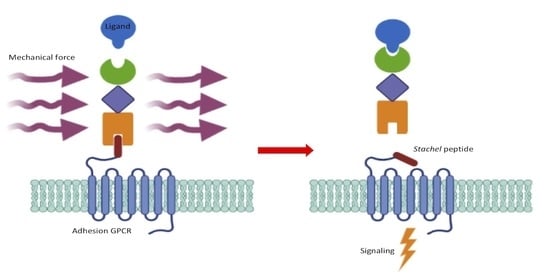
Ligands and Beyond: Mechanosensitive Adhesion GPCRs
Abstract
:1. Introduction
2. Adhesion GPCRs: Structural Characteristics and Activation Mechanisms
3. Mechanosensitive Adhesion GPCRs
3.1. ADGRE2/EMR2
3.2. ADGRE5/CD97
3.3. ADGRG1/GPR56
3.4. ADGRG5/GPR114
3.5. ADGRG6/GPR126
3.6. ADGRL1/LPHN1/CIRL
3.7. ADGRV1/VLGR1
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marinissen, M.J.; Gutkind, J.S. G-protein-coupled receptors and signaling networks: Emerging paradigms. Trends Pharmacol. Sci. 2001, 22, 368–376. [Google Scholar] [CrossRef]
- Kobilka, B.K. G protein coupled receptor structure and activation. Biochim. Biophys. Acta 2007, 1768, 794–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenbaum, D.M.; Rasmussen, S.G.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marullo, S.; Doly, S.; Saha, K.; Enslen, H.; Scott, M.G.H.; Coureuil, M. Mechanical GPCR Activation by Traction Forces Exerted on Receptor N-Glycans. ACS Pharmacol. Transl. Sci. 2020, 3, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Akazawa, H.; Qin, Y.; Sano, M.; Takano, H.; Minamino, T.; Makita, N.; Iwanaga, K.; Zhu, W.; Kudoh, S.; et al. Mechanical stress activates angiotensin II type 1 receptor without the involvement of angiotensin II. Nat. Cell Biol. 2004, 6, 499–506. [Google Scholar] [CrossRef]
- Xu, J.; Mathur, J.; Vessieres, E.; Hammack, S.; Nonomura, K.; Favre, J.; Grimaud, L.; Petrus, M.; Francisco, A.; Li, J.; et al. GPR68 Senses Flow and Is Essential for Vascular Physiology. Cell 2018, 173, 762–775.e716. [Google Scholar] [CrossRef] [Green Version]
- Erdogmus, S.; Storch, U.; Danner, L.; Becker, J.; Winter, M.; Ziegler, N.; Wirth, A.; Offermanns, S.; Hoffmann, C.; Gudermann, T.; et al. Helix 8 is the essential structural motif of mechanosensitive GPCRs. Nat. Commun. 2019, 10, 5784. [Google Scholar] [CrossRef]
- Scholz, N.; Monk, K.R.; Kittel, R.J.; Langenhan, T. Adhesion GPCRs as a Putative Class of Metabotropic Mechanosensors. Handb. Exp. Pharmacol. 2016, 234, 221–247. [Google Scholar] [CrossRef]
- Boyden, S.E.; Desai, A.; Cruse, G.; Young, M.L.; Bolan, H.C.; Scott, L.M.; Eisch, A.R.; Long, R.D.; Lee, C.C.; Satorius, C.L.; et al. Vibratory Urticaria Associated with a Missense Variant in ADGRE2. N. Engl. J. Med. 2016, 374, 656–663. [Google Scholar] [CrossRef]
- Karpus, O.N.; Veninga, H.; Hoek, R.M.; Flierman, D.; van Buul, J.D.; Vandenakker, C.C.; vanBavel, E.; Medof, M.E.; van Lier, R.A.; Reedquist, K.A.; et al. Shear stress-dependent downregulation of the adhesion-G protein-coupled receptor CD97 on circulating leukocytes upon contact with its ligand CD55. J. Immunol. 2013, 190, 3740–3748. [Google Scholar] [CrossRef] [Green Version]
- Wilde, C.; Fischer, L.; Lede, V.; Kirchberger, J.; Rothemund, S.; Schoneberg, T.; Liebscher, I. The constitutive activity of the adhesion GPCR GPR114/ADGRG5 is mediated by its tethered agonist. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2016, 30, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Yeung, J.; Adili, R.; Stringham, E.N.; Luo, R.; Vizurraga, A.; Rosselli-Murai, L.K.; Stoveken, H.M.; Yu, M.; Piao, X.; Holinstat, M.; et al. GPR56/ADGRG1 is a platelet collagen-responsive GPCR and hemostatic sensor of shear force. Proc. Natl. Acad. Sci. USA 2020, 117, 28275–28286. [Google Scholar] [CrossRef] [PubMed]
- Kusuluri, D.K.; Guler, B.E.; Knapp, B.; Horn, N.; Boldt, K.; Ueffing, M.; Aust, G.; Wolfrum, U. Adhesion G protein-coupled receptor VLGR1/ADGRV1 regulates cell spreading and migration by mechanosensing at focal adhesions. iScience 2021, 24, 102283. [Google Scholar] [CrossRef] [PubMed]
- Scholz, N.; Gehring, J.; Guan, C.; Ljaschenko, D.; Fischer, R.; Lakshmanan, V.; Kittel, R.J.; Langenhan, T. The adhesion GPCR latrophilin/CIRL shapes mechanosensation. Cell Rep. 2015, 11, 866–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fredriksson, R.; Lagerstrom, M.C.; Lundin, L.G.; Schioth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef] [Green Version]
- Nordstrom, K.J.; Lagerstrom, M.C.; Waller, L.M.; Fredriksson, R.; Schioth, H.B. The Secretin GPCRs descended from the family of Adhesion GPCRs. Mol. Biol. Evol. 2009, 26, 71–84. [Google Scholar] [CrossRef]
- Piao, X.; Hill, R.S.; Bodell, A.; Chang, B.S.; Basel-Vanagaite, L.; Straussberg, R.; Dobyns, W.B.; Qasrawi, B.; Winter, R.M.; Innes, A.M.; et al. G protein-coupled receptor-dependent development of human frontal cortex. Science 2004, 303, 2033–2036. [Google Scholar] [CrossRef] [Green Version]
- Chang, G.W.; Hsiao, C.C.; Peng, Y.M.; Vieira Braga, F.A.; Kragten, N.A.; Remmerswaal, E.B.; van de Garde, M.D.; Straussberg, R.; Konig, G.M.; Kostenis, E.; et al. The Adhesion G Protein-Coupled Receptor GPR56/ADGRG1 Is an Inhibitory Receptor on Human NK Cells. Cell Rep. 2016, 15, 1757–1770. [Google Scholar] [CrossRef] [Green Version]
- I, K.Y.; Tseng, W.Y.; Wang, W.C.; Gordon, S.; Ng, K.F.; Lin, H.H. Stimulation of Vibratory Urticaria-Associated Adhesion-GPCR, EMR2/ADGRE2, Triggers the NLRP3 Inflammasome Activation Signal in Human Monocytes. Front. Immunol. 2020, 11, 602016. [Google Scholar] [CrossRef]
- Capasso, M.; Durrant, L.G.; Stacey, M.; Gordon, S.; Ramage, J.; Spendlove, I. Costimulation via CD55 on human CD4+ T cells mediated by CD97. J. Immunol. 2006, 177, 1070–1077. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Yang, L.; Begum, S.; Xu, L. GPR56 is essential for testis development and male fertility in mice. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2010, 239, 3358–3367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Osuka, S.; Zhang, Z.; Reichert, Z.R.; Yang, L.; Kanemura, Y.; Jiang, Y.; You, S.; Zhang, H.; Devi, N.S.; et al. BAI1 Suppresses Medulloblastoma Formation by Protecting p53 from Mdm2-Mediated Degradation. Cancer Cell 2018, 33, 1004–1016.e1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, Y.; Lake, R.; Martin, P.L.; Killian, K.; Salerno, P.; Wang, T.; Meltzer, P.; Merino, M.; Cheng, S.Y.; Santoro, M.; et al. CD97 amplifies LPA receptor signaling and promotes thyroid cancer progression in a mouse model. Oncogene 2013, 32, 2726–2738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamann, J.; Aust, G.; Arac, D.; Engel, F.B.; Formstone, C.; Fredriksson, R.; Hall, R.A.; Harty, B.L.; Kirchhoff, C.; Knapp, B.; et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 2015, 67, 338–367. [Google Scholar] [CrossRef]
- Liebscher, I.; Cevheroglu, O.; Hsiao, C.C.; Maia, A.F.; Schihada, H.; Scholz, N.; Soave, M.; Spiess, K.; Trajkovic, K.; Kosloff, M.; et al. A guide to adhesion GPCR research. FEBS J. 2021. [Google Scholar] [CrossRef]
- Lagerstrom, M.C.; Schioth, H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008, 7, 339–357. [Google Scholar] [CrossRef]
- Yona, S.; Lin, H.H.; Siu, W.O.; Gordon, S.; Stacey, M. Adhesion-GPCRs: Emerging roles for novel receptors. Trends Biochem. Sci. 2008, 33, 491–500. [Google Scholar] [CrossRef]
- Arac, D.; Boucard, A.A.; Bolliger, M.F.; Nguyen, J.; Soltis, S.M.; Sudhof, T.C.; Brunger, A.T. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012, 31, 1364–1378. [Google Scholar] [CrossRef] [Green Version]
- Liebscher, I.; Schon, J.; Petersen, S.C.; Fischer, L.; Auerbach, N.; Demberg, L.M.; Mogha, A.; Coster, M.; Simon, K.U.; Rothemund, S.; et al. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 2014, 9, 2018–2026. [Google Scholar] [CrossRef] [Green Version]
- Stoveken, H.M.; Hajduczok, A.G.; Xu, L.; Tall, G.G. Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc. Natl. Acad. Sci. USA 2015, 112, 6194–6199. [Google Scholar] [CrossRef] [Green Version]
- Promel, S.; Langenhan, T.; Arac, D. Matching structure with function: The GAIN domain of adhesion-GPCR and PKD1-like proteins. Trends Pharmacol. Sci. 2013, 34, 470–478. [Google Scholar] [CrossRef]
- Lin, H.H.; Chang, G.W.; Davies, J.Q.; Stacey, M.; Harris, J.; Gordon, S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 2004, 279, 31823–31832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.H.; Stacey, M.; Yona, S.; Chang, G.W. GPS proteolytic cleavage of adhesion-GPCRs. Adv. Exp. Med. Biol. 2010, 706, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Nieberler, M.; Kittel, R.J.; Petrenko, A.G.; Lin, H.H.; Langenhan, T. Control of Adhesion GPCR Function Through Proteolytic Processing. Handb. Exp. Pharmacol. 2016, 234, 83–109. [Google Scholar] [CrossRef]
- Paavola, K.J.; Hall, R.A. Adhesion G protein-coupled receptors: Signaling, pharmacology, and mechanisms of activation. Mol. Pharmacol. 2012, 82, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Vizurraga, A.; Adhikari, R.; Yeung, J.; Yu, M.; Tall, G.G. Mechanisms of adhesion G protein-coupled receptor activation. J. Biol. Chem. 2020, 295, 14065–14083. [Google Scholar] [CrossRef]
- Kishore, A.; Hall, R.A. Versatile Signaling Activity of Adhesion GPCRs. Handb. Exp. Pharmacol. 2016, 234, 127–146. [Google Scholar] [CrossRef] [PubMed]
- Paavola, K.J.; Stephenson, J.R.; Ritter, S.L.; Alter, S.P.; Hall, R.A. The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J. Biol. Chem. 2011, 286, 28914–28921. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, R.; Noorbakhsh, F.; Defea, K.; Hollenberg, M.D. Targeting proteinase-activated receptors: Therapeutic potential and challenges. Nat. Rev. Drug Discov. 2012, 11, 69–86. [Google Scholar] [CrossRef]
- Schoneberg, T.; Liebscher, I.; Luo, R.; Monk, K.R.; Piao, X. Tethered agonists: A new mechanism underlying adhesion G protein-coupled receptor activation. J. Recept. Signal Transduct. Res. 2015, 35, 220–223. [Google Scholar] [CrossRef]
- Liebscher, I.; Schoneberg, T. Tethered Agonism: A Common Activation Mechanism of Adhesion GPCRs. Handb. Exp. Pharmacol. 2016, 234, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Sharif-Naeini, R.; Folgering, J.H.; Bichet, D.; Duprat, F.; Lauritzen, I.; Arhatte, M.; Jodar, M.; Dedman, A.; Chatelain, F.C.; Schulte, U.; et al. Polycystin-1 and -2 dosage regulates pressure sensing. Cell 2009, 139, 587–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Ng, C.; Liu, X.; Wang, Y.; Li, B.; Kashyap, P.; Chaudhry, H.A.; Castro, A.; Kalontar, E.M.; Ilyayev, L.; et al. The ion channel function of polycystin-1 in the polycystin-1/polycystin-2 complex. EMBO Rep. 2019, 20, e48336. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Hackmann, K.; Gao, J.; He, X.; Piontek, K.; Garcia-Gonzalez, M.A.; Menezes, L.F.; Xu, H.; Germino, G.G.; Zuo, J.; et al. Essential role of cleavage of Polycystin-1 at G protein-coupled receptor proteolytic site for kidney tubular structure. Proc. Natl. Acad. Sci. USA 2007, 104, 18688–18693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurbegovic, A.; Kim, H.; Xu, H.; Yu, S.; Cruanes, J.; Maser, R.L.; Boletta, A.; Trudel, M.; Qian, F. Novel functional complexity of polycystin-1 by GPS cleavage in vivo: Role in polycystic kidney disease. Mol. Cell. Biol. 2014, 34, 3341–3353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trudel, M.; Yao, Q.; Qian, F. The Role of G-Protein-Coupled Receptor Proteolysis Site Cleavage of Polycystin-1 in Renal Physiology and Polycystic Kidney Disease. Cells 2016, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Bjarnadottir, T.K.; Fredriksson, R.; Hoglund, P.J.; Gloriam, D.E.; Lagerstrom, M.C.; Schioth, H.B. The human and mouse repertoire of the adhesion family of G-protein-coupled receptors. Genomics 2004, 84, 23–33. [Google Scholar] [CrossRef]
- McKnight, A.J.; Macfarlane, A.J.; Dri, P.; Turley, L.; Willis, A.C.; Gordon, S. Molecular cloning of F4/80, a murine macrophage-restricted cell surface glycoprotein with homology to the G-protein-linked transmembrane 7 hormone receptor family. J. Biol. Chem. 1996, 271, 486–489. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.H.; Stubbs, L.J.; Mucenski, M.L. Identification and characterization of a seven transmembrane hormone receptor using differential display. Genomics 1997, 41, 301–308. [Google Scholar] [CrossRef]
- Beliu, G.; Altrichter, S.; Guixa-Gonzalez, R.; Hemberger, M.; Brauer, I.; Dahse, A.K.; Scholz, N.; Wieduwild, R.; Kuhlemann, A.; Batebi, H.; et al. Tethered agonist exposure in intact adhesion/class B2 GPCRs through intrinsic structural flexibility of the GAIN domain. Mol. Cell 2021, 81, 905–921.e905. [Google Scholar] [CrossRef]
- Zhu, B.; Luo, R.; Jin, P.; Li, T.; Oak, H.C.; Giera, S.; Monk, K.R.; Lak, P.; Shoichet, B.K.; Piao, X. GAIN domain-mediated cleavage is required for activation of G protein-coupled receptor 56 (GPR56) by its natural ligands and a small-molecule agonist. J. Biol. Chem. 2019, 294, 19246–19254. [Google Scholar] [CrossRef] [PubMed]
- Petersen, S.C.; Luo, R.; Liebscher, I.; Giera, S.; Jeong, S.J.; Mogha, A.; Ghidinelli, M.; Feltri, M.L.; Schoneberg, T.; Piao, X.; et al. The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with laminin-211. Neuron 2015, 85, 755–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwakkenbos, M.J.; Kop, E.N.; Stacey, M.; Matmati, M.; Gordon, S.; Lin, H.H.; Hamann, J. The EGF-TM7 family: A postgenomic view. Immunogenetics 2004, 55, 655–666. [Google Scholar] [CrossRef] [PubMed]
- McKnight, A.J.; Gordon, S. EGF-TM7: A novel subfamily of seven-transmembrane-region leukocyte cell-surface molecules. Immunol. Today 1996, 17, 283–287. [Google Scholar] [CrossRef]
- Lin, H.H.; Stacey, M.; Hamann, J.; Gordon, S.; McKnight, A.J. Human EMR2, a novel EGF-TM7 molecule on chromosome 19p13.1, is closely related to CD97. Genomics 2000, 67, 188–200. [Google Scholar] [CrossRef]
- Lin, H.H.; Hsiao, C.C.; Pabst, C.; Hebert, J.; Schoneberg, T.; Hamann, J. Adhesion GPCRs in Regulating Immune Responses and Inflammation. Adv. Immunol. 2017, 136, 163–201. [Google Scholar] [CrossRef]
- Davies, J.Q.; Chang, G.W.; Yona, S.; Gordon, S.; Stacey, M.; Lin, H.H. The role of receptor oligomerization in modulating the expression and function of leukocyte adhesion-G protein-coupled receptors. J. Biol. Chem. 2007, 282, 27343–27353. [Google Scholar] [CrossRef] [Green Version]
- Chang, G.W.; Davies, J.Q.; Stacey, M.; Yona, S.; Bowdish, D.M.; Hamann, J.; Chen, T.C.; Lin, C.Y.; Gordon, S.; Lin, H.H. CD312, the human adhesion-GPCR EMR2, is differentially expressed during differentiation, maturation, and activation of myeloid cells. Biochem. Biophys. Res. Commun. 2007, 353, 133–138. [Google Scholar] [CrossRef]
- Lewis, S.M.; Treacher, D.F.; Edgeworth, J.; Mahalingam, G.; Brown, C.S.; Mare, T.A.; Stacey, M.; Beale, R.; Brown, K.A. Expression of CD11c and EMR2 on neutrophils: Potential diagnostic biomarkers for sepsis and systemic inflammation. Clin. Exp. Immunol. 2015, 182, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.H.; Jeng, W.J.; Ho, Y.P.; Teng, W.; Hsieh, Y.C.; Chen, W.T.; Chen, Y.C.; Lin, H.H.; Sheen, I.S.; Lin, C.Y. Increased EMR2 expression on neutrophils correlates with disease severity and predicts overall mortality in cirrhotic patients. Sci. Rep. 2016, 6, 38250. [Google Scholar] [CrossRef] [Green Version]
- Stacey, M.; Chang, G.W.; Davies, J.Q.; Kwakkenbos, M.J.; Sanderson, R.D.; Hamann, J.; Gordon, S.; Lin, H.H. The epidermal growth factor-like domains of the human EMR2 receptor mediate cell attachment through chondroitin sulfate glycosaminoglycans. Blood 2003, 102, 2916–2924. [Google Scholar] [CrossRef] [PubMed]
- Yona, S.; Lin, H.H.; Dri, P.; Davies, J.Q.; Hayhoe, R.P.; Lewis, S.M.; Heinsbroek, S.E.; Brown, K.A.; Perretti, M.; Hamann, J.; et al. Ligation of the adhesion-GPCR EMR2 regulates human neutrophil function. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2008, 22, 741–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.Y.; Hwang, T.L.; Lin, C.Y.; Lin, T.N.; Lai, H.Y.; Tsai, W.P.; Lin, H.H. EMR2 receptor ligation modulates cytokine secretion profiles and cell survival of lipopolysaccharide-treated neutrophils. Chang. Gung Med. J. 2011, 34, 468–477. [Google Scholar] [PubMed]
- Kop, E.N.; Kwakkenbos, M.J.; Teske, G.J.; Kraan, M.C.; Smeets, T.J.; Stacey, M.; Lin, H.H.; Tak, P.P.; Hamann, J. Identification of the epidermal growth factor-TM7 receptor EMR2 and its ligand dermatan sulfate in rheumatoid synovial tissue. Arthritis Rheum. 2005, 52, 442–450. [Google Scholar] [CrossRef] [PubMed]
- I, K.Y.; Huang, Y.S.; Hu, C.H.; Tseng, W.Y.; Cheng, C.H.; Stacey, M.; Gordon, S.; Chang, G.W.; Lin, H.H. Activation of Adhesion GPCR EMR2/ADGRE2 Induces Macrophage Differentiation and Inflammatory Responses via Galpha16/Akt/MAPK/NF-kappaB Signaling Pathways. Front. Immunol. 2017, 8, 373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irmscher, S.; Brix, S.R.; Zipfel, S.L.H.; Halder, L.D.; Mutluturk, S.; Wulf, S.; Girdauskas, E.; Reichenspurner, H.; Stahl, R.A.K.; Jungnickel, B.; et al. Serum FHR1 binding to necrotic-type cells activates monocytic inflammasome and marks necrotic sites in vasculopathies. Nat. Commun. 2019, 10, 2961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamann, J.; Vogel, B.; van Schijndel, G.M.; van Lier, R.A. The seven-span transmembrane receptor CD97 has a cellular ligand (CD55, DAF). J. Exp. Med. 1996, 184, 1185–1189. [Google Scholar] [CrossRef]
- Kwakkenbos, M.J.; Pouwels, W.; Matmati, M.; Stacey, M.; Lin, H.H.; Gordon, S.; van Lier, R.A.; Hamann, J. Expression of the largest CD97 and EMR2 isoforms on leukocytes facilitates a specific interaction with chondroitin sulfate on B cells. J. Leukoc. Biol. 2005, 77, 112–119. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Ward, Y.; Tian, L.; Lake, R.; Guedez, L.; Stetler-Stevenson, W.G.; Kelly, K. CD97, an adhesion receptor on inflammatory cells, stimulates angiogenesis through binding integrin counterreceptors on endothelial cells. Blood 2005, 105, 2836–2844. [Google Scholar] [CrossRef]
- Wandel, E.; Saalbach, A.; Sittig, D.; Gebhardt, C.; Aust, G. Thy-1 (CD90) is an interacting partner for CD97 on activated endothelial cells. J. Immunol. 2012, 188, 1442–1450. [Google Scholar] [CrossRef] [Green Version]
- Leemans, J.C.; te Velde, A.A.; Florquin, S.; Bennink, R.J.; de Bruin, K.; van Lier, R.A.; van der Poll, T.; Hamann, J. The epidermal growth factor-seven transmembrane (EGF-TM7) receptor CD97 is required for neutrophil migration and host defense. J. Immunol. 2004, 172, 1125–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spendlove, I.; Sutavani, R. The role of CD97 in regulating adaptive T-cell responses. Adv. Exp. Med. Biol. 2010, 706, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Kop, E.N.; Adriaansen, J.; Smeets, T.J.; Vervoordeldonk, M.J.; van Lier, R.A.; Hamann, J.; Tak, P.P. CD97 neutralisation increases resistance to collagen-induced arthritis in mice. Arthritis Res. Ther. 2006, 8, R155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aust, G.; Zhu, D.; Van Meir, E.G.; Xu, L. Adhesion GPCRs in Tumorigenesis. Handb. Exp. Pharmacol. 2016, 234, 369–396. [Google Scholar] [CrossRef] [Green Version]
- Niu, M.; Xu, S.; Yang, J.; Yao, D.; Li, N.; Yan, J.; Zhong, G.; Song, G. Structural basis for CD97 recognition of the decay-accelerating factor CD55 suggests mechanosensitive activation of adhesion GPCRs. J. Biol. Chem. 2021, 296, 100776. [Google Scholar] [CrossRef]
- Hilbig, D.; Sittig, D.; Hoffmann, F.; Rothemund, S.; Warmt, E.; Quaas, M.; Sturmer, J.; Seiler, L.; Liebscher, I.; Hoang, N.A.; et al. Mechano-Dependent Phosphorylation of the PDZ-Binding Motif of CD97/ADGRE5 Modulates Cellular Detachment. Cell Rep. 2018, 24, 1986–1995. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Chen, W.; Lou, J.; Rittase, W.; Li, K. Mechanosensing through immunoreceptors. Nat. Immunol. 2019, 20, 1269–1278. [Google Scholar] [CrossRef]
- Cerny, O.; Godlee, C.; Tocci, R.; Cross, N.E.; Shi, H.; Williamson, J.C.; Alix, E.; Lehner, P.J.; Holden, D.W. CD97 stabilises the immunological synapse between dendritic cells and T cells and is targeted for degradation by the Salmonella effector SteD. PLoS Pathog. 2021, 17, e1009771. [Google Scholar] [CrossRef]
- Salzman, G.S.; Ackerman, S.D.; Ding, C.; Koide, A.; Leon, K.; Luo, R.; Stoveken, H.M.; Fernandez, C.G.; Tall, G.G.; Piao, X.; et al. Structural Basis for Regulation of GPR56/ADGRG1 by Its Alternatively Spliced Extracellular Domains. Neuron 2016, 91, 1292–1304. [Google Scholar] [CrossRef] [Green Version]
- Little, K.D.; Hemler, M.E.; Stipp, C.S. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: Central role of CD81 in facilitating GPR56-Galpha q/11 association. Mol. Biol. Cell 2004, 15, 2375–2387. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Begum, S.; Hearn, J.D.; Hynes, R.O. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2006, 103, 9023–9028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, R.; Jeong, S.J.; Jin, Z.; Strokes, N.; Li, S.; Piao, X. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc. Natl. Acad. Sci. USA 2011, 108, 12925–12930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giera, S.; Luo, R.; Ying, Y.; Ackerman, S.D.; Jeong, S.J.; Stoveken, H.M.; Folts, C.J.; Welsh, C.A.; Tall, G.G.; Stevens, B.; et al. Microglial transglutaminase-2 drives myelination and myelin repair via GPR56/ADGRG1 in oligodendrocyte precursor cells. eLife 2018, 7, e33385. [Google Scholar] [CrossRef] [PubMed]
- Chiang, N.Y.; Chang, G.W.; Huang, Y.S.; Peng, Y.M.; Hsiao, C.C.; Kuo, M.L.; Lin, H.H. Heparin interacts with the adhesion GPCR GPR56, reduces receptor shedding, and promotes cell adhesion and motility. J. Cell Sci. 2016, 129, 2156–2169. [Google Scholar] [CrossRef] [Green Version]
- Jin, G.; Sakitani, K.; Wang, H.; Jin, Y.; Dubeykovskiy, A.; Worthley, D.L.; Tailor, Y.; Wang, T.C. The G-protein coupled receptor 56, expressed in colonic stem and cancer cells, binds progastrin to promote proliferation and carcinogenesis. Oncotarget 2017, 8, 40606–40619. [Google Scholar] [CrossRef]
- Chen, H.; Nwe, P.K.; Yang, Y.; Rosen, C.E.; Bielecka, A.A.; Kuchroo, M.; Cline, G.W.; Kruse, A.C.; Ring, A.M.; Crawford, J.M.; et al. A Forward Chemical Genetic Screen Reveals Gut Microbiota Metabolites That Modulate Host Physiology. Cell 2019, 177, 1217–1231.e1218. [Google Scholar] [CrossRef]
- Li, T.; Chiou, B.; Gilman, C.K.; Luo, R.; Koshi, T.; Yu, D.; Oak, H.C.; Giera, S.; Johnson-Venkatesh, E.; Muthukumar, A.K.; et al. A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. EMBO J. 2020, 39, e104136. [Google Scholar] [CrossRef]
- Singh, A.K.; Lin, H.H. The role of GPR56/ADGRG1 in health and disease. Biomed. J. 2021, 44, 534–547. [Google Scholar] [CrossRef]
- Budday, S.; Steinmann, P.; Kuhl, E. Physical biology of human brain development. Front. Cell. Neurosci. 2015, 9, 257. [Google Scholar] [CrossRef] [Green Version]
- Luo, R.; Jeong, S.J.; Yang, A.; Wen, M.; Saslowsky, D.E.; Lencer, W.I.; Arac, D.; Piao, X. Mechanism for adhesion G protein-coupled receptor GPR56-mediated RhoA activation induced by collagen III stimulation. PLoS ONE 2014, 9, e100043. [Google Scholar] [CrossRef] [Green Version]
- Kuivaniemi, H.; Tromp, G. Type III collagen (COL3A1): Gene and protein structure, tissue distribution, and associated diseases. Gene 2019, 707, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Chiang, N.Y.; Peng, Y.M.; Juang, H.H.; Chen, T.C.; Pan, H.L.; Chang, G.W.; Lin, H.H. GPR56/ADGRG1 Activation Promotes Melanoma Cell Migration via NTF Dissociation and CTF-Mediated Galpha12/13/RhoA Signaling. J. Investig. Dermatol. 2017, 137, 727–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, J.P.; Wrann, C.D.; Rao, R.R.; Nair, S.K.; Jedrychowski, M.P.; You, J.S.; Martinez-Redondo, V.; Gygi, S.P.; Ruas, J.L.; Hornberger, T.A.; et al. G protein-coupled receptor 56 regulates mechanical overload-induced muscle hypertrophy. Proc. Natl. Acad. Sci. USA 2014, 111, 15756–15761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, K.; Hunter, I.; Engel, J. Structure and function of laminin: Anatomy of a multidomain glycoprotein. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 1990, 4, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Fesus, L.; Piacentini, M. Transglutaminase 2: An enigmatic enzyme with diverse functions. Trends Biochem. Sci. 2002, 27, 534–539. [Google Scholar] [CrossRef]
- Peng, Y.M.; van de Garde, M.D.; Cheng, K.F.; Baars, P.A.; Remmerswaal, E.B.; van Lier, R.A.; Mackay, C.R.; Lin, H.H.; Hamann, J. Specific expression of GPR56 by human cytotoxic lymphocytes. J. Leukoc. Biol. 2011, 90, 735–740. [Google Scholar] [CrossRef] [Green Version]
- Stoveken, H.M.; Bahr, L.L.; Anders, M.W.; Wojtovich, A.P.; Smrcka, A.V.; Tall, G.G. Dihydromunduletone Is a Small-Molecule Selective Adhesion G Protein-Coupled Receptor Antagonist. Mol. Pharmacol. 2016, 90, 214–224. [Google Scholar] [CrossRef]
- Stoveken, H.M.; Larsen, S.D.; Smrcka, A.V.; Tall, G.G. Gedunin- and Khivorin-Derivatives Are Small-Molecule Partial Agonists for Adhesion G Protein-Coupled Receptors GPR56/ADGRG1 and GPR114/ADGRG5. Mol. Pharmacol. 2018, 93, 477–488. [Google Scholar] [CrossRef]
- Leon, K.; Cunningham, R.L.; Riback, J.A.; Feldman, E.; Li, J.; Sosnick, T.R.; Zhao, M.; Monk, K.R.; Arac, D. Structural basis for adhesion G protein-coupled receptor Gpr126 function. Nat. Commun. 2020, 11, 194. [Google Scholar] [CrossRef] [Green Version]
- Musa, G.; Cazorla-Vazquez, S.; van Amerongen, M.J.; Stemmler, M.P.; Eckstein, M.; Hartmann, A.; Braun, T.; Brabletz, T.; Engel, F.B. Gpr126 (Adgrg6) is expressed in cell types known to be exposed to mechanical stimuli. Ann. N. Y. Acad. Sci. 2019, 1456, 96–108. [Google Scholar] [CrossRef]
- Paavola, K.J.; Sidik, H.; Zuchero, J.B.; Eckart, M.; Talbot, W.S. Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci. Signal. 2014, 7, ra76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuffer, A.; Lakkaraju, A.K.; Mogha, A.; Petersen, S.C.; Airich, K.; Doucerain, C.; Marpakwar, R.; Bakirci, P.; Senatore, A.; Monnard, A.; et al. The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature 2016, 536, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Patra, C.; van Amerongen, M.J.; Ghosh, S.; Ricciardi, F.; Sajjad, A.; Novoyatleva, T.; Mogha, A.; Monk, K.R.; Muhlfeld, C.; Engel, F.B. Organ-specific function of adhesion G protein-coupled receptor GPR126 is domain-dependent. Proc. Natl. Acad. Sci. USA 2013, 110, 16898–16903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monk, K.R.; Oshima, K.; Jors, S.; Heller, S.; Talbot, W.S. Gpr126 is essential for peripheral nerve development and myelination in mammals. Development 2011, 138, 2673–2680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxendale, S.; Asad, A.; Shahidan, N.O.; Wiggin, G.R.; Whitfield, T.T. The adhesion GPCR Adgrg6 (Gpr126): Insights from the zebrafish model. Genesis 2021, 59, e23417. [Google Scholar] [CrossRef] [PubMed]
- Torregrosa-Carrion, R.; Pineiro-Sabaris, R.; Siguero-Alvarez, M.; Grego-Bessa, J.; Luna-Zurita, L.; Fernandes, V.S.; MacGrogan, D.; Stainier, D.Y.R.; de la Pompa, J.L. Adhesion G protein-coupled receptor Gpr126/Adgrg6 is essential for placental development. Sci. Adv. 2021, 7, eabj5445. [Google Scholar] [CrossRef] [PubMed]
- Mogha, A.; Benesh, A.E.; Patra, C.; Engel, F.B.; Schoneberg, T.; Liebscher, I.; Monk, K.R. Gpr126 functions in Schwann cells to control differentiation and myelination via G-protein activation. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 17976–17985. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; He, L.; Jia, K.; Yue, Z.; Li, S.; Jin, Y.; Li, Z.; Siwko, S.; Xue, F.; Su, J.; et al. Regulation of body length and bone mass by Gpr126/Adgrg6. Sci. Adv. 2020, 6, eaaz0368. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Hussien, A.A.; Wang, Y.; Heckmann, T.; Gonzalez, R.; Karner, C.M.; Snedeker, J.G.; Gray, R.S. An adhesion G protein-coupled receptor is required in cartilaginous and dense connective tissues to maintain spine alignment. eLife 2021, 10, e67781. [Google Scholar] [CrossRef]
- Kou, I.; Takahashi, Y.; Johnson, T.A.; Takahashi, A.; Guo, L.; Dai, J.; Qiu, X.; Sharma, S.; Takimoto, A.; Ogura, Y.; et al. Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis. Nat. Genet. 2013, 45, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Ravenscroft, G.; Nolent, F.; Rajagopalan, S.; Meireles, A.M.; Paavola, K.J.; Gaillard, D.; Alanio, E.; Buckland, M.; Arbuckle, S.; Krivanek, M.; et al. Mutations of GPR126 are responsible for severe arthrogryposis multiplex congenita. Am. J. Hum. Genet. 2015, 96, 955–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseini, M.; Fattahi, Z.; Abedini, S.S.; Hu, H.; Ropers, H.H.; Kalscheuer, V.M.; Najmabadi, H.; Kahrizi, K. GPR126: A novel candidate gene implicated in autosomal recessive intellectual disability. Am. J. Med. Genet. Part A 2019, 179, 13–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKee, K.K.; Yang, D.H.; Patel, R.; Chen, Z.L.; Strickland, S.; Takagi, J.; Sekiguchi, K.; Yurchenco, P.D. Schwann cell myelination requires integration of laminin activities. J. Cell Sci. 2012, 125, 4609–4619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davletov, B.A.; Shamotienko, O.G.; Lelianova, V.G.; Grishin, E.V.; Ushkaryov, Y.A. Isolation and biochemical characterization of a Ca2+-independent alpha-latrotoxin-binding protein. J. Biol. Chem. 1996, 271, 23239–23245. [Google Scholar] [CrossRef] [Green Version]
- Krasnoperov, V.G.; Beavis, R.; Chepurny, O.G.; Little, A.R.; Plotnikov, A.N.; Petrenko, A.G. The calcium-independent receptor of alpha-latrotoxin is not a neurexin. Biochem. Biophys. Res. Commun. 1996, 227, 868–875. [Google Scholar] [CrossRef]
- Lelianova, V.G.; Davletov, B.A.; Sterling, A.; Rahman, M.A.; Grishin, E.V.; Totty, N.F.; Ushkaryov, Y.A. Alpha-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. J. Biol. Chem. 1997, 272, 21504–21508. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Salinas, A.L.; Avila-Zozaya, M.; Ugalde-Silva, P.; Hernandez-Guzman, D.A.; Missirlis, F.; Boucard, A.A. Latrophilins: A Neuro-Centric View of an Evolutionary Conserved Adhesion G Protein-Coupled Receptor Subfamily. Front. Neurosci. 2019, 13, 700. [Google Scholar] [CrossRef]
- Scholz, N.; Guan, C.; Nieberler, M.; Grotemeyer, A.; Maiellaro, I.; Gao, S.; Beck, S.; Pawlak, M.; Sauer, M.; Asan, E.; et al. Mechano-dependent signaling by Latrophilin/CIRL quenches cAMP in proprioceptive neurons. eLife 2017, 6, e28360. [Google Scholar] [CrossRef]
- Dannhauser, S.; Lux, T.J.; Hu, C.; Selcho, M.; Chen, J.T.; Ehmann, N.; Sachidanandan, D.; Stopp, S.; Pauls, D.; Pawlak, M.; et al. Antinociceptive modulation by the adhesion GPCR CIRL promotes mechanosensory signal discrimination. eLife 2020, 9, e56738. [Google Scholar] [CrossRef]
- Gibert, Y.; McMillan, D.R.; Kayes-Wandover, K.; Meyer, A.; Begemann, G.; White, P.C. Analysis of the very large G-protein coupled receptor gene (Vlgr1/Mass1/USH2C) in zebrafish. Gene 2005, 353, 200–206. [Google Scholar] [CrossRef] [Green Version]
- McMillan, D.R.; Kayes-Wandover, K.M.; Richardson, J.A.; White, P.C. Very large G protein-coupled receptor-1, the largest known cell surface protein, is highly expressed in the developing central nervous system. J. Biol. Chem. 2002, 277, 785–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, J.; Fu, Y.H.; Clark, A.M.; Nakahara, S.; Hamano, K.; Iwasaki, N.; Matsui, A.; Arinami, T.; Ptacek, L.J. A nonsense mutation of the MASS1 gene in a family with febrile and afebrile seizures. Ann. Neurol. 2002, 52, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Weston, M.D.; Luijendijk, M.W.; Humphrey, K.D.; Moller, C.; Kimberling, W.J. Mutations in the VLGR1 gene implicate G-protein signaling in the pathogenesis of Usher syndrome type II. Am. J. Hum. Genet. 2004, 74, 357–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skradski, S.L.; Clark, A.M.; Jiang, H.; White, H.S.; Fu, Y.H.; Ptacek, L.J. A novel gene causing a mendelian audiogenic mouse epilepsy. Neuron 2001, 31, 537–544. [Google Scholar] [CrossRef] [Green Version]
- Yagi, H.; Takamura, Y.; Yoneda, T.; Konno, D.; Akagi, Y.; Yoshida, K.; Sato, M. Vlgr1 knockout mice show audiogenic seizure susceptibility. J. Neurochem. 2005, 92, 191–202. [Google Scholar] [CrossRef]
- McGee, J.; Goodyear, R.J.; McMillan, D.R.; Stauffer, E.A.; Holt, J.R.; Locke, K.G.; Birch, D.G.; Legan, P.K.; White, P.C.; Walsh, E.J.; et al. The very large G-protein-coupled receptor VLGR1: A component of the ankle link complex required for the normal development of auditory hair bundles. J. Neurosci. Off. J. Soc. Neurosci. 2006, 26, 6543–6553. [Google Scholar] [CrossRef]
- Maerker, T.; van Wijk, E.; Overlack, N.; Kersten, F.F.; McGee, J.; Goldmann, T.; Sehn, E.; Roepman, R.; Walsh, E.J.; Kremer, H.; et al. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 2008, 17, 71–86. [Google Scholar] [CrossRef]
- Reiners, J.; van Wijk, E.; Marker, T.; Zimmermann, U.; Jurgens, K.; te Brinke, H.; Overlack, N.; Roepman, R.; Knipper, M.; Kremer, H.; et al. Scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2. Hum. Mol. Genet. 2005, 14, 3933–3943. [Google Scholar] [CrossRef] [Green Version]
- Mathur, P.; Yang, J. Usher syndrome: Hearing loss, retinal degeneration and associated abnormalities. Biochim. Biophys. Acta 2015, 1852, 406–420. [Google Scholar] [CrossRef] [Green Version]
- Geiger, B.; Spatz, J.P.; Bershadsky, A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009, 10, 21–33. [Google Scholar] [CrossRef]
- Shen, B.; Delaney, M.K.; Du, X. Inside-out, outside-in, and inside-outside-in: G protein signaling in integrin-mediated cell adhesion, spreading, and retraction. Curr. Opin. Cell Biol. 2012, 24, 600–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Guo, S.S.; Fassler, R. Integrin-mediated mechanotransduction. J. Cell Biol. 2016, 215, 445–456. [Google Scholar] [CrossRef] [PubMed]



| Receptor | Ligand | GPS Cleavage | Mechanical Force | In Vitro/In Vivo Evidence | References |
|---|---|---|---|---|---|
| ADGRE2/EMR2 | DS, Ab | Dependent | Vibratory shaking | Vibratory urticaria, in vitro cell-based system | [9] |
| ADGRE5/CD97 | CD55 | Dependent | Shear stress | Blood flow/animal model | [10] |
| ADGRG1/GPR56 | Collagen III TG2/laminin, Ab | Dependent Dependent | Shear stress N/A | Blood flow/animal model in vitro cell-based system | [12] [51] |
| ADGRG5/GPR114 | N/A | Independent | Vibratory shaking | in vitro cell-based system | [11] |
| ADGRG6/GPR126 | laminin-211 | Dependent | Ligand polymerization | Zebrafish model, in vitro cell-based system | [52] |
| ADGRL1/LPHN1/CIRL | N/A | Independent | Touch, sound, stretch | Drosophila model | [14] |
| ADGRV1/VLGR1 | N/A | N/A | mechanical stretch | in vitro cell-based system | [13] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.-H.; Ng, K.-F.; Chen, T.-C.; Tseng, W.-Y. Ligands and Beyond: Mechanosensitive Adhesion GPCRs. Pharmaceuticals 2022, 15, 219. https://doi.org/10.3390/ph15020219
Lin H-H, Ng K-F, Chen T-C, Tseng W-Y. Ligands and Beyond: Mechanosensitive Adhesion GPCRs. Pharmaceuticals. 2022; 15(2):219. https://doi.org/10.3390/ph15020219
Chicago/Turabian StyleLin, Hsi-Hsien, Kwai-Fong Ng, Tse-Ching Chen, and Wen-Yi Tseng. 2022. "Ligands and Beyond: Mechanosensitive Adhesion GPCRs" Pharmaceuticals 15, no. 2: 219. https://doi.org/10.3390/ph15020219
APA StyleLin, H.-H., Ng, K.-F., Chen, T.-C., & Tseng, W.-Y. (2022). Ligands and Beyond: Mechanosensitive Adhesion GPCRs. Pharmaceuticals, 15(2), 219. https://doi.org/10.3390/ph15020219