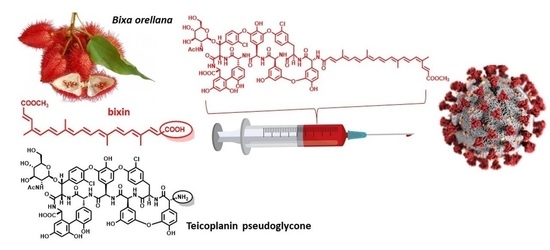
Natural Apocarotenoids and Their Synthetic Glycopeptide Conjugates Inhibit SARS-CoV-2 Replication
Abstract
:1. Introduction
2. Results
2.1. Chemical Synthesis
2.2. Antiviral Evaluations
2.3. Mechanism of Action
2.4. Antibacterial Evaluation
3. Discussion
4. Materials and Methods
4.1. Chemical Synthesis
4.1.1. General Information
4.1.2. Isolation of Bixin (1a)
4.1.3. Synthesis of Crocetin Monomethyl Ester (1b)
4.1.4. Synthesis of β-Apo-8′-carotenoic Acid (1c)
4.1.5. Synthesis of Norbixin (1d)
4.1.6. General Procedure for the Synthesis of Active Esters 1a-ester, 1b-ester and 1c-ester
4.1.7. General Procedure for the Synthesis of Apocarotenoid–Glycopeptide Conjugates 5a, 5b, 5c, 6 and 7
4.1.8. Synthesis of 9
4.2. Biological Studies
4.2.1. Antiviral Activity Screening Using Viral RNA Reduction Assay
4.2.2. Antiviral Activity Determination Using CPE-Based Assay
4.2.3. Antiviral Activity Determination Using Immunofluorescence Assay (IFA)
4.2.4. Determination of Compound Cytotoxicity in Vero E6 Cells
4.2.5. Cathepsin Inhibition Assays
Enzyme Kinetics
Determination of Relative Inhibition
CLPro Inhibition Assay, Cloning and Expression
Purification of Recombinant 3CLPro Protein
Enzyme Activity Assays
4.2.6. Antibacterial Evaluations
4.3. Low-Mode Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Gomez., C.E.; Perdiguero, B.; Esteban, M. Emerging SARS-CoV-2 variants and impact in global vaccination programs against SARS-CoV-2/COVID-19. Vaccines 2021, 9, 243. [Google Scholar] [CrossRef]
- Jean, S.-S.; Hsueh, P.-R. Old and repurposed drugs for the treatment of COVID-19. Expert Rev. Anti-Infect. Ther. 2020, 18, 843–847. [Google Scholar] [CrossRef]
- Xiao, X.; Wang, C.; Chang, D.; Wang, Y.; Dong, X.; Jiao, T.; Zhao, Z.; Ren, L.; Dela Cruz, C.S.; Sharma, L.; et al. Identification of Potent and Safe Antiviral Therapeutic Candidates against SARS-CoV-2. Front. Immunol. 2020, 11, 586572. [Google Scholar] [CrossRef]
- Furuta, Y.; Takahashi, K.; Shiraki, K.; Sakamoto, K.; Smee, D.F.; Barnard, D.L.; Gowen, B.B.; Julander, J.G.; Morrey, J.D. T-705 (favipiravir) and related compounds: Novel broad-spectrum inhibitors of RNA viral infections. Antivir. Res. 2009, 82, 95–102. [Google Scholar] [CrossRef]
- Sanders, J.M.; Monogue, M.L.; Jodlowski, T.Z.; Cutrell, J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. J. Am. Med. Assoc. 2020, 323, 1824–1836. [Google Scholar] [CrossRef]
- Bansal, V.; Mahapure, K.S.; Bhurwal, A.; Gupta, I.; Hassanain, S.; Makadia, J.; Madas, N.; Armaly, P.; Singh, R.; Mehra, I.; et al. Mortality benefit of remdesivir in COVID-19: A systematic review and meta-analysis. Front. Med. 2020, 7, 606429. [Google Scholar] [CrossRef]
- Chodera, J.; Lee, A.A.; London, N.; von Delft, F. Crowdsourcing drug discovery for pandemics. Nat. Chem. 2020, 12, 581. [Google Scholar] [CrossRef]
- Douangamath, A.; Fearon, D.; Gehrtz, P.; Krojer, T.; Lukacik, P.; Owen, C.D.; Resnick, E.; Strain-Damerell, C.; Aimon, A.; Ábrányi-Balogh, P.; et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020, 11, 5047. [Google Scholar] [CrossRef]
- Schuller, M.; Correy, G.J.; Gahbauer, S.; Fearon, D.; Wu, T.; Díaz, R.E.; Young, I.D.; Martins, L.C.; Smith, D.H.; Schulze-Gahmen, U.; et al. Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci. Adv. 2021, 7, eabf8711. [Google Scholar] [CrossRef]
- Cox, O.B.; Krojer, T.; Collins, P.; Monteiro, O.; Talon, R.; Bradley, A.; Fedorov, O.; Amin, J.; Marsden, B.D.; Spencer, J.; et al. A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain. Chem. Sci. 2016, 7, 2322–2330. [Google Scholar] [CrossRef] [Green Version]
- Bajusz, D.; Wade, W.S.; Satała, G.; Bojarski, A.J.; Ilaš, J.; Ebner, J.; Grebien, F.; Papp, H.; Jakab, F.; Douangamath, A.; et al. Exploring protein hotspots by optimized fragment pharmacophores. Nat. Commun. 2021, 12, 3201. [Google Scholar] [CrossRef] [PubMed]
- Bakowski, M.A.; Beutler, N.; Wolff, K.C.; Kirkpatrick, M.G.; Chen, E.; Nguyen, T.-T.H.; Riva, L.; Shaabani, N.; Parren, M.; Ricketts, J.; et al. Drug repurposing screens identify chemical entities for the development of COVID-19 interventions. Nat. Commun. 2021, 12, 3309. [Google Scholar] [CrossRef] [PubMed]
- Xiang, R.; Yu, Z.; Wang, Y.; Wang, L.; Huo, S.; Li, Y.; Liang, R.; Hao, Q.; Ying, T.; Gao, Y.; et al. Recent advances in developing small-molecule inhibitors against SARS-CoV-2. Acta Pharm. Sin. B 2021. [Google Scholar] [CrossRef]
- Gomes, C.P.; Fernandes, D.E.; Casimiro, F.; da Mata, G.F.; Passos, M.T.; Varela, P.; Mastroianni-Kirsztajn, G.; Pesquero, J.B. Cathepsin L in COVID-19: From pharmacological evidences to genetics. Front. Cell. Infect. Microbiol. 2020, 10, 589505. [Google Scholar] [CrossRef]
- Liu, T.; Luo, S.; Libby, P.; Shi, G.-P. Cathepsin L-selective inhibitors: A potentially promising treatment for COVID-19 patients. Pharmacol. Ther. 2020, 213, 107587. [Google Scholar] [CrossRef]
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020, 178, 104792. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiegens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterisation of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.-M.; Yang, W.-L.; Yang, F.-Y.; Zhang, L.; Huang, W.-J.; Hou, W.; Fan, C.-F.; Jin, R.-H.; Feng, Y.-M.; Wang, Y.-C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 2021, 6, 134. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Pan, T.; Zhang, J.; Li, Q.; Zhang, X.; Bai, C.; Huang, F.; Peng, T.; Zhang, J.; Liu, C.; et al. Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of Ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV). J. Biol. Chem. 2016, 291, 9218–9232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Ma, X.; Yu, F.; Liu, J.; Zou, F.; Pan, T.; Zhang, H. Teicoplanin potently blocks the cell entry of 2019-nCoV. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Baron, S.A.; Devaux, C.; Colson, P.; Raoult, D.; Rolain, J.M. Teicoplanin: An alternative drug for the treatment of COVID-19? Int. J. Antimicrob. Agents 2020, 55, 105944. [Google Scholar] [CrossRef]
- Ceccarelli, G.; Alessandri, F.; Oliva, A.; Borrazzo, C.; Dell’Isola, S.; Ialungo, A.M.; Rastrelli, E.; Pelli, M.; Raponi, G.; Turriziani, O.; et al. The role of teicoplanin in the treatment of SARS-CoV-2 infection: A retrospective study in critically ill COVID-19 patients (Tei-COVID study). J. Med. Virol. 2021, 93, 4319–4325. [Google Scholar] [CrossRef]
- Balzarini, J.; Keyaerts, E.; Vijgen, L.; Egberink, H.; De Clercq, E.; Van Ranst, M.; Printsevskaya, S.S.; Olsufyeva, E.N.; Solovieva, S.E.; Preobrazhenskaya, M.N. Inhibition of feline (FIPV) and human (SARS) coronavirus by semisynthetic derivatives of glycopeptide antibiotics. Antivir. Res. 2006, 72, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Szűcs, Z.; Naesens, L.; Stevaert, A.; Ostorházi, E.; Batta, G.; Herczegh, P.; Borbás, A. Reprogramming of the antibacterial drug vancomycin results in potent antiviral agents devoid of antibacterial activity. Pharmaceuticals 2020, 13, 139. [Google Scholar] [CrossRef] [PubMed]
- Bereczki, I.; Csávás, M.; Szűcs, Z.; Rőth, E.; Batta, G.; Ostorházi, E.; Naesens, L.; Borbás, A.; Herczegh, P. Synthesis of antiviral perfluoroalkyl derivatives of teicoplanin and vancomycin. ChemMedChem 2020, 15, 1661–1671. [Google Scholar] [CrossRef]
- Balzarini, J.; Pannecouque, C.; De Clercq, E.; Pavlov, A.Y.; Printsevskaya, S.S.; Miroshnikova, O.V.; Reznikova, M.I.; Preobrazhenskaya, M.N. Antiretroviral Activity of Semisynthetic Derivatives of Glycopeptide Antibiotics. J. Med. Chem. 2003, 46, 2755–2764. [Google Scholar] [CrossRef] [PubMed]
- De Burghgraeve, T.; Kaptein, S.J.; Ayala-Nunez, N.V.; Mondotte, J.A.; Pastorino, B.; Printsevskaya, S.S.; de Lamballerie, X.; Jacobs, M.; Preobrazhenskaya, M.; Gamarnik, A.V.; et al. An analogue of the antibiotic teicoplanin prevents Flavivirus entry in vitro. PLoS ONE 2012, 7, e37244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naesens, L.; Vanderlinden, E.; Rőth, E.; Jekő, J.; Andrei, G.; Snoeck, R.; Pannecouque, C.; Illyés, E.; Batta, G.; Herczegh, P.; et al. Anti-influenza virus activity and structure–activity relationship of aglycoristocetin derivatives with cyclobutenedione carrying hydrophobic chains. Antivir. Res. 2009, 82, 89–94. [Google Scholar] [CrossRef]
- Pintér, G.; Batta, G.; Kéki, S.; Mándi, A.; Komáromi, I.; Takács-Novák, K.; Sztaricskai, F.; Rőth, E.; Ostorházi, E.; Rozgonyi, F.; et al. Diazo transfer-click reaction route to new, lipophilic teicoplanin and ristocetin aglycone derivatives with high antibacterial and anti-influenza virus activity: An aggregation and receptor binding study. J. Med. Chem. 2009, 52, 6053–6061. [Google Scholar] [CrossRef]
- Sipos, A.; Máté, G.; Rőth, E.; Borbás, A.; Batta, G.; Bereczki, I.; Kéki, S.; Jóna, I.; Ostorházi, E.; Rozgonyi, F.; et al. Synthesis of fluorescent ristocetin aglycone derivatives with remarkable antibacterial and antiviral activities. Eur. J. Med. Chem. 2012, 58, 361–367. [Google Scholar] [CrossRef]
- Bereczki, I.; Kicsák, M.; Dobray, L.; Borbás, A.; Batta, G.; Kéki, S.; Nemes Nikodém, É.; Ostorházi, E.; Rozgonyi, F.; Vanderlinden, E.; et al. Semisynthetic teicoplanin derivatives as new influenza virus binding inhibitors: Synthesis and antiviral studies. Bioorg. Med. Chem. Lett. 2014, 24, 3251–3254. [Google Scholar] [CrossRef]
- Szűcs, Z.; Kelemen, V.; Thai, S.L.; Csávás, M.; Rőth, E.; Batta, G.; Stevaert, A.; Vanderlinden, E.; Naesens, L.; Herczegh, P.; et al. Structure-activity relationship studies of lipophilic teicoplanin pseudoaglycon derivatives as new anti-influenza virus agents. Eur. J. Med. Chem. 2018, 157, 1017–1030. [Google Scholar] [CrossRef]
- Hammond, B.R.; Renzi-Hammond, L.M. Carotenoids. Adv. Nutr. 2013, 4, 474–476. [Google Scholar] [CrossRef] [Green Version]
- Kaulmann, A.; Bohn, T. Carotenoids, inflammation, and oxidative stress—Implications of cellular signaling pathways and relation to chronic disease prevention. Nutr. Res. 2014, 34, 907–929. [Google Scholar] [CrossRef] [PubMed]
- Meléndez-Martínez, A.J. An Overview of Carotenoids, Apocarotenoids, and Vitamin a in Agro-Food, Nutrition, Health, and Disease. Mol. Nutr. Food Res. 2019, 63, 1801045. [Google Scholar] [CrossRef] [Green Version]
- Eroglu, A.; Harrison, E.H. Carotenoid metabolism in mammals, including man: Formation, occurrence, and function of apocarotenoids. J. Lipid Res. 2013, 54, 1719–1730. [Google Scholar] [CrossRef] [Green Version]
- Rivera-Madrid, R.; Aguilar-Espinosa, M.; Cárdenas-Conejo, Y.; Garza-Caligaris, L.E. Carotenoid Derivates in Achiote (Bixa orellana) Seeds: Synthesis and Health Promoting Properties. Front. Plant Sci. 2016, 7, 1406. [Google Scholar] [CrossRef] [Green Version]
- Nassiri-Asl, M.; Hosseinzadeh, H. Neuropharmacology Effects of Saffron (Crocus sativus) and Its Active Constituents. In Bioactive Nutraceuticals and Dietary Supplements in Neurological and Brain Disease, Prevention and Therapy; Watson, R.R., Preedy, V.R., Eds.; Academic Press, Elsevier: London, UK, 2015; pp. 29–39. ISBN 978-0-12-411462-3. [Google Scholar] [CrossRef]
- Ebert, D.H.; Deussing, J.; Peters, C.; Dermody, T.S. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 2002, 277, 24609–24617. [Google Scholar] [CrossRef] [Green Version]
- Simmons, G.; Gosalia, D.N.; Rennekamp, A.J.; Reeves, J.D.; Diamond, S.L.; Bates, P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 2005, 102, 11876–11881. [Google Scholar] [CrossRef] [Green Version]
- Ryabchikova, E.I.; Kolesnikova, L.V.; Luchko, S.V. An analysis of features of pathogenesis in two animal models of Ebola virus infection. J. Infect. Dis. 1999, 179, S199–S202. [Google Scholar] [CrossRef]
- Chandran, K.; Sullivan, N.J.; Felbor, U.; Whelan, S.P.; Cunningham, J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 2005, 308, 1643–1645. [Google Scholar] [CrossRef] [Green Version]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Tripathi, P.K.; Upadhyay, S.; Singh, M.; Raghavendhar, S.; Bhardwaj, M.; Sharma, P.; Patel, A.K. Screening and evaluation of approved drugs as inhibitors of main protease of SARS-CoV-2. Int. J. Biol. Macromol. 2020, 164, 2622–2631. [Google Scholar] [CrossRef]
- Keserű, G.M.; Kolossváry, I. Fully flexible low-mode docking: Application to induced fit in HIV integrase. J. Am. Chem. Soc. 2001, 123, 12708–12709. [Google Scholar] [CrossRef]
- Kolossváry, I.; Guida, W.C. Low mode search. An efficient, automated computational method for conformational analysis: Application to cyclic and acyclic alkanes and cyclic peptides. J. Am. Chem. Soc. 1996, 118, 5011–5019. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Xian, Y.; Zhang, J.; Bian, Z.; Zhou, H.; Zhang, Z.; Lin, Z.; Xu, H. Bioactive natural compounds against human coronaviruses: A review and perspective. Acta Pharm. Sin. B 2020, 10, 1163–1174. [Google Scholar] [CrossRef]
- Vougogiannopoulou, K.; Corona, A.; Tramontano, E.; Alexis, M.N.; Skaltsounis, A.L. Natural and Nature-Derived Products Targeting Human Coronaviruses. Molecules 2021, 26, 448. [Google Scholar] [CrossRef]
- Umesh, U.; Kundu, D.; Selvaraj, C.; Singh, S.K.; Dubey, V.K. Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. J. Biomol. Struct. Dyn. 2020, 39, 3428–3434. [Google Scholar] [CrossRef]
- Andrault, P.M.; Samsonov, S.A.; Weber, G.; Coquet, L.; Nazmi, K.; Bolscher, J.G.; Lalmanach, A.C.; Jouenne, T.; Brömme, D.; Pisabarro, M.T.; et al. Antimicrobial peptide LL-37 is both a substrate of cathepsins S and K and a selective inhibitor of Cathepsin L. Biochemistry 2015, 54, 2785–2798. [Google Scholar] [CrossRef]
- Scharinger, F.; Pálvölgyi, A.M.; Zeindlhofer, V.; Schnürch, M.; Schröder, C.; Bica-Schröder, K. Counterion Enhanced Organocatalysis: A Novel Approach for the Asymmetric Transfer Hydrogenation of Enone. ChemCatChem 2020, 12, 3776–3782. [Google Scholar] [CrossRef]
- Sztaricskai, F.; Batta, G.; Herczegh, P.; Balázs, A.; Jekő, J.; Rőth, E.; Szabó, P.T.; Kardos, S.; Rozgonyi, F.; Boda, Z. A new series of glycopeptide antibiotics incorporating a squaric acid moiety. Synthesis, structural and antibacterial studies. J. Antibiot. 2006, 59, 564–582. [Google Scholar] [CrossRef]
- Malabarba, A.; Ferrari, P.; Gallo, G.G.; Kettenring, J.; Cavalleri, B. Teicoplanin, antibiotics from Actinoplanes teichomyceticus nov. sp. VII. Preparation and NMR characteristics of the aglycone of teicoplanin. J. Antibiot. 1986, 39, 1430–1442. [Google Scholar] [CrossRef] [Green Version]
- Häberli, A.; Pfander, H. Synthesis of Bixin and Three Minor Carotenoids from Annatto (Bixa orellana). Chim. Acta 1999, 82, 696–706. [Google Scholar] [CrossRef]
- Frederico, D.; Donate, P.M.; Constantino, M.G.; Bronze, E.S.; Sairre, M.I. A short and efficient synthesis of crocetin-dimethylester and crocetindial. J. Org. Chem. 2003, 68, 9126–9128. [Google Scholar] [CrossRef]
- Scotter, M. The chemistry and analysis of annatto food colouring: A review. Food Addit. Contam. Part A 2009, 26, 1123–1145. [Google Scholar] [CrossRef]
- Kuhelj, R.; Dolinar, M.; Pungerčar, J.; Turk, V. The Preparation of Catalytically Active Human Cathepsin B from Its Precursor Expressed in Escherichia coli in the Form of Inclusion Bodies. Eur. J. Biochem. 1995, 229, 533–539. [Google Scholar] [CrossRef]
- Dolinar, M.; Maganja, D.B.; Turk, V. Expression of Full-Length Human Procathepsin L cDNA in Escherichia coli and Refolding of the Expression Product. Biol. Chem. Hoppe. Seyler. 1995, 376, 385–388. [Google Scholar] [CrossRef]
- Feng, B.Y.; Shoichet, B.K. A detergent-based assay for the detection of promiscuous inhibitors. Nat. Protoc. 2006, 1, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Tomar, S.; Johnston, M.L.; St John, S.E.; Osswald, H.L.; Nyalapatla, P.R.; Paul, L.N.; Ghosh, A.K.; Denison, M.R.; Mesecar, A.D. Ligand-induced dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 protease (3CLpro): Implications for nsp5 regulation and the development of antivirals. J. Biol. Chem. 2015, 290, 19403–19422. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Pettersson, H.I.; Huitema, C.; Niu, C.; Yin, J.; James, N.M.G.; Eltis, L.D.; Vederas, J.C. Design, synthesis, and evaluation of inhibitors for severe acute respiratory syndrome 3C-like protease based on phthalhydrazide ketones or heteroaromatic esters. J. Med. Chem. 2007, 50, 1850–1864. [Google Scholar] [CrossRef]
- Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020.
- Wei, B.; Gunzner-Toste, J.; Yao, H.; Wang, T.; Wang, J.; Xu, Z.; Chen, J.; Wai, J.; Nonomiya, J.; Tsai, S.P.; et al. Discovery of Peptidomimetic Antibody-Drug Conjugate Linkers with Enhanced Protease Specificity. J. Med. Chem. 2018, 61, 989–1000. [Google Scholar] [CrossRef]
- Shenoy, R.T.; Chowdhury, S.F.; Kumar, S.; Joseph, L.; Purisima, E.O.; Sivaraman, J. A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors. J. Med. Chem. 2009, 52, 6335–6346. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- EFSA Panel on Food Additives and Nutrient Sources added to Food (ANS). The safety of annatto extracts (E 160b) as a food additive. EFSA J. 2016, 14, e04544. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority (EFSA); Tard, A. Exposure assessment of annatto colouring principles bixin and norbixin (E 160b) when as food additives. EFSA J. 2017, 15, e04966. [Google Scholar] [CrossRef]





| Compound | SARS-CoV-2 RNA Reduction EC50 (µM) | SARS-CoV-2 CPE, EC50 (µM) | SARS-CoV-2 IFA, EC50 (µM) | CC50 (µM) | Therapeutic Index *** CC50/EC50 |
|---|---|---|---|---|---|
| 1a | 5.9 ± 1.7 | 14 ± 3.5 | 28 ± 8.8 | >100 | >17 |
| 1b | not active at 50 µM | n.d. | n.d. | n.d. | n.d. |
| 1c | 15 ± 2 | 14 ± 2.2 | 31 ± 7.9 * | >100 ** | >6.7 |
| 1d (norbixin) | not active at 50 µM | n.d. | n.d. | >100 | n.d. |
| Annatto (E-160b) | 9.2 ± 0.1 | n.d. | n.d. | >100 | >10.9 |
| 2 | not active at 50 µM | n.d. | n.d. | n.d. | n.d. |
| 3 | 5.6 ± 1.3 | 16 ± 1.7 | 18 ± 5.4 | >100 | >17.9 |
| 4 | not active at 50 µM | n.d. | n.d. | n.d. | n.d. |
| 5a | 5.9 ± 1.3 | 91 ± 88 | 74 ± 20 | >100 | >17 |
| 5b | 5.2 ± 0.2 | 19 ± 1.8 | 8.8 ± 1.4 | >100 | >19.2 |
| 5c | 4.4 ± 0.4 | 56 ± 10 | 24 ± 5.3 | >100 | >22.7 |
| 6 | 1.8 ± 0.2 | 56 ± 6.2 | 51 ± 9.8 | >100 | >55.6 |
| 7 | 6.7 ± 0.4 | n.d. | n.d. | n.d. | n.d. |
| 9 | n.d. | 31 ± 4.7 | n.d. | >100 | n.d. |
| Compound | Exopeptidase Cathepsin B IC50 (µM) | Cathepsin L IC50 (µM) | 3CLPro IC50 (µM) |
|---|---|---|---|
| 1a (bixin) | 14.56 ± 1.71 | 41.92 ± 1.12 | 56% at 100 μM |
| 1b | n.d. | n.d. | 24% at 100 μM |
| 1c | 21.31 ± 1.20 | 76.04 ± 1.74 | 73.00 ± 7.10 |
| 1d (norbixin) | n.d. | n.d. | 36.11 ± 11.71 |
| 2 | 7.80% at 50 μM | 0.04% at 50 μM | 34% at 200 μM |
| 3 | 0.45% at 50 μM | 5.07% at 50 μM | 13% at 200 μM |
| 5a | 47.95 ± 1.07 | 42.39 ± 1.86 | 13.86 ± 1.73 |
| 5b | 99.54 ± 1.16 | 103.0 ± 1.12 | 50% at 200 μM |
| 5c | 56.53 ± 1.06 | 47.05 ± 1.05 | 34.52 ± 12.49 |
| 6 | 64.42 ± 1.05 | 56.51 ± 1.08 | 53% at 200 μM |
| 9 | 23.78% at 50 μM | 22.08% at 50 μM | 42% at 100 μM |
| 1a | Teico (3) | 5a | 5b | 5c | 6 | 7 | 9 | |
|---|---|---|---|---|---|---|---|---|
| MIC [g] (μg/mL) | ||||||||
| Bacillus subtilis ATCC [a] 6633 | 512 | 0.5 | 32 | 16 | 16 | 4 | 32 | 512 |
| Staphylococcus aureus MSSA [b] ATCC 29213 | 512 | 0.5 | 16 | 8 | 8 | 16 | 64 | 512 |
| Staphylococcus aureus MRSA [c] ATCC 33591 | 512 | 0.5 | 0.5 | 2 | 1 | 16 | 64 | 512 |
| Staphylococcus epidermidis ATCC 35984 biofilm | 512 | 2 | 0.5 | 0.5 | 0.5 | 32 | 64 | 512 |
| Staphylococcus epidermidis mecA [d] | 512 | 16 | 16 | 2 | 0.5 | 256 | 64 | 512 |
| Enterococcus faecalis ATCC 29212 | 512 | 2 | 16 | 4 | 4 | 4 | 32 | 512 |
| Enterococcus faecalis 15376 VanA [e] | 512 | 256 | 16 | 8 | 8 | 4 | 256 | 512 |
| Enterococcus faecalis ATCC 51299 VanB [f] | 512 | 4 | 16 | 8 | 8 | 4 | 64 | 512 |
 | ||
|---|---|---|
| Annotation | 1H Shift (ppm) | 13C Shift (ppm) |
| 19 | 7.899 | 140.42 |
| 1b | 6.788 | 119.07 |
| 1e | 6.993 | 119.09 |
| 1f | 7.039 | 125.61 |
| 20 | 5.959 | 118.11 |
| 21 (OMe) | 3.707 | 51.84 |
| 22,23 | 1.976 | 13.07 |
| 24 | 1.897 | 13.07 |
| 25 | 1.940 | 20.33 |
| 2b | 7.226 | 131.46 |
| 2e | 7.269 | 125.33 |
| 2f | 7.706 | 130.68 |
| 3 | 6.269 | 120.85 |
| 3b | 6.360 | 110.56 |
| 3d | 6.375 | 105.48 |
| 3f | 6.323 | 102.23 |
| 4 | 7.246 | 144.29 |
| 4b | 5.563 | 108.03 |
| 4f | 5.113 | 104.97 |
| 5b | 7.120 | 136.49 |
| 5e | 6.667 | 116.99 |
| 5f | 6.673 | 125.82 |
| 6b | 7.872 | 128.98 |
| 6e | 7.265 | 123.66 |
| 6f | 7.256 | 128.61 |
| 7d | 6.420 | 103.70 |
| 7f | 6.495 | 108.28 |
| DMSO | 2.505 | 40.24 |
| G1 | 4.396 | 99.67 |
| G2 | 3.510 | 56.32 |
| G3 | 3.218 | 70.59 |
| G4 | 3.412 | 73.82 |
| G5 | 3.104 | 77.26 |
| G6a | 3.641 | 60.84 |
| G6b | 3.582 | 60.85 |
| X1 | 4.340 | 59.57 |
| X2 | 4.947 | 55.17 |
| X3 | 5.360 | 58.54 |
| X4 | 5.629 | 55.33 |
| X5 | 4.359 | 54.13 |
| X6 | 4.160 | 61.39 |
| X7 | 5.769 | 56.32 |
| z2a | 3.327 | 37.19 |
| z2b | 2.842 | 37.19 |
| z6 | 5.393 | 76.54 |
 | ||
|---|---|---|
| Annotation | 1H Shift (ppm) | 13C Shift (ppm) |
| 1b | 6.789 | 119.70 |
| 1e | 7.043 | 118.97 |
| 1f | 7.043 | 125.86 |
| 2b | 7.214 | 131.42 |
| 2e | 7.265 | 125.40 |
| 2f | 7.670 | 130.69 |
| 3b | 6.353 | 110.47 |
| 3d | 6.390 | 105.40 |
| 3f | 6.366 | 102.41 |
| 3xMe | 1.963 | 13.06 |
| 4? | 7.235 | 139.13 |
| 4b | 5.562 | 108.09 |
| 4f | 5.108 | 105.05 |
| 5b | 7.125 | 136.50 |
| 5e | 6.675 | 116.99 |
| 5f | 6.674 | 125.87 |
| 6b | 7.863 | 128.99 |
| 6e | 7.247 | 123.67 |
| 6f | 7.251 | 128.51 |
| 7d | 6.386 | 103.42 |
| 7f | 6.440 | 108.05 |
| DMSO | 2.510 | 40.28 |
| G1 | 4.375 | 100.12 |
| G2 | 3.525 | 56.13 |
| G3 | 3.215 | 70.45 |
| G4 | 3.408 | 73.78 |
| G5 | 3.088 | 77.15 |
| G6a | 3.644 | 60.67 |
| G6b | 3.588 | 60.66 |
| Me | 1.967 | 13.52 |
| OMe | 3.694 | 52.12 |
| X1 | 4.330 | 59.23 |
| X2 | 4.953 | 55.13 |
| X3 | 5.334 | 58.51 |
| X4 | 5.621 | 55.33 |
| X5 | 4.363 | 54.11 |
| X6 | 4.173 | 61.35 |
| X7 | 5.710 | 56.90 |
| z2a | 3.310 | 37.30 |
| z2b | 2.828 | 37.31 |
| z6 | 5.333 | 77.00 |
 | ||
|---|---|---|
| Annotation | 1H Shift (ppm) | 13C Shift (ppm) |
| 1b | 6.792 | 119.57 |
| 1e | 6.976 | 119.05 |
| 1f | 7.060 | 125.92 |
| 21 | 2.018 | 33.10 |
| 22 | 1.589 | 19.23 |
| 23 | 1.455 | 39.75 |
| 25, 26, 27 | 1.959 | 13.01 |
| 28 | 1.974 | 13.51 |
| 29 | 1.701 | 21.98 |
| 2b | 7.231 | 131.47 |
| 2e | 7.285 | 125.36 |
| 2f | 7.678 | 130.63 |
| 30, 31 | 1.029 | 29.30 |
| 3b | 6.365 | 110.51 |
| 3d | 6.368 | 105.39 |
| 3f | 6.418 | 103.39 |
| 4b | 5.568 | 107.94 |
| 4f | 5.117 | 104.94 |
| 5b | 7.122 | 136.49 |
| 5e | 6.642 | 116.93 |
| 5f | 6.682 | 125.95 |
| 6b | 7.879 | 128.97 |
| 6e | 7.257 | 123.66 |
| 6f | 7.259 | 128.64 |
| 7d | 6.300 | 102.09 |
| 7f | 6.511 | 108.42 |
| DMSO | 2.513 | 40.31 |
| G1 | 4.423 | 99.26 |
| G2 | 3.515 | 56.47 |
| G3 | 3.241 | 70.58 |
| G4 | 3.412 | 73.91 |
| G5 | 3.114 | 77.34 |
| G6 | 3.603 | 60.89 |
| X1 | 4.337 | 59.69 |
| X2 | 4.976 | 55.12 |
| X3 | 5.357 | 58.50 |
| X4 | 5.631 | 55.38 |
| X5 | 4.364 | 54.13 |
| X6 | 4.152 | 61.43 |
| X7 | 5.731 | 56.87 |
| z2a | 2.845 | 37.32 |
| z2b | 3.333 | 37.31 |
| z6 | 5.442 | 76.09 |
 | ||
|---|---|---|
| Annotation | 1H Shift (ppm) | 13C Shift (ppm) |
| 1b | 6.716 | 118.01 |
| 1e | 6.956 | 118.87 |
| 1f | 7.013 | 124.98 |
| 20 | 5.961 | 118.15 |
| 3 | 6.293 | 121.15 |
| 3b | 6.416 | 109.95 |
| 3f | 6.470 | 104.05 |
| 4b | 5.655 | 107.36 |
| 4f | 5.308 | 106.17 |
| 5b | 7.113 | 136.00 |
| 5e | 6.623 | 117.25 |
| 5f | 6.678 | 126.12 |
| 6b | 7.843 | 128.64 |
| 6c | 7.191 | 123.30 |
| 7d | 6.385 | 103.29 |
| 7f | 6.066 | 105.68 |
| Bix21 (OMe) | 3.710 | 51.84 |
| Me (3d) | 1.971 | 8.79 |
| OMe-7 | 3.699 | 55.41 |
| x1 | 4.509 | 57.10 |
| x2 | 5.946 | 55.65 |
| x3 | 5.261 | 58.51 |
| x4 | 5.579 | 55.11 |
| x5 | 4.418 | 53.99 |
| x6 | 4.182 | 62.49 |
| x7 | 5.075 | 60.89 |
| z2 | 5.145 | 71.72 |
| z6 | 5.129 | 72.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bereczki, I.; Papp, H.; Kuczmog, A.; Madai, M.; Nagy, V.; Agócs, A.; Batta, G.; Milánkovits, M.; Ostorházi, E.; Mitrović, A.; et al. Natural Apocarotenoids and Their Synthetic Glycopeptide Conjugates Inhibit SARS-CoV-2 Replication. Pharmaceuticals 2021, 14, 1111. https://doi.org/10.3390/ph14111111
Bereczki I, Papp H, Kuczmog A, Madai M, Nagy V, Agócs A, Batta G, Milánkovits M, Ostorházi E, Mitrović A, et al. Natural Apocarotenoids and Their Synthetic Glycopeptide Conjugates Inhibit SARS-CoV-2 Replication. Pharmaceuticals. 2021; 14(11):1111. https://doi.org/10.3390/ph14111111
Chicago/Turabian StyleBereczki, Ilona, Henrietta Papp, Anett Kuczmog, Mónika Madai, Veronika Nagy, Attila Agócs, Gyula Batta, Márton Milánkovits, Eszter Ostorházi, Ana Mitrović, and et al. 2021. "Natural Apocarotenoids and Their Synthetic Glycopeptide Conjugates Inhibit SARS-CoV-2 Replication" Pharmaceuticals 14, no. 11: 1111. https://doi.org/10.3390/ph14111111
APA StyleBereczki, I., Papp, H., Kuczmog, A., Madai, M., Nagy, V., Agócs, A., Batta, G., Milánkovits, M., Ostorházi, E., Mitrović, A., Kos, J., Zsigmond, Á., Hajdú, I., Lőrincz, Z., Bajusz, D., Keserű, G. M., Hodek, J., Weber, J., Jakab, F., ... Borbás, A. (2021). Natural Apocarotenoids and Their Synthetic Glycopeptide Conjugates Inhibit SARS-CoV-2 Replication. Pharmaceuticals, 14(11), 1111. https://doi.org/10.3390/ph14111111