influence of Aβ-dependent and independent pathways on TDP-43 proteinopathy in Alzheimer’s disease: A possible connection to LATE-NC. Neurobiol. Aging 2020, 95, 161–167. [Google Scholar] [CrossRef]
- Xing, S.; Lu, Y. P3-199: Comprehensive understanding of tau exon 10 alternative splicing regulation. Alzheimer’s Dement. 2018, 14, 1142–1143. [Google Scholar] [CrossRef]
- Isik, A.T. Late onset Alzheimer’s disease in older people. Clin. Interv. Aging 2010, 5, 307–311. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.P.; Xie, Y.; Meng, X.Y.; Kang, J.S. History and progress of hypotheses and clinical trials for Alzheimer’s disease. Signal Transd. Targ. Ther. 2019, 4, 1–22. [Google Scholar] [CrossRef]
- Viles, J.H. Metal ions and amyloid fiber formation in neurodegenerative diseases. Copper, zinc and iron in Alzheimer’s, Parkinson’s and prion diseases. Coord. Chem. Rev. 2012, 256, 2271–2284. [Google Scholar] [CrossRef]
- Ben-Shushan, S.; Miller, Y. Neuropeptides: Roles and activities as metal chelators in neurodegenerative diseases. J. Phys. Chem. B 2021, 125, 2796–2811. [Google Scholar] [CrossRef]
- Morris, D.R.; Levenson, C.W. Ion channels and zinc: Mechanisms of neurotoxicity and neurodegeneration. J. Toxicol. 2012, 2012, 785647. [Google Scholar] [CrossRef]
- Datki, Z.; Galik-Olah, Z.; Janosi-Mozes, E.; Szegedi, V.; Kalman, J.; Hunya, Á.G.; Fulop, L.; Tamano, H.; Takeda, A.; Adlard, P.A.; et al. Alzheimer risk factors age and female sex induce cortical Aβ aggregation by raising extracellular zinc. Mol. Psychiatr. 2020, 25, 2728–2741. [Google Scholar] [CrossRef] [PubMed]
- Ayton, S.; Lei, P.; Bush, A.I. Metallostasis in Alzheimer’s disease. Free Radic. Biol. Med. 2013, 62, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Chasapis, C.T.; Ntoupa, P.S.A.; Spiliopoulou, C.A.; Stefanidou, M.E. Recent aspects of the effects of zinc on human health. Arch. Toxicol. 2020, 94, 1443–1460. [Google Scholar] [CrossRef]
- Olechnowicz, J.; Tinkov, A.; Skalny, A.; Suliburska, J. Zinc status is associated with inflammation, oxidative stress, lipid, and glucose metabolism. J. Physiol. Sci. 2018, 68, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Kulikova, A.A.; Makarov, A.A.; Kozin, S.A. Roles of zinc ions and structural polymorphism of β-amyloid in the development of Alzheimer’s disease. Mol. Biol. 2015, 49, 217–230. [Google Scholar] [CrossRef]
- Eisele, Y.S.; Bolmont, T.; Heikenwalder, M.; Langer, F.; Jacobson, L.H.; Yan, Z.X.; Roth, K.; Aguzzi, A.; Staufenbiel, M.; Walker, L.C.; et al. Induction of cerebral β-amyloidosis: Intracerebral versus systemic Aβ inoculation. Proc. Natl. Acad. Sci. USA 2009, 106, 12926–12931. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Wu, H.; Zhao, J. Multifunctional roles of zinc in Alzheimer’s disease. Neurotoxicology 2020, 80, 112–123. [Google Scholar] [CrossRef]
- Isaev, N.K.; Stelmashook, E.V.; Genrikhs, E.E. Role of zinc and copper ions in the pathogenetic mechanisms of traumatic brain injury and Alzheimer’s disease. Rev. Neurosci. 2020, 31, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Gozes, I. Activity-dependent neuroprotective protein (ADNP)/NAP (CP201): Autism, schizophrenia, and Alzheimer’s disease. In Neuroprotection in Autism, Schizophrenia and Alzheimer’s Disease; Academic Press: Cambridge, MA, USA, 2019; pp. 3–20. [Google Scholar] [CrossRef]
- Cacabelos, R.; Cacabelos, P.; Torrellas, C.; Tellado, I.; Carril, J.C. Pharmacogenomics of Alzheimer’s disease: Novel therapeutic strategies for drug development. In Pharmacogenomics in Drug Discovery and Development; Yan, Q., Ed.; Springer: New York, NY, USA, 2014; Volume 1175, pp. 323–556. [Google Scholar] [CrossRef]
- Heimesaat, M.M.; Mousavi, S.; Kløve, S.; Genger, C.; Weschka, D.; Giladi, E. Immune-modulatory properties of the octapeptide nap in campylobacter jejuni infected mice suffering from acute enterocolitis. Microorganisms 2020, 8, 802. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Gray, A.J.; Hirata-Fukae, C.; Minami, S.S.; Waterhouse, E.G.; Mattson, M.P.; LaFerla, F.M.; Gozes, I.; Aisen, P.S. Intranasal NAP administration reduces accumulation of amyloid peptide and tau hyperphosphorylation in a transgenic mouse model of Alzheimer’s disease at early pathological stage. J. Mol. Neurosci. 2007, 31, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Divinski, I.; Mittelman, L.; Gozes, I. A femtomolar acting octapeptide interacts with tubulin and protects astrocytes against zinc intoxication. J. Biol. Chem. 2004, 279, 28531–28538. [Google Scholar] [CrossRef] [Green Version]
- Kitai, S.T.; Shepard, P.D.; Callaway, J.C.; Scroggs, R. Afferent modulation of dopamine neuron firing patterns. Curr. Opin. Neurobiol. 1999, 9, 690–697. [Google Scholar] [CrossRef]
- Ciobanu, C.I.; Stefanescu, R.; Niculaua, M.; Teslaru, T.; Gradinaru, R.; Drochioiu, G. Mass spectrometric evidence for iron binding to the neuroprotective peptide NAP and its Cys5 mutant. Eur. J. Mass Spectrom. 2016, 22, 97–104. [Google Scholar] [CrossRef]
- Lupaescu, A.V.; Jureschi, M.; Ciobanu, C.I.; Ion, L.; Zbancioc, G.; Petre, B.A.; Drochioiu, G. FTIR and MS evidence for heavy metal binding to anti-amyloidal NAP-like peptides. Int. J. Pept. Res. Ther. 2019, 25, 303–309. [Google Scholar] [CrossRef]
- Lupaescu, A.V.; Sandu, I.; Petre, B.A.; Ion, L.; Ciobanu, C.I.; Drochioiu, G. Nap neuroprotective peptide and its analogs: Simultaneously copper and iron binding and reduction. Rev. Chim. 2019, 70, 1784–1790. [Google Scholar] [CrossRef]
- Moriyasu, K.; Ichinose, T.; Nakahata, A.; Tanaka, M.; Matsui, T.; Furuya, S. The dipeptides Ile-Tyr and Ser-Tyr exert distinct effects on catecholamine metabolism in the mouse brainstem. Int. J. Pep. 2016, 2016, 6020786. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Kiyohara, H.; Yoshino, A.; Nakano, A.; Takata, F.; Dohgu, S.; Kataoka, Y.; Matsui, T. Brain-transportable soy dipeptide, Tyr-Pro, attenuates amyloid β peptide 25-35-induced memory impairment in mice. NPJ Sci. Food 2020, 4, 1–4. [Google Scholar] [CrossRef]
- Suprun, E.V.; Zaryanov, N.V.; Radko, S.P.; Kulikova, A.A.; Kozin, S.A.; Makarov, A.A.; Archakov, A.I.; Shumyantseva, V.V. Tyrosine based electrochemical analysis of amyloid-β fragment (1-16) binding to metal (II) ions. Electrochim. Acta 2015, 179, 93–99. [Google Scholar] [CrossRef]
- Yamauchi, O.; Odani, A.; Takani, M. Metal-amino acid chemistry. Weak interactions and related functions of side chain groups. J. Chem. Soc. Dalton Trans. 2002, 18, 3411–3421. [Google Scholar] [CrossRef]
- Ohata, J.; Miller, M.K.; Mountain, C.M.; Vohidov, F.; Ball, Z.T. A three-component organometallic tyrosine bioconjugation. Angew. Chem. Int. Ed. 2018, 57, 2827–2830. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, L.; Fernandez, F.; Johnson, J.B.; Naiker, M.; Owoola, A.G.; Broszczak, D.A. Oxidative stress in alzheimer’s disease: A review on emergent natural polyphenolic therapeutics. Complement. Ther. Med. 2020, 49, 102294. [Google Scholar] [CrossRef]
- Mocanu, C.S.; Bocec, A.S.; Gradinaru, V.R.; Anton-Paduraru, D.T. A biochemical method for tyrosine determination in phenylketonuria using a colorimetric enzymatic approach. Rev. Chim. 2020, 71, 285–294. [Google Scholar] [CrossRef]
- Mocanu, C.S.; Drochioiu, G. The interaction of possible anti-AD ASA-NAP peptide conjugate with tubulin: A theoretical and experimental insight. Int. J. Pept. Res. Ther. 2021, 1–17. [Google Scholar] [CrossRef]
- Mocanu, C.S.; Jureschi, M.; Drochioiu, G. Aluminium binding to modified amyloid-β peptides: Implications for alzheimer’s disease. Molecules 2020, 25, 4536. [Google Scholar] [CrossRef]
- Lupaescu, A.V.; Ciobanu, C.I.; Humelnicu, I.; Petre, B.A.; Murariu, M.; Drochioiu, G. Design and synthesis of new anti-amyloid NAP-based/like peptides. Rev. Roum. Chim. 2019, 64, 535–546. [Google Scholar] [CrossRef]
- Ciobanu, C.I.; Lupaescu, A.V.; Drochioiu, G. Study of zinc binding to neuroprotective peptides. In Proceedings of the 19th International Multidisciplinary Scientific GeoConference (SGEM 2019), Albena, Bulgaria, 28 June–7 July 2019; pp. 905–912. [Google Scholar] [CrossRef]
- Ekblad, B.; Kristiansen, P.E. NMR structures and mutational analysis of the two peptides constituting the bacteriocin plantaricin S. Sci. Rep. 2019, 9, 2333. [Google Scholar] [CrossRef] [Green Version]
- Danielsson, J.; Pierattelli, R.; Banci, L.; Gräslund, A. High-resolution NMR studies of the zinc-binding site of the Alzheimer’s amyloid beta-peptide. FEBS J. 2007, 274, 46–59. [Google Scholar] [CrossRef]
- Syme, C.D.; Viles, J.H. Solution 1H NMR investigation of Zn2+ and Cd2+ binding to amyloid-beta peptide (Abeta) of Alzheimer’s disease. Biochim. Biophys. Acta 2006, 1764, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Barranco, E.; Martín, N.; Segura, J.L.; Seoane, C.; De la Cruz, P.; Langa, F.; Gonzalez, A.; Pingarrón, J.M. Syntheses, electrochemistry and molecular modeling of N,N’-dicyanoquinonediimine (DCNQI) derivatives of substituted 1,4-anthracenediones: Precursors for organic metals. Tetrahedron 1993, 49, 4881–4892. [Google Scholar] [CrossRef]
- Auchus, R.J.; Miller, W.L. Molecular modeling of human P450c17 (17α-Hydroxylase/ 17,20-Lyase): Insights into reaction mechanisms and effects of mutations. J. Mol. Endocrinol. 1999, 13, 1169–1182. [Google Scholar] [CrossRef] [PubMed]
- Leker, R.R.; Teichner, A.; Grigoriadis, N.; Ovadia, H.; Brenneman, D.E.; Fridkin, M.; Giladi, E.; Romano, J.; Gozes, I. NAP, a femtomolar-acting peptide, protects the brain against ischemic injury by reducing apoptotic death. Stroke 2002, 33, 1085–1092. [Google Scholar] [CrossRef] [Green Version]
- Sragovich, S.; Ziv, Y.; Vaisvaser, S.; Shomron, N.; Hendler, T.; Gozes, I. The autism-mutated ADNP plays a key role in stress response. Transl. Psychiatry 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Roychaudhuri, R.; Yang, M.; Hoshi, M.M.; Teplow, D.B. Amyloid β-protein assembly and Alzheimer disease. J. Biol. Chem. 2009, 284, 4749–4753. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Yin, Y.L.; Liu, X.Z.; Shen, P.; Zheng, Y.G.; Lan, X.R.; Lu, C.B.; Wang, J.Z. Current understanding of metal ions in the pathogenesis of Alzheimer’s disease. Transl. Neurodegener. 2020, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sensi, S.L.; Granzotto, A.; Siotto, M.; Squitti, R. Copper and zinc dysregulation in alzheimer’s disease. Trends Pharmacol. Sci. 2018, 39, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Al-Shammari, N.; Savva, L.; Kennedy-Britten, O.; Platts, J.A. Forcefield evaluation and accelerated molecular dynamics simulation of Zn(II) binding to N-terminus of amyloid-β. Comput. Biol. Chem. 2021, 93, 107540. [Google Scholar] [CrossRef]
- Zirah, S.; Kozin, S.A.; Mazur, A.K.; Blond, A.; Cheminant, M.; Ségalas-Milazzo, I.; Debey, P.; Rebuffat, S. Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging. J. Biol. Chem. 2006, 281, 2151–2161. [Google Scholar] [CrossRef] [Green Version]
- Gaggelli, E.; Janicka-Klos, A.; Jankowska, E.; Kozlowski, H.; Migliorini, C.; Molteni, E.; Valensin, D.; Valensin, G.; Wieczerzak, E. NMR studies of the Zn2+ interactions with rat and human beta-amyloid (1–28) peptides in water-micelle environment. J. Phys. Chem. B 2008, 112, 100–109. [Google Scholar] [CrossRef]
- Trivunac, K.; Stevanovic, S. Effects of operating parameters on efficiency of cadmium and zinc removal by the complexation–filtration process. Desalination 2006, 198, 282–287. [Google Scholar] [CrossRef]
- Suprun, E.V.; Shumyantseva, V.V.; Archakov, A.I. Protein electrochemistry: Application in medicine. A review. Electrochim. Acta 2014, 140, 72–82. [Google Scholar] [CrossRef]
- Ben-Naim, A. The role of hydrogen bonds in protein folding and protein association. J. Phys. Chem. 1991, 95, 1437–1444. [Google Scholar] [CrossRef]
- Mallamace, D.; Fazio, E.; Mallamace, F.; Corsaro, C. The role of hydrogen bonding in the folding/unfolding process of hydrated lysozyme: A review of recent NMR and FTIR results. Int. J. Mol. Sci. 2018, 19, 3825. [Google Scholar] [CrossRef] [Green Version]
- Ganesan, S.J.; Matysiak, S. Interplay between the hydrophobic effect and dipole interactions in peptide aggregation at interfaces. Phys. Chem. Chem. Phys. 2016, 18, 2449–2458. [Google Scholar] [CrossRef]
- Festa, G.; Mallamace, F.; Sancesario, G.M.; Corsaro, C.; Mallamace, D.; Fazio, E. Aggregation states of Aβ1–40, Aβ1–42 and Aβp3–42 amyloid beta peptides: A SANS study. Int. J. Mol. Sci. 2019, 20, 4126. [Google Scholar] [CrossRef] [Green Version]
- Thirumalai, D.; Reddy, G.; Straub, J.E. Role of water in protein aggregation and amyloid polymorphism. ACC. Chem. Res. 2012, 45, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.W.; Knowles, T.P.; Waudby, C.A.; Vendruscolo, M.; Dobson, C.M. Inversion of the balance between hydrophobic and hydrogen bonding interactions in protein folding and aggregation. PLoS Comput. Biol. 2011, 10, 1002169. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Ju, M.; Hyun Cho, O.; Kim, Y.; Nam, K. Tyrosine-rich peptides as a platform for assembly and material synthesis. Adv. Sci. 2019, 6, 1801255. [Google Scholar] [CrossRef] [Green Version]
- Pu, S.-Z.; Zhang, W.-H.; Shi, B. Effect of pH on structure and stability of collagen-like peptide: Insight from molecular dynamics simulation. J. Theor. Comput. Chem. 2011, 10, 245–259. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE); 2013.08; Chemical Computing Group ULC: Montreal, QC, Canada, 2018.
- Bomble, Y.J. Amsterdam Density Functional 2005 Scientific Computing and Modelling NV, Vrije Universiteit, Theoretical Chemistry, De Boelelaan 1083, 1081 HV Amsterdam, the Netherlands. J. Am. Chem. Soc. 2006, 128, 3103. [Google Scholar] [CrossRef]
- Schmidt, M.W.; Baldridge, K.K.; Boatz, J.A.; Elbert, S.T.; Gordon, M.S.; Jensen, J.H.; Koseki, S.; Matsunaga, N.; Nguyen, K.A.; Su, S.; et al. General atomic and molecular electronic structure system. J. Comp. Chem. 1993, 14, 1347–1363. [Google Scholar] [CrossRef]
- Gill, P.; Murray, W.; Wright, M. Practical Optimization; Academic Press: London, UK, 1981. [Google Scholar]
- Chan, S.L.; Labute, P. Training a scoring function for the alignment of small molecules. J. Chem. Inf. Model. 2010, 50, 1724–1735. [Google Scholar] [CrossRef] [PubMed]
- Kearsley, S.K.; Smith, G.M. An alternative method for the alignment of molecular structures: Maximizing electrostatic and steric overlap. Tetrahedron Comput. Methodol. 1990, 3, 615–633. [Google Scholar] [CrossRef]
- Labute, P.; Williams, C.; Feher, M.; Sourial, E.; Schmidt, J.M. Flexible alignment of small molecules. J. Med. Chem. 2001, 44, 1483–1490. [Google Scholar] [CrossRef]
- Clark, A.M.; Labute, P. 2D depiction of protein−ligand complexes. J. Chem. Inf. Model. 2007, 47, 1933–1944. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.M.; Labute, P.; Santavy, M. 2D structure depiction. J. Chem. Inf. Model. 2006, 46, 1107–1123. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.; Postma, J.V.; van Gunsteren, W.F.; DiNola, A.R.H.J.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Bond, S.D.; Leimkuhler, B.J.; Laird, B.B. The Nosé–Poincaré method for constant temperature molecular dynamics. J. Comput. Phys. 1999, 151, 114–134. [Google Scholar] [CrossRef] [Green Version]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| Peptide | Molecular Ion | Theoretical (m/z) | Experimental (m/z) |
|---|---|---|---|
| NAP (H2N–NAPVSIPQ-COOH) | [M-16+H]+ | 809.4 | 809.6 |
| [M+H]+ | 825.4 | 825.6 | |
| [M+Na]+ | 847.4 | 847.6 | |
| [M+K]+ | 863.4 | 863.6 | |
| [M+2Na-H]+ | 869.4 | 869.6 | |
| [M+Na+K-H]+ | 885.4 | 885.6 | |
| [M+Zn-H]+ | 887.4 | 887.6 | |
| NAPY (H2N–NAPVYIPQ-COOH) | [M-16+H]+ | 885.5 | 885.7 |
| [M+H]+ | 901.5 | 901.7 | |
| [M+Na]+ | 923.5 | 923.7 | |
| [M+K]+ | 939.4 | 939.6 | |
| [M+2Na-H]+ | 945.4 | 945.6 | |
| [M+Na+K-H]+ | 961.4 | 961.6 | |
| [M+Zn-H]+ | 963.4 | 963.6 |
| Chemical Shift (ppm) | NAP pH 5 | NAP pH 7.5 | NAP-Zn pH 7.5 | NAP-Zn pH 9.5 |
|---|---|---|---|---|
| 1Hα Asn | 4.34 | 4.10 | 4.04 | 3.77 |
| 1Hβ Asn | 2.99 | 2.87 | 2.82 | 2.65 |
| 2.85 | 2.72 | 2.69 | 2.51 | |
| 1Hα Gln | 4.36 | 4.17 | 4.17 | 4.17 |
| 1Hβ Gln | 2.20 | 2.13 | 2.13 | 2.13 |
| 1Hγ Gln | 2.41 | 2.36 | 2.36 | 2.36 |
| Chemical Shift (ppm) | NAPY pH 5 | NAPY pH 7.5 | NAPY-Zn pH 7.5 | NAPY-Zn pH 9.5 |
|---|---|---|---|---|
| 1Hα Asn | 4.33 | 3.39 | 3.87 | 3.75 |
| 1Hβ Asn | 2.99 | 2.74 | 2.73 | 2.65 |
| 2.85 | 2.64 | 2.59 | 2.51 | |
| 1Hα Gln | 4.34 | 4.15 | 4.15 | 4.15 |
| 1Hβ Gln | 2.22 | 2.13 | 2.13 | 2.13 |
| 1Hγ Gln | 2.41 | 2.34 | 2.34 | 2.34 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lupaescu, A.-V.; Mocanu, C.S.; Drochioiu, G.; Ciobanu, C.-I. Zinc Binding to NAP-Type Neuroprotective Peptides: Nuclear Magnetic Resonance Studies and Molecular Modeling. Pharmaceuticals 2021, 14, 1011. https://doi.org/10.3390/ph14101011
Lupaescu A-V, Mocanu CS, Drochioiu G, Ciobanu C-I. Zinc Binding to NAP-Type Neuroprotective Peptides: Nuclear Magnetic Resonance Studies and Molecular Modeling. Pharmaceuticals. 2021; 14(10):1011. https://doi.org/10.3390/ph14101011
Chicago/Turabian StyleLupaescu, Ancuta-Veronica, Cosmin Stefan Mocanu, Gabi Drochioiu, and Catalina-Ionica Ciobanu. 2021. "Zinc Binding to NAP-Type Neuroprotective Peptides: Nuclear Magnetic Resonance Studies and Molecular Modeling" Pharmaceuticals 14, no. 10: 1011. https://doi.org/10.3390/ph14101011
APA StyleLupaescu, A. -V., Mocanu, C. S., Drochioiu, G., & Ciobanu, C. -I. (2021). Zinc Binding to NAP-Type Neuroprotective Peptides: Nuclear Magnetic Resonance Studies and Molecular Modeling. Pharmaceuticals, 14(10), 1011. https://doi.org/10.3390/ph14101011