Exploiting the Metabolism of the Gut Microbiome as a Vehicle for Targeted Drug Delivery to the Colon
Abstract
:1. Introduction
1.1. Gut Microbiome Metabolism Specific to the Colon
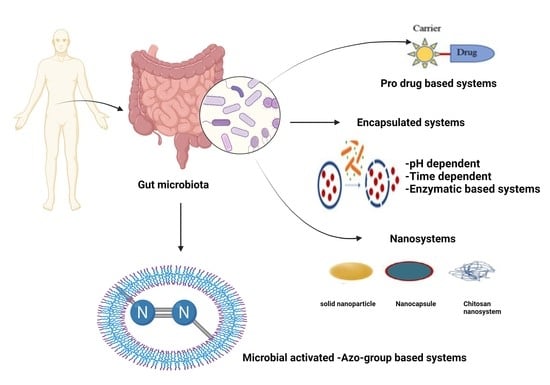
1.2. Prodrugs
1.3. Bacterial Gene Therapy
1.4. Potential of the Azo Polymer-Based Hydrogel Drug Delivery System
1.5. Encapsulation
2. Merits and Demerits of Colon Drug Delivery Systems
2.1. Probiotic-Aided Colon-Specific Drug Delivery
2.2. Pharmaco-Microbiomes
2.3. Conclusions and Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global cancer observatory: Cancer today. Lyon France IARC. 2018, pp. 1–6. Available online: https://gco.iarc.fr/today (accessed on 27 January 2021).
- Alatab, S.; Sepanlou, S.G.; Ikuta, K.; Vahedi, H.; Bisignano, C.; Safiri, S.; Sadeghi, A.; Nixon, M.R.; Abdoli, A.; Abolhassani, H.; et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Philip, A.; Philip, B. Colon Targeted Drug Delivery Systems: A Review on Primary and Novel Approaches. Oman Med. J. 2010, 25, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Kotla, N.G.; Rana, S.; Sivaraman, G.; Sunnapu, O.; Vemula, P.K.; Pandit, A.; Rochev, Y. Bio-responsive drug delivery systems in intestinal inflammation: State-of-the-art and future perspectives. Adv. Drug Deliv. Rev. 2019, 146, 248–266. [Google Scholar] [CrossRef] [PubMed]
- WHO. “Noncommunicable Diseases”. Retrieved 23 September 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/noncommunicable-diseases (accessed on 21 May 2021).
- Chen, C.K.; Huang, P.K.; Law, W.C.; Chu, C.H.; Chen, N.T.; Lo, L.W. Biodegradable polymers for gene-delivery applications. Int. J. Nanomed. 2020, 15, 2131. [Google Scholar] [CrossRef] [Green Version]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.; del Pilar Rodriguez-Torres, M.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 1–33. [Google Scholar] [CrossRef] [Green Version]
- Pircalabioru, G.G.; Chifiriuc, M.C. Nanoparticulate drug-delivery systems for fighting microbial biofilms: From bench to bedside. Future Microbiol. 2020, 15, 679–698. [Google Scholar] [CrossRef]
- Chang, W.W.; Lee, C.H. Salmonella as an innovative therapeutic antitumor agent. Int. J. Mol. Sci. 2014, 15, 14546–14554. [Google Scholar] [CrossRef] [Green Version]
- Bissell, T.; Steele, L. Human Anatomy & Physiology. In Human Anatomy and Physiology, 8th ed.; Pearson Benjamin Cummings: San Francisco, CA, USA, 2010. [Google Scholar]
- Reinus, J.F.; Simon, D. (Eds.) Gastrointestinal Anatomy and Physiology: The Essentials; John Wiley & Sons: Chichester, UK, 2014. [Google Scholar]
- Sartor, R.B. Microbial influences in inflammatory bowel diseases. Gastroenterology 2008, 134, 577–594. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank, D.N.; Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [Green Version]
- Sartor, R.B. Genetics and environmental interactions shape the intestinal microbiome to promote inflammatory bowel disease versus mucosal homeostasis. Gastroenterology 2010, 139, 1816–1819. [Google Scholar] [CrossRef] [PubMed]
- Sinha, V.R.; Kumria, R. Polysaccharides in colon-specific drug delivery. Int. J. Pharm. 2001, 224, 19–38. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S. Fermentation in the human large intestine: Its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 2011, 45, S120–S127. [Google Scholar] [CrossRef] [Green Version]
- El Aidy, S.; Van Den Bogert, B.; Kleerebezem, M. The small intestine microbiota, nutritional modulation and relevance for health. Curr. Opin. Biotechnol. 2015, 32, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Rinta-Kanto, J.M.; Sun, S.; Sharma, S.; Kiene, R.P.; Moran, M.A. Bacterial community transcription patterns during a marine phytoplankton bloom. Environ. Microbiol. 2012, 14, 228–239. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Boyd, S.D.; Liu, Y.; Wang, C.; Martin, V.; Dunn-Walters, D.K. Human lymphocyte repertoires in ageing. Curr. Opin. Immunol. 2013, 25, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Macfarlane, G.T.; Macfarlane, S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012, 95, 50–60. [Google Scholar] [CrossRef]
- Zhang, T.; Yang, Y.; Liang, Y.; Jiao, X.; Zhao, C. Beneficial effect of intestinal fermentation of natural polysaccharides. Nutrients 2018, 10, 1055. [Google Scholar] [CrossRef] [Green Version]
- Fluitman, K.S.; De Clercq, N.C.; Keijser, B.J.; Visser, M.; Nieuwdorp, M.; IJzerman, R.G. The intestinal microbiota, energy balance, and malnutrition: Emphasis on the role of short-chain fatty acids. Expert Rev. Endocrinol. Metab. 2017, 12, 215–226. [Google Scholar] [CrossRef] [Green Version]
- Conterno, L.; Fava, F.; Viola, R.; Tuohy, K.M. Obesity and the gut microbiota: Does up-regulating colonic fermentation protect against obesity and metabolic disease? Genes Nutr. 2011, 6, 241–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, A.C.; Hoffmann, C.; Mota, J.F. The human gut microbiota: Metabolism and perspective in obesity. Gut Microbes 2018, 9, 308–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steliou, K.; Boosalis, M.S.; Perrine, S.P.; Sangerman, J.; Faller, D.V. Butyrate histone deacetylase inhibitors. BioRes. Open Access 2012, 1, 192–198. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Testa, B. Prodrugs: Bridging pharmacodynamic/pharmacokinetic gaps. Curr. Opin. Chem. Biol. 2009, 13, 338–344. [Google Scholar] [CrossRef]
- Sinha, V.R.; Kumria, R. Microbially triggered drug delivery to the colon. Eur. J. Pharm. Sci. 2003, 18, 3–18. [Google Scholar] [CrossRef]
- Rautio, J.; Meanwell, N.A.; Di, L.; Hageman, M.J. The expanding role of prodrugs in contemporary drug design and development. Nat. Rev. Drug Discov. 2018, 17, 559–587. [Google Scholar] [CrossRef]
- Sinha, V.R.; Kumria, R. Colonic drug delivery: Prodrug approach. Pharm. Res. 2001, 18, 557–564. [Google Scholar] [CrossRef]
- Zou, M.J.; Cheng, G.; Okamoto, H.; Hao, X.H.; An, F.; Cui, F.D.; Danjo, K. Colon-specific drug delivery systems based on cyclodextrin prodrugs: In vivo evaluation of 5-aminosalicylic acid from its cyclodextrin conjugates. World J. Gastroenterol. 2005, 11, 7457. [Google Scholar] [CrossRef]
- Yang, Y.; Kim, W.; Kim, D.; Jeong, S.; Yoo, J.W.; Jung, Y. A colon-specific prodrug of metoclopramide ameliorates colitis in an experimental rat model. Drug Des. Del. 2019, 13, 231. [Google Scholar] [CrossRef] [Green Version]
- Ansari, M.; Sadarani, B.; Majumdar, A. Colon targeted beads loaded with pterostilbene: Formulation, optimization, characterization and in vivo evaluation. Saudi Pharm. J. 2019, 27, 71–81. [Google Scholar] [CrossRef]
- Barros, P.D.; Dias, I.F.; Zanin, G.D.; Bunhak, É.J. Development and evaluation of dapsone tablets coated for specific colon release. Drug Dev. Ind. Pharm. 2020, 46, 246–252. [Google Scholar] [CrossRef]
- Afkhami-Poostchi, A.; Mashreghi, M.; Iranshahi, M.; Matin, M.M. Use of a genetically engineered E. coli overexpressing β-glucuronidase accompanied by glycyrrhizic acid, a natural and anti-inflammatory agent, for directed treatment of colon carcinoma in a mouse model. Int. J. Pharm. 2020, 579, 119159. [Google Scholar] [CrossRef]
- Available online: https://www.bbc.co.uk/news/uk-wales-south-west-wales-36712240 (accessed on 15 August 2021).
- Jain, A.; Gupta, Y.; Jain, S.K. Azo chemistry and its potential for colonic delivery. Crit. Rev. Ther. Drug Carrier. Syst. 2006, 23. [Google Scholar] [CrossRef]
- Roldo, M.; Barbu, E.; Brown, J.F.; Laight, D.W.; Smart, J.D.; Tsibouklis, J. Azo compounds in colon-specific drug delivery. Expert Opin. Drug Deliv. 2007, 4, 547–560. [Google Scholar] [CrossRef]
- Hou, L.; Shi, Y.; Jiang, G.; Liu, W.; Han, H.; Feng, Q.; Ren, J.; Yuan, Y.; Wang, Y.; Shi, J.; et al. Smart nanocomposite hydrogels based on azo crosslinked graphene oxide for oral colon-specific drug delivery. Nanotechnology 2016, 27, 315105. [Google Scholar] [CrossRef]
- Ray, S. Advanced colon-specific delivery systems for treating local disorders. In Polysaccharide Carriers for Drug Delivery; Woodhead Publishing: Sawston, UK, 2019; pp. 737–762. [Google Scholar]
- Akala, E.O.; Elekwachi, O.; Chase, V.; Johnson, H.; Lazarre, M.; Scott, K. Organic redox-initiated polymerization process for the fabrication of hydrogels for colon-specific drug delivery. Drug Dev. Ind. Pharm. 2003, 29, 375–386. [Google Scholar] [CrossRef]
- Shen, M.Y.; Liu, T.I.; Yu, T.W.; Kv, R.; Chiang, W.H.; Tsai, Y.C.; Chen, H.H.; Lin, S.C.; Chiu, H.C. Hierarchically targetable polysaccharide-coated solid lipid nanoparticles as an oral chemo/thermotherapy delivery system for local treatment of colon cancer. Biomaterials 2019, 197, 86–100. [Google Scholar] [CrossRef]
- Kumar, B.; Murali, A.; Bharath, A.B.; Giri, S. Guar gum modified upconversion nanocomposites for colorectal cancer treatment through enzyme-responsive drug release and NIR-triggered photodynamic therapy. Nanotechnology 2019, 30, 315102. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, Y.; Cai, P.; Yuan, S.; Ma, Q.; Song, Y.; Wei, H.; Wu, Z.; Wu, Z.; Qi, X. Highly specific colon-targeted transformable capsules containing indomethacin immediate-release pellets for colon cancers therapy. J. Drug Target. 2020, 28, 102–110. [Google Scholar] [CrossRef]
- Kaur, R.; Gulati, M.; Singh, S.K. Role of synbiotics in polysaccharide assisted colon targeted microspheres of mesalamine for the treatment of ulcerative colitis. Int. J. Biol. Macromol. 2017, 95, 438–450. [Google Scholar] [CrossRef]
- Kumar, B.; Kulanthaivel, S.; Mondal, A.; Mishra, S.; Banerjee, B.; Bhaumik, A.; Banerjee, I.; Giri, S. Mesoporous silica nano-particle based enzyme responsive system for colon specific drug delivery through guar gum capping. Colloids Surf. B 2017, 150, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Meng, Y.J.; Li, J.; Liu, J.P.; Liu, Z.Q.; Li, D.Q. A novel and simple oral colon-specific drug delivery system based on the pectin/modified nano-carbon sphere nanocomposite gel films. Int. J. Biol. Macromol. 2020, 157, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Gadalla, H.H.; Mohammed, F.A.; El-Sayed, A.M.; Soliman, G.M. Colon-targeting of progesterone using hybrid polymeric microspheres improves its bioavailability and in vivo biological efficacy. Int. J. Pharm. 2020, 577, 119070. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.; Hu, T.G.; Li, L.; Zong, M.H.; Wu, H. A colon-specific delivery system for quercetin with enhanced cancer prevention based on co-axial electrospinning. Food Funct. 2018, 9, 5999–6009. [Google Scholar] [CrossRef]
- Duan, H.; Lü, S.; Gao, C.; Bai, X.; Qin, H.; Wei, Y.; Wu, X.A.; Liu, M. Mucoadhesive microparticulates based on polysaccharide for target dual drug delivery of 5-aminosalicylic acid and curcumin to inflamed colon. Colloids Surf. B 2016, 145, 510–519. [Google Scholar] [CrossRef]
- Sinha, P.; Udhumansha, U.; Rathnam, G.; Ganesh, M.; Jang, H.T. Capecitabine encapsulated chitosan succinate-sodium alginate macromolecular complex beads for colon cancer targeted delivery: In vitro evaluation. Int. J. Biol. Macromol. 2018, 117, 840–850. [Google Scholar] [CrossRef]
- Cao, J.; Cheng, J.; Xi, S.; Qi, X.; Shen, S.; Ge, Y. Alginate/chitosan microcapsules for in-situ delivery of the protein, interleukin-1 receptor antagonist (IL-1Ra), for the treatment of dextran sulfate sodium (DSS)-induced colitis in a mouse model. Eur. J. Pharm. Biopharm. 2019, 137, 112–121. [Google Scholar] [CrossRef]
- Sabra, R.; Billa, N.; Roberts, C.J. Cetuximab-conjugated chitosan-pectinate (modified) composite nanoparticles for targeting colon cancer. Int. J. Pharm. 2019, 572, 118775. [Google Scholar] [CrossRef]
- Asnani, G.P.; Bahekar, J.; Kokare, C.R. Development of novel pH–responsive dual crosslinked hydrogel beads based on Portulaca oleracea polysaccharide-alginate-borax for colon specific delivery of 5-fluorouracil. J. Drug Deliv. Sci. Technol. 2018, 48, 200–208. [Google Scholar] [CrossRef]
- Mohanta, S.; Singh, S.K.; Kumar, B.; Gulati, M.; Kumar, R.; Yadav, A.K.; Wadhwa, S.; Jyoti, J.; Som, S.; Dua, K.; et al. Efficacy of co-administration of modified apple polysaccharide and probiotics in guar gum-Eudragit S100 based mesalamine mini tablets: A novel approach in treating ulcerative colitis. Int. J. Biol. Macromol. 2019, 126, 427–435. [Google Scholar] [CrossRef]
- Jain, A.; Jain, S.K.; Ganesh, N.; Barve, J.; Beg, A.M. Design and development of ligand-appended polysaccharidic nanoparticles for the delivery of oxaliplatin in colorectal cancer. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 179–190. [Google Scholar] [CrossRef]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of Lactobacillus reuteri in human health and diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef]
- Nishida, K.; Sawada, D.; Kuwano, Y.; Tanaka, H.; Rokutan, K. Health benefits of Lactobacillus gasseri CP2305 tablets in young adults exposed to chronic stress: A randomized, double-blind, placebo-controlled study. Nutrients 2019, 11, 1859. [Google Scholar] [CrossRef] [Green Version]
- Chourasia, M.K.; Jain, S.K. Pharmaceutical approaches to colon targeted drug delivery systems. J. Pharm. Pharm. Sci. 2003, 6, 33–66. [Google Scholar] [PubMed]
- Tuohy, K.M.; Kolida, S.; Lustenberger, A.M.; Gibson, G.R. The prebiotic effects of biscuits containing partially hydrolysed guar gum and fructo-oligosaccharides–a human volunteer study. Br. J. Nutr. 2001, 86, 341–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, S. Physiological and pharmaceutical considerations for rectal drug formulations. Front. Pharmacol. 2019, 10, 1196. [Google Scholar] [CrossRef] [PubMed]
- Beery, R.M.; Kane, S. Current approaches to the management of new-onset ulcerative colitis. Clin. Exp. Gastroenterol. 2014, 7, 111. [Google Scholar]
- Prasad, Y.R.; Krishnaiah, Y.S.; Satyanarayana, S. In vitro evaluation of guar gum as a carrier for colon-specific drug delivery. J Control. Release 1998, 51, 281–287. [Google Scholar] [CrossRef]
- Lilly, D.M.; Stillwell, R.H. Probiotics: Growth-promoting factors produced by microorganisms. Science 1965, 147, 747–748. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.; Bishop, N.C.; Oliveira, M.; Tauler, P. Daily probiotic’s (Lactobacillus casei Shirota) reduction of infection incidence in athletes. Int. J. Sport Nutr. Exerc. Metab. 2011, 21, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, M.C.; Golner, B.B.; Goldin, B.R.; Barakat, S.; Dallal, G.E.; Russell, R.M. Survival of yogurt-containing organisms and Lactobacillus gasseri (ADH) and their effect on bacterial enzyme activity in the gastrointestinal tract of healthy and hypochlorhydric elderly subjects. Am. J. Clin. Nutr. 1995, 61, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.T.; Xu, H.; Ye, J.Z.; Wu, W.R.; Shi, D.; Fang, D.Q.; Liu, Y.; Li, L.J. Efficacy of Lactobacillus rhamnosus GG in treatment of acute pediatric diarrhea: A systematic review with meta-analysis. World J. Gastroenterol. 2019, 25, 4999. [Google Scholar] [CrossRef] [PubMed]
- Kourelis, A.; Kotzamanidis, C.; Litopoulou-Tzanetaki, E.; Papaconstantinou, J.; Tzanetakis, N.; Yiangou, M. Immunostimulatory activity of potential probiotic yeast strains in the dorsal air pouch system and the gut mucosa. J. Appl. Microbiol. 2010, 109, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, S.; Lee, J.H.; Kim, J.H.; Che, X.; Ma, H.W.; Seo, D.H.; Kim, T.I.; Kim, W.H.; Kim, S.W.; et al. Lactobacillus acidophilus suppresses intestinal inflammation by inhibiting endoplasmic reticulum stress. J. Gastroenterol. Hepatol. 2019, 34, 178–185. [Google Scholar] [CrossRef] [Green Version]
- Takada, M.; Nishida, K.; Kataoka-Kato, A.; Gondo, Y.; Ishikawa, H.; Suda, K.; Kawai, M.; Hoshi, R.; Watanabe, O.; Igarashi, T.; et al. Probiotic Lactobacillus casei strain Shirota relieves stress-associated symptoms by modulating the gut–brain interaction in human and animal models. Neurogastroenterol. Motil. 2016, 28, 1027–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bull, M.; Plummer, S.; Marchesi, J.; Mahenthiralingam, E. The life history of Lactobacillus acidophilus as a probiotic: A tale of revisionary taxonomy, misidentification and commercial success. FEMS Microbiol. Lett. 2013, 349, 77–87. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Hu, W.; Liu, W.X.; Zhao, L.Y.; Huang, D.; Liu, X.D.; Chan, H.; Zhang, Y.; Zeng, J.D.; Coker, O.O.; et al. Streptococcus thermophilus inhibits colorectal tumorigenesis through secreting β-galactosidase. Gastroenterology 2021, 160, 1179–1193. [Google Scholar] [CrossRef]
- Lee, S.H.; Cho, S.Y.; Yoon, Y.; Park, C.; Sohn, J.; Jeong, J.J.; Jeon, B.N.; Jang, M.; An, C.; Lee, S.; et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat. Microbiol. 2021, 6, 277–288. [Google Scholar] [CrossRef]
- Zhang, M.; Sun, K.; Wu, Y.; Yang, Y.; Tso, P.; Wu, Z. Interactions between intestinal microbiota and host immune response in inflammatory bowel disease. Front. Immunol. 2017, 8, 942. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.; Polk, D.B. Probiotics: Progress toward novel therapies for intestinal diseases. Curr. Opin. Gastroenterol. 2010, 26, 95. [Google Scholar] [CrossRef] [Green Version]
- Vanderpool, C.; Yan, F.; Polk, B.D. Mechanisms of probiotic action: Implications for therapeutic applications in inflammatory bowel diseases. Inflamm. Bowel Dis. 2008, 14, 1585–1596. [Google Scholar] [CrossRef]
- Land, M.H.; Rouster-Stevens, K.; Woods, C.R.; Cannon, M.L.; Cnota, J.; Shetty, A.K. Lactobacillus sepsis associated with probiotic therapy. Pediatrics 2005, 115, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.; Yan, F.; Polk, D.B.; Rao, R.K. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC-and MAP kinase-dependent mechanism. Am. J. Physiol. Gastrointest. 2008, 294, G1060–G1069. [Google Scholar] [CrossRef] [Green Version]
- Fijan, S. Microorganisms with claimed probiotic properties: An overview of recent literature. Int. J. Environ. Res. Public Health 2014, 11, 4745–4767. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Cao, H.; Cover, T.L.; Washington, M.K.; Shi, Y.; Liu, L.; Chaturvedi, R.; Peek, R.M.; Wilson, K.T.; Polk, D.B. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J. Clin. Investig. 2011, 121, 2242–2253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ElRakaiby, M.; Dutilh, B.E.; Rizkallah, M.R.; Boleij, A.; Cole, J.N.; Aziz, R.K. Pharmacomicrobiomics: The impact of human microbiome variations on systems pharmacology and personalized therapeutics. Omics 2014, 18, 402–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70, S38–S44. [Google Scholar] [CrossRef] [Green Version]
- Haiser, H.J.; Gootenberg, D.B.; Chatman, K.; Sirasani, G.; Balskus, E.P.; Turnbaugh, P.J. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 2013, 341, 295–298. [Google Scholar] [CrossRef] [Green Version]
- Available online: http://www.pharmacomicrobiomics.org (accessed on 25 June 2021).
- Oates, J.T.; Lopez, D. Pharmacogenetics: An Important Part of Drug Development with a Focus on Its Application. Int. J. Biomed. Adv. 2018, 1, 111. [Google Scholar] [CrossRef]
- Spanogiannopoulos, P.; Bess, E.N.; Carmody, R.N.; Turnbaugh, P.J. The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 2016, 14, 273–287. [Google Scholar] [CrossRef]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between drugs and the gut microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Clayton, T.A.; Baker, D.; Lindon, J.C.; Everett, J.R.; Nicholson, J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14728–14733. [Google Scholar] [CrossRef] [Green Version]
- Wallace, B.D.; Roberts, A.B.; Pollet, R.M.; Ingle, J.D.; Biernat, K.A.; Pellock, S.J.; Venkatesh, M.K.; Guthrie, L.; O’Neal, S.K.; Robinson, S.J.; et al. Structure and inhibition of microbiome β-glucuronidases essential to the alleviation of cancer drug toxicity. Chem. Biol. 2015, 22, 1238–1249. [Google Scholar] [CrossRef] [Green Version]
- Hylander, B.L.; Repasky, E.A. Temperature as a modulator of the gut microbiome: What are the implications and opportunities for thermal medicine? Int. J. Hyperth. 2019, 36, 83–89. [Google Scholar] [CrossRef] [Green Version]
- Rosean, C.B.; Feng, T.Y.; Azar, F.N.; Rutkowski, M.R. Impact of the microbiome on cancer progression and response to anti-cancer therapies. Adv. Cancer Res. 2019, 143, 255–294. [Google Scholar]
- Grzelak, E.M.; Choules, M.P.; Gao, W.; Cai, G.; Wan, B.; Wang, Y.; McAlpine, J.B.; Cheng, J.; Jin, Y.; Lee, H.; et al. Strategies in anti-Mycobacterium tuberculosis drug discovery based on phenotypic screening. J. Antibiot. Res. 2019, 72, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef] [Green Version]
- Strandwitz, P.; Kim, K.H.; Terekhova, D.; Liu, J.K.; Sharma, A.; Levering, J.; McDonald, D.; Dietrich, D.; Ramadhar, T.R.; Lekbua, A.; et al. GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 2019, 4, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Mimee, M.; Nadeau, P.; Hayward, A.; Carim, S.; Flanagan, S.; Jerger, L.; Collins, J.; McDonnell, S.; Swartwout, R.; Citorik, R.J.; et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 2018, 360, 915–918. [Google Scholar] [CrossRef] [Green Version]
- Lorenzo-Lamosa, M.L.; Remunan-Lopez, C.; Vila-Jato, J.L.; Alonso, M.J. Design of microencapsulated chitosan microspheres for colonic drug delivery. J. Control. Release 1998, 52, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Sood, A.; Dev, A.; Mohanbhai, S.J.; Shrimali, N.; Kapasiya, M.; Kushwaha, A.C.; Choudhury, S.R.; Guchhait, P.; Karmakar, S. Disulfide-bridged chitosan-Eudragit S-100 nanoparticles for colorectal cancer. ACS Appl. Nano Mater. 2019, 2, 6409–6417. [Google Scholar] [CrossRef]


| Polysaccharide | Delivery System | Drug Molecule | Therapeutic Application | Feature | Ref. |
|---|---|---|---|---|---|
| Chitosan | Eudragit S-100 and chitosan-based nanoparticles | Paclitaxel | Colorectal cancer | Sustained-release, pH-responsive, bacterial enzyme sensitive, and cancer-targeted | [44] |
| Dextran | The doxorubicin and superparamagnetic iron oxide nanoparticles-loaded solid lipid nanoparticle coated with folate and dextran | Doxorubicin and superparamagnetic iron oxide nanoparticles | Colon cancer | The microbial enzyme sensitive and tumor-targeted delivery system used for chemo/magnetothermal combination therapy | [45] |
| Guar gum | The guar gum modified upconversion nanocomposite | 5-Fluorouracil | Colorectal cancer | Bacterial enzyme-sensitive and NIR-triggered | [46] |
| Guar gum | Transformable capsules containing indomethacin immediate-release pellets | Indomethacin | Colon cancer | Bacterial enzyme-sensitive | [47] |
| Guar gum | Microspheres | Mesalamine and symbiotic | Ulcerative colitis | Bacterial enzyme-sensitive | [48] |
| Guar gum | 5-Fluorouracil-containing mesoporous silica nanoparticles with guar gum capping | 5-Fluorouracil | Colon cancer | Bacterial enzyme-sensitive | [49] |
| Pectin | The pectin/modified nano-carbon sphere nanocomposite gel films | 5-Fluorouracil | Colon cancer | Bacterial enzyme-sensitive | [50] |
| Pectin | Pectin–zinc acetate beads coated with Eudragit S100 | Pterostilbene | Colorectal cancer | pH-responsive and bacterial enzyme-sensitive | [51] |
| Chitosan and alginate | Thiolated chitosan/alginate composite microparticulate coated by Eudragit S-100 | 5-Aminosalicylic acid and curcumin | Colitis | pH-responsive, bacterial enzyme-sensitive, and mucoadhesive | [52] |
| Chitosan and sodium alginate | The sodium alginate-coated electrospun fiber mat containing quercetin-loaded chitosan nanoparticles and prebiotics | Quercetin and prebiotics | Colon cancer | Bacterial enzyme-sensitive | [53] |
| Chitosan succinate and sodium alginate | Capecitabine encapsulated chitosan succinate–sodium alginate macromolecular complex beads | Capecitabine | Colon cancer | pH-responsive, bacterial enzyme-sensitive, and mucoadhesive | [54] |
| Chitosan and alginate | Microcapsules | Interleukin-1 receptor antagonist | Inflammatory bowel disease | pH-responsive and bacterial enzyme-sensitive | [55] |
| Chitosan and pectin | Modified citrus pectinate–chitosan nanoparticles | Cetuximab and curcumin | Colon cancer | Bacterial enzyme-sensitive, mucoadhesive, and tumor-targeted | [56] |
| Sodium alginate and Portulaca polysaccharide | Polymeric beads encapsulating5-fluorouracil | 5-Fluorouracil | Colorectal cancer | pH-responsive and bacterial enzyme-sensitive | [57] |
| Guar gum and pectin | Tablets coated with guar gum and Eudragit S100 | Modified apple polysaccharide and mesalamine | Ulcerative colitis | Bacterial enzyme-sensitive | [58] |
| Hyaluronic acid and chitosan | Hyaluronic acid-coupled chitosan nanoparticles bearing oxaliplatin encapsulated in Eudragit S100-coated pellets | Oxaliplatin | Colon cancer | Bacterial enzyme-sensitive | [59,60] |
| Bacterial Strain | Effects in Clinical Trials | References |
|---|---|---|
| Lactobacillus reuteri | Colonizing the intestines, primarily animal experiments thus far, perhaps a potential human probiotic | [67] |
| Lactobacillus gasseri (ADH-) | Fecal decreased enzyme and intestinal tract survival | [68] |
| Lactobacillus casei Shirota | Disease prevention, treatment of rotavirus diarrhea, balancing intestinal flora, reduction in the functioning of the fecal enzyme activities, beneficial effects on surface bladder cancer therapy, enhanced immune system in early colon cancer, and immune-boosting | [69,70] |
| Lactobacillus GG (ATCC 53013) | Preventing diarrhea linked with antibiotics, treatment, and the prevention of rotaviruses diarrhea; Clostridium difficile diarrhea therapy; prevention of acute diarrhea; Crohn’s disease; antagonistic against carcinogenic bacteria; vaccine adjuvant; and vaccination adjuvant | [71] |
| Lactobacillus acidophilus NCFB 1748- | Decreased colonic enzyme activity, decreased fecal mutagenicity, avoidance of diarrhea associated radiation, and constipation treatment | [72] |
| Lactobacillus acidophilus LA1 | An immune-stimulating adjuvant attaching to human intestinal cells and the microflora in the intestines | [73] |
| Streptococcus thermophilus | No rotavirus diarrhea impact, no immune enhancement of rotavirus diarrhea, and no fecal enzyme activity | [74] |
| Bifidobacterium bifidum | Rotavirus diarrhea therapy, micro-flora of the intestines, and viral diarrhea treatment | [75] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakshi, H.A.; Quinn, G.A.; Aljabali, A.A.A.; Hakkim, F.L.; Farzand, R.; Nasef, M.M.; Abuglela, N.; Ansari, P.; Mishra, V.; Serrano-Aroca, Á.; et al. Exploiting the Metabolism of the Gut Microbiome as a Vehicle for Targeted Drug Delivery to the Colon. Pharmaceuticals 2021, 14, 1211. https://doi.org/10.3390/ph14121211
Bakshi HA, Quinn GA, Aljabali AAA, Hakkim FL, Farzand R, Nasef MM, Abuglela N, Ansari P, Mishra V, Serrano-Aroca Á, et al. Exploiting the Metabolism of the Gut Microbiome as a Vehicle for Targeted Drug Delivery to the Colon. Pharmaceuticals. 2021; 14(12):1211. https://doi.org/10.3390/ph14121211
Chicago/Turabian StyleBakshi, Hamid A., Gerry A. Quinn, Alaa A. A. Aljabali, Faruck L. Hakkim, Rabia Farzand, Mohamed M. Nasef, Naji Abuglela, Prawej Ansari, Vijay Mishra, Ángel Serrano-Aroca, and et al. 2021. "Exploiting the Metabolism of the Gut Microbiome as a Vehicle for Targeted Drug Delivery to the Colon" Pharmaceuticals 14, no. 12: 1211. https://doi.org/10.3390/ph14121211
APA StyleBakshi, H. A., Quinn, G. A., Aljabali, A. A. A., Hakkim, F. L., Farzand, R., Nasef, M. M., Abuglela, N., Ansari, P., Mishra, V., Serrano-Aroca, Á., & Tambuwala, M. M. (2021). Exploiting the Metabolism of the Gut Microbiome as a Vehicle for Targeted Drug Delivery to the Colon. Pharmaceuticals, 14(12), 1211. https://doi.org/10.3390/ph14121211