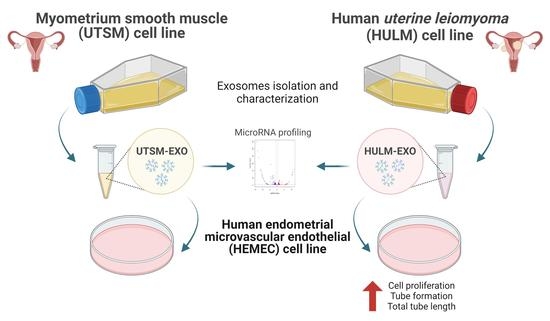
Report of Exosomes Isolated from a Human Uterine Leiomyoma Cell Line and Their Impact on Endometrial Vascular Endothelial Cells
Abstract
:1. Introduction
2. Results
2.1. Isolation and Depiction of Exosomes Derived from Human Uterine Leiomyoma Cell Line
2.2. Uptake of Exosomes from Uterine Leiomyoma Cell Line by Human Endometrial Microvascular Endothelial Cells
2.3. Exosomes from Human Uterine Leiomyoma Cell Line Increase Cell Proliferation
2.4. Exosomes from Human Uterine Leiomyoma Cell Line Cocultured with HEMEC Demonstrate Angiogenic Properties
2.5. MicroRNA Profiling of HULM- and UTSM-Derived Exosomes
2.6. Pathway Analysis of Differentially Regulated miRNA Target Genes
3. Discussion
4. Materials and Methods
4.1. Cell Culture Conditions
4.2. Isolation of Exosomes and Treatment
4.3. Characterization of Exosomes from HULM and UTSM Cell Lines
4.4. Exosome Labeling
4.5. Cell Proliferation Assay
4.6. RNA Extraction and Gene Expression
4.7. Endothelial Tube Formation Assay
4.8. miRNA Extraction, Small RNA-Sequencing, and Bioinformatic Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersen, J.; Barbieri, R.L. Abnormal gene expression in uterine leiomyomas. J. Soc. Gynecol. Investig. 1995, 2, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.A. Uterine fibroids. Lancet 2001, 357, 293–298. [Google Scholar] [CrossRef]
- Wallach, E.E.; Vlahos, N.F. Uterine myomas: An overview of development, clinical features, and management. Obstet. Gynecol. 2004, 104, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.L.; Stewart, E.A. Uterine fibroids: The elephant in the room. Science 2005, 308, 1589–1592. [Google Scholar] [CrossRef] [Green Version]
- Pavone, D.; Clemenza, S.; Sorbi, F.; Fambrini, M.; Petraglia, F. Epidemiology and Risk Factors of Uterine Fibroids. Best Pract Res. Clin. Obstet. Gynaecol. 2018, 46, 3–11. [Google Scholar] [CrossRef]
- Ali, M.; Shahin, S.M.; Sabri, N.A.; Al-Hendy, A.; Yang, Q. Hypovitaminosis D exacerbates the DNA damage load in human uterine fibroids, which is ameliorated by vitamin D3 treatment. Acta Pharmacol. Sin. 2019, 40, 957–970. [Google Scholar] [CrossRef]
- Bariani, M.V.; Rangaswamy, R.; Siblini, H.; Yang, Q.; Al-Hendy, A.; Zota, A.R. The role of endocrine-disrupting chemicals in uterine fibroid pathogenesis. Curr. Opin. Endocrinol. Diabetes 2020, 27, 380–387. [Google Scholar] [CrossRef]
- Bulun, S.E. Uterine fibroids. N. Engl. J. Med. 2013, 369, 1344–1355. [Google Scholar] [CrossRef] [Green Version]
- Cardozo, E.R.; Clark, A.D.; Banks, N.K.; Henne, M.B.; Stegmann, B.J.; Segars, J.H. The estimated annual cost of uterine leiomyomata in the United States. Am. J. Obstet. Gynecol. 2012, 206, 211.e1–211.e2119. [Google Scholar] [CrossRef] [Green Version]
- Norian, J.M.; Malik, M.; Parker, C.Y.; Joseph, D.; Leppert, P.C.; Segars, J.H.; Catherino, W.H. Transforming growth factor beta3 regulates the versican variants in the extracellular matrix-rich uterine leiomyomas. Reprod. Sci. 2009, 16, 1153–1164. [Google Scholar] [CrossRef] [Green Version]
- Sozen, I.; Olive, D.L.; Arici, A. Expression and hormonal regulation of monocyte chemotactic protein-1 in myometrium and leiomyomata. Fertil. Steril. 1998, 69, 1095–1102. [Google Scholar] [CrossRef]
- Senturk, L.M.; Sozen, I.; Gutierrez, L.; Arici, A. Interleukin 8 production and interleukin 8 receptor expression in human myometrium and leiomyoma. Am. J. Obstet. Gynecol. 2001, 184, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Elkafas, H.; Ali, M.; Elmorsy, E.; Kamel, R.; Thompson, W.E.; Badary, O.; Al-Hendy, A.; Yang, Q.; Yang, Q. Vitamin D3 Ameliorates DNA Damage Caused by Developmental Exposure to Endocrine Disruptors in the Uterine Myometrial Stem Cells of Eker Rats. Cells 2020, 9, 1459. [Google Scholar] [CrossRef] [PubMed]
- Kitaya, K.; Yasuo, T. Leukocyte density and composition in human cycling endometrium with uterine fibroids. Hum. Immunol. 2010, 71, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Yin, P.; Ono, M.; Monsivais, D.; Moravek, M.B.; Coon, J.S., 5th; Dyson, M.T.; Wei, J.J.; Bulun, S.E. 5-Hydroxymethylcytosine promotes proliferation of human uterine leiomyoma: A biological link to a new epigenetic modification in benign tumors. J. Clin. Endocrinol. Metab. 2014, 99, E2437–E2445. [Google Scholar] [CrossRef]
- Ali, M.; Esfandyari, S.; Al-Hendy, A. Evolving role of microRNAs in uterine fibroid pathogenesis: Filling the gap! Fertil. Steril. 2020, 113, 1167–1168. [Google Scholar] [CrossRef]
- Zavadil, J.; Ye, H.; Liu, Z.; Wu, J.; Lee, P.; Hernando, E.; Soteropoulos, P.; Toruner, G.A.; Wei, J.-J. Profiling and functional analyses of microRNAs and their target gene products in human uterine leiomyomas. PLoS ONE 2010, 5, e12362. [Google Scholar] [CrossRef]
- Navarro, A.; Bariani, M.V.; Yang, Q.; Al-Hendy, A. Understanding the Impact of Uterine Fibroids on Human Endometrium Function. Front. Cell Dev. Biol. 2021, 9, 633180. [Google Scholar] [CrossRef]
- Li, M.; Zeringer, E.; Barta, T.; Schageman, J.; Cheng, A.; Vlassov, A.V. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R Soc. Lond B Biol Sci. 2014, 369, 20130502. [Google Scholar] [CrossRef]
- Sharma, A.; Khatun, Z.; Shiras, A. Tumor exosomes: Cellular postmen of cancer diagnosis and personalized therapy. Nanomedicine 2016, 11, 421–437. [Google Scholar] [CrossRef]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, S.; Sanderson, M.P.; Stoeck, A.; Altevogt, P. Exosomes: From biogenesis and secretion to biological function. Immunol. Lett. 2006, 107, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Frydrychowicz, M.; Kolecka-Bednarczyk, A.; Madejczyk, M.; Yasar, S.; Dworacki, G. Exosomes-structure, biogenesis and biological role in non-small-cell lung cancer. Scand. J. Immunol. 2015, 81, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Sluijter, J.P.; Verhage, V.; Deddens, J.C.; van den Akker, F.; Doevendans, P.A. Microvesicles and exosomes for intracardiac communication. Cardiovasc. Res. 2014, 102, 302–311. [Google Scholar] [CrossRef] [Green Version]
- Di Pietro, C. Exosome-mediated communication in the ovarian follicle. J. Assist. Reprod. Genet. 2016, 33, 303–311. [Google Scholar] [CrossRef] [Green Version]
- Aliotta, J.M.; Pereira, M.; Johnson, K.W.; De Paz, N.; Dooner, M.S.; Puente, N.; Ayala, C.; Brilliant, K.; Berz, D.; Lee, D. Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Exp. Hematol. 2010, 38, 233–245. [Google Scholar] [CrossRef] [Green Version]
- Ng, Y.H.; Rome, S.; Jalabert, A.; Forterre, A.; Singh, H.; Hincks, C.L.; Salamonsen, L.A. Endometrial exosomes/microvesicles in the uterine microenvironment: A new paradigm for embryo-endometrial cross talk at implantation. PLoS ONE 2013, 8, e58502. [Google Scholar] [CrossRef] [Green Version]
- Harp, D.; Driss, A.; Mehrabi, S.; Chowdhury, I.; Xu, W.; Liu, D.; Garcia-Barrio, M.; Taylor, R.N.; Gold, B.; Jefferson, S.; et al. Exosomes derived from endometriotic stromal cells have enhanced angiogenic effects in vitro. Cell Tissue Res. 2016, 365, 187–196. [Google Scholar] [CrossRef] [Green Version]
- Khalaj, K.; Miller, J.E.; Lingegowda, H.; Fazleabas, A.T.; Young, S.L.; Lessey, B.A.; Koti, M.; Tayade, C. Extracellular vesicles from endometriosis patients are characterized by a unique miRNA-lncRNA signature. JCI Insight. 2019, 4, e128846. [Google Scholar] [CrossRef]
- Mineo, M.; Garfield, S.H.; Taverna, S.; Flugy, A.; De Leo, G.; Alessandro, R.; Kohn, E.C. Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a Src-dependent fashion. Angiogenesis 2012, 15, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Shifren, J.L.; Tseng, J.F.; Zaloudek, C.J.; Ryan, I.P.; Meng, Y.G.; Ferrara, N.; Jaffe, R.B.; Taylor, R.N. Ovarian steroid regulation of vascular endothelial growth factor in the human endometrium: Implications for angiogenesis during the menstrual cycle and in the pathogenesis of endometriosis. J. Clin. Endocrinol. Metab. 1996, 81, 3112–3118. [Google Scholar] [CrossRef] [Green Version]
- Shabbir, A.; Cox, A.; Rodriguez-Menocal, L.; Salgado, M.; Van Badiavas, E. Mesenchymal Stem Cell Exosomes Induce Proliferation and Migration of Normal and Chronic Wound Fibroblasts, and Enhance Angiogenesis In Vitro. Stem. Cells Dev. 2015, 24, 1635–1647. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef] [Green Version]
- Zota, A.R.; Geller, R.J.; Vannoy, B.N.; Marfori, C.Q.; Tabbara, S.; Hu, L.Y.; Baccarelli, A.A.; Moawad, G.N. Phthalate Exposures and MicroRNA Expression in Uterine Fibroids: The FORGE Study. Epigenet. Insights 2020, 13, 2516865720904057. [Google Scholar] [CrossRef] [Green Version]
- Chuang, T.D.; Khorram, O. Expression Profiling of lncRNAs, miRNAs, and mRNAs and Their Differential Expression in Leiomyoma Using Next-Generation RNA Sequencing. Reprod. Sci. 2018, 25, 246–255. [Google Scholar] [CrossRef]
- Kim, Y.J.; Kim, Y.Y.; Shin, J.H.; Kim, H.; Ku, S.Y.; Suh, C.S. Variation in MicroRNA Expression Profile of Uterine Leiomyoma with Endometrial Cavity Distortion and Endometrial Cavity Non-Distortion. Int. J. Mol. Sci. 2018, 19, 2524. [Google Scholar] [CrossRef] [Green Version]
- Marsh, E.E.; Lin, Z.; Yin, P.; Milad, M.; Chakravarti, D.; Bulun, S.E. Differential expression of microRNA species in human uterine leiomyoma versus normal myometrium. Fertil. Steril. 2008, 89, 1771–1776. [Google Scholar] [CrossRef] [Green Version]
- Yang, E.; Xue, L.; Li, Z.; Yi, T. Lnc-AL445665.1-4 may be involved in the development of multiple uterine leiomyoma through interacting with miR-146b-5p. BMC Cancer 2019, 19, 709. [Google Scholar] [CrossRef]
- Marsh, E.E.; Steinberg, M.L.; Parker, J.B.; Wu, J.; Chakravarti, D.; Bulun, S.E. Decreased expression of microRNA-29 family in leiomyoma contributes to increased major fibrillar collagen production. Fertil. Steril. 2016, 106, 766–772. [Google Scholar] [CrossRef] [Green Version]
- Ciebiera, M.; Włodarczyk, M.; Zgliczyński, S.; Łoziński, T.; Walczak, K.; Czekierdowski, A. The Role of miRNA and Related Pathways in Pathophysiology of Uterine Fibroids-From Bench to Bedside. Int. J. Mol. Sci. 2020, 21, 3016. [Google Scholar] [CrossRef] [Green Version]
- Lazzarini, R.; Caffarini, M.; Delli Carpini, G.; Ciavattini, A.; Di Primio, R.; Orciani, M. From 2646 to 15: Differentially regulated microRNAs between progenitors from normal myometrium and leiomyoma. Am. J. Obstet. Gynecol. 2020, 222, 596.e1–596.e9. [Google Scholar] [CrossRef]
- Wei, S.; Li, Q.; Li, Z.; Wang, L.; Zhang, L.; Xu, Z. miR-424-5p promotes proliferation of gastric cancer by targeting Smad3 through TGF-β signaling pathway. Oncotarget 2016, 7, 75185–75196. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Ma, N.; Zhao, R.; Wu, G.; Zhang, Y.; Qiao, Y.; Han, D.; Xu, Y.; Xiang, Y.; Yan, B.; et al. Overexpression of miR-483-5p/3p cooperate to inhibit mouse liver fibrosis by suppressing the TGF-β stimulated HSCs in transgenic mice. J. Cell Mol. Med. 2014, 18, 966–974. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Q.; Zhou, Y.; Yang, Z.; Tan, M. Inhibition of miR-182-5p attenuates pulmonary fibrosis via TGF-β/Smad pathway. Hum. Exp. Toxicol. 2020, 39, 683–695. [Google Scholar] [CrossRef]
- Li, X.M.; Yu, W.Y.; Chen, Q.; Zhuang, H.R.; Gao, S.Y.; Zhao, T.L. LncRNA TUG1 exhibits pro-fibrosis activity in hypertrophic scar through TAK1/YAP/TAZ pathway via miR-27b-3p. Mol. Cell Biochem. 2021, 476, 3009–3020. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Jiang, B.; Zhang, Q.; Zhou, R.; Zhang, L.; Wang, C. Study on the role of Hsa-miR-31-5p in hypertrophic scar formation and the mechanism. Exp. Cell Res. 2017, 361, 201–209. [Google Scholar] [CrossRef]
- Moghaddam, A.S.; Afshari, J.T.; Esmaeili, S.A.; Saburi, E.; Joneidi, Z.; Momtazi-Borojeni, A.A. Cardioprotective microRNAs: Lessons from stem cell-derived exosomal microRNAs to treat cardiovascular disease. Atherosclerosis 2019, 285, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Yan, T.; Huang, C.; Xu, Z.; Wang, L.; Jiang, E.; Wang, H.; Chen, Y.; Liu, K.; Shao, Z.; et al. Melanoma cell-secreted exosomal miR-155-5p induce proangiogenic switch of cancer-associated fibroblasts via SOCS1/JAK2/STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2018, 37, 242. [Google Scholar] [CrossRef] [Green Version]
- Kolkova, Z.; Holubekova, V.; Grendar, M.; Nachajova, M.; Zubor, P.; Pribulova, T.; Loderer, D.; Zigo, I.; Biringer, K.; Hornakova, A. Association of Circulating miRNA Expression with Preeclampsia, Its Onset, and Severity. Diagnostics 2021, 11, 476. [Google Scholar] [CrossRef]
- Dong, X.; Sui, C.; Huang, K.; Wang, L.; Hu, D.; Xiong, T.; Wang, R.; Zhang, H. MicroRNA-223-3p suppresses leukemia inhibitory factor expression and pinopodes formation during embryo implantation in mice. Am. J. Transl. Res. 2016, 8, 1155–1163. [Google Scholar]
- An, J.; Li, L.; Zhang, X.; Liu, L.; Wang, L.; Zhang, X. A clinical and basic study of optimal endometrial preparation protocols for patients with infertility undergoing frozen-thawed embryo transfer. Exp. Ther. Med. 2020, 20, 2191–2199. [Google Scholar] [CrossRef]
- Butler, A.E.; Ramachandran, V.; Hayat, S.; Dargham, S.; Cunningham, T.K.; Benurwar, M.; Sathyapalan, T.; Najafi-Shoushtari, S.H.; Atkin, S.L. Expression of microRNA in follicular fluid in women with and without PCOS. Sci. Rep. 2019, 9, 16306. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Y.; Wang, L.; Gong, C.; Sun, J.; Chen, X.; Chen, Y.; Yang, L.; Zhang, Y.; Yang, X.; et al. MicroRNAome in decidua: A new approach to assess the maintenance of pregnancy. Fertil. Steril. 2015, 103, 980–989.e6. [Google Scholar] [CrossRef]
- Machtinger, R.; Rodosthenous, R.; Adir, M.; Mansour, A.; Racowsky, C.; Baccarelli, A.A.; Hauser, R. Extracellular microRNAs in follicular fluid and their potential association with oocyte fertilization and embryo quality: An exploratory study. J. Assist. Reprod. Genet. 2017, 34, 525–533. [Google Scholar] [CrossRef] [Green Version]
- Vidal-Gómez, X.; Pérez-Cremades, D.; Mompeón, A.; Dantas, A.P.; Novella, S.; Hermenegildo, C. MicroRNA as Crucial Regulators of Gene Expression in Estradiol-Treated Human Endothelial Cells. Cell Physiol. Biochem. 2018, 45, 1878–1892. [Google Scholar] [CrossRef]
- Akbar, R.; Ullah, K.; Rahman, T.U.; Cheng, Y.; Pang, H.-Y.; Jin, L.Y.; Wang, Q.-J.; Huang, H.-F.; Sheng, J.-Z. miR-183-5p regulates uterine receptivity and enhances embryo implantation. J. Mol. Endocrinol. 2020, 64, 43–52. [Google Scholar] [CrossRef]
- Suo, M.; Sun, Y.; Yang, H.; Ji, J.; He, Y.; Dong, L.; Wang, Y.; Zhang, Y.; Zhang, Y.; Hao, M. miR-183-5p suppressed the invasion and migration of HTR-8/SVneo trophoblast cells partly via targeting MMP-9 in preeclampsia. Biosci. Rep. 2020, 40, BSR20192575. [Google Scholar] [CrossRef]
- Carney, S.A.; Tahara, H.; Swartz, C.D.; Risinger, J.I.; He, H.; Moore, A.B.; Haseman, J.K.; Barrett, J.C.; Dixon, D. Immortalization of human uterine leiomyoma and myometrial cell lines after induction of telomerase activity: Molecular and phenotypic characteristics. Lab. Investig. 2002, 82, 719–728. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarro, A.; Bariani, M.V.; Park, H.-S.; Zota, A.R.; Al-Hendy, A. Report of Exosomes Isolated from a Human Uterine Leiomyoma Cell Line and Their Impact on Endometrial Vascular Endothelial Cells. Pharmaceuticals 2022, 15, 577. https://doi.org/10.3390/ph15050577
Navarro A, Bariani MV, Park H-S, Zota AR, Al-Hendy A. Report of Exosomes Isolated from a Human Uterine Leiomyoma Cell Line and Their Impact on Endometrial Vascular Endothelial Cells. Pharmaceuticals. 2022; 15(5):577. https://doi.org/10.3390/ph15050577
Chicago/Turabian StyleNavarro, Antonia, Maria Victoria Bariani, Hang-Soo Park, Ami R. Zota, and Ayman Al-Hendy. 2022. "Report of Exosomes Isolated from a Human Uterine Leiomyoma Cell Line and Their Impact on Endometrial Vascular Endothelial Cells" Pharmaceuticals 15, no. 5: 577. https://doi.org/10.3390/ph15050577
APA StyleNavarro, A., Bariani, M. V., Park, H. -S., Zota, A. R., & Al-Hendy, A. (2022). Report of Exosomes Isolated from a Human Uterine Leiomyoma Cell Line and Their Impact on Endometrial Vascular Endothelial Cells. Pharmaceuticals, 15(5), 577. https://doi.org/10.3390/ph15050577