Actinomycetes as Producers of Biologically Active Terpenoids: Current Trends and Patents
Abstract
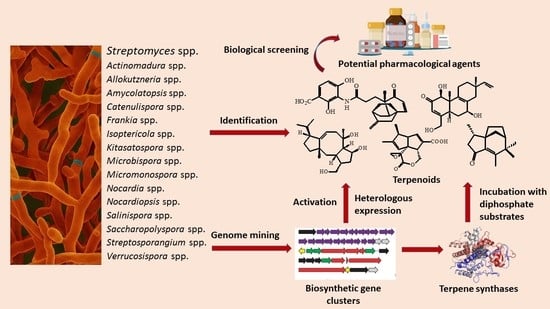
:1. Introduction
2. Terpene Biosynthesis by Actinomycetes
2.1. Terpene Derivatives Produced by Streptomycetes and Their Enzymes
2.1.1. Mono- and Sesquiterpenes and Their Derivatives
 |  |  |  | |
| 1 | 2 | 3 | 4 | |
 |  |  |  | |
| 5 | 6 | 7 | 8 | |
 |  |  |  |  |
| 9 | 10 | 11 | 12 | 13 |
 |  |  |  |
| 14 R1=R2=H, R3=OH, R4=H 15 R1=R2=R3=H, R4=OH 16 R1=R3=H, R2=R4=OH 17 R1=OH, R2=R3=H, R4=OH | 18 | 19 | 20 |
 |  |  |  |
| 21 | 22 | 23 | 24 |
 |  |  |  |
| 25 | 26 | 27 | 28 |
 |  |  |  |
| 29 | 30 | 31 R1=R2=R3=H 32 R1=OH, R2=R3=H 33 R1=R3=H, R2=OH 34 R1=R2=H, R3=OH | 35 |
 |  |  |  |
| 36 R=H 37 R=OH | 38 R=H 39 R=OH | 40 | 41 |
 |  |  | |||
| 42 | 43 | 44 | |||
 |  |  |  | ||
| 45 | 46 R1=OH, R2=R3=H 47 R1=H, R2=R3=OH | 48 R1=R2=H, R3=OH 49 R1=OH, R2=R3=H 50 R2=OH, R1=R3=H | 51 | ||
 |  |  |  | ||
| 52 | 53 | 54 | 55 R1=OH, R2=R3=H 56 R1=OH, R2=H, R3=OH 57 R1=H, R2=OH, R3=H | ||
 |  |  |  |
| 58 | 59 R1=R2=R3=H 60 R1=OH, R2=R3=H 61 R1=R2=H, R3=OH 62 R2=R3=OH, R1=H | 63 | 64 |
 |  |  |  |
| 65 | 66 | 67 | 68 |
 |  |  | |
| 69 | 70 | 71 |
 |  |  |  |
| 72 | 73 | 74 | 75 |
 |  |  |  |
| 76 | 77 | 78 | 79 |
 |  |  |  |
| 80 | 81 | 82 | 83 R=H 84 R=OH |
 |  |  |  |
| 85 | 86 | 87 | 88 |
 |  |  |  |
| 89 | 90 | 91 | 92 |
 |  |  |  |
| 93 | 94 R=H 95 R1=H, R2=αOH 96 R1=H, R2=βOH 97 R1=αOH, R2=H | 98 R1=OH, R2=H 99 R2=OH, R1=H | 100 |
 |  |  |  |
101 R=αH 102 R=βH | 103 R1=R2=H 104 R1=αOH, R2=H 105 R1=βOH, R2=H 106 R1=αOH, R2=OH | 107 | 108 |
 |  |  |  |
| 109 | 110 | 111 | 112 |
 |  |  |  |
| 113 | 114 | 115 | 116 |
 |  |  |  |
| 117 | 118 | 119 | 120 |
 |  |  |  |
| 121 | 122 | 123 | 124 |
 |  |  |  |
| 125 | 126 | 127 | 128 |
 |  |  | |
| 129 | 130 | 131 |
 |  |  |  |
| 132 | 133 R1=H, R2=CH2OH 134 R1=H, R2=CHO 135 R1=OH, R2=COOH | 136 R=H 137 R=OH | 138 R=H 139 R=OH |
 |  |  |  |
| 140 | 141 | 142 | 143 |
 |  |  |  |
| 144 | 145 | 146 | 147 |
 |  |  | |
| 148 R=H 149 R=OH | 150 | 151 |
 |  |  |  |
| 152 | 153 | 154 | 155 R=prenyl 156 R=geranyl 157 R=ipent |
 |  |  | |
| 158 | 159 R=prenyl | 160 m=0, n=2 161 m=1, n=0 | |
2.1.2. Diterpenes and Their Derivatives
 |  |  |  | |
| 162 | 163 | 164 | 165 | |
 |  |  |  | |
| 166 | 167 | 168 | 169 | |
 |  |  |  |
| 170 R=H 171 R=OH | 172 | 173 | 174 |
 |  |  |  |
| 175 R=H 176 R=OH | 177 R1=H, R2=CH2OH 178 R1=OH, R2=CH3 | 179 | 180 |
 |  | ||
| 181 R=OH 182 R=H | 183 | ||
 |  |  |  |
| 184 R1=R2=CH3 185 R1=R2=CH2OH 186 R1=CH3, R2=CH2OH | 187 | 188 | 189 R=H 190 R=βOH |
 |  |  |  |
| 191 R1=OH, R2=COCH3 192 R1=R2=H | 193 | 194 R1=αOH, R2=H 195 R1=βOH, R2=H 196 R1=H, R2=αOH | 197 |
 |  |  |
| 198 | 199 | 200 |
 |  | |
| 201 | 202 |
 |  |  | ||
| 203 | 204 | 205 | ||
 |  |  |  | |
| 206 | 207 | 208 | 209 | |
 |  |  |  | |
| 210 | 211 | 212 | 213 | |
2.1.3. Sester-, Tri-, and Tetraterpenes and Their Derivatives
 |  | |||
| 214 | 215 | |||
 |  |  | ||
| 216 | 217 | 218 | ||
 |  |  | ||
| 219 | 220 | 221 | ||
 | ||||
| 222 | ||||
 |  |
| 223 | 224 |
 | |
| 225 |
2.1.4. Hybrid Metabolites (Meroterpenoids)
 | R1 | R2 | R3 | |||||||
| 226 | OH | CH3 | OH | |||||||
| 227 | OH | CH2OH | H | |||||||
| 228 | H | CH3 | H | |||||||
| 229 | OH | CH3 | H | |||||||
| 230 | H | CH2OH | H | |||||||
| 231 | OH | CH2OH | OH | |||||||
| 232 | OH | COOH | H | |||||||
| 233 | OH | CONH2 | H | |||||||
 |  | |||||||||
| 234 | 235 | |||||||||
 |  | |||||||||
| 236 | 237 | |||||||||
 |  |  | ||||||||
| 238 | 239 | 240 | ||||||||
 |  |  | ||||||||
| 241 | 242 | 243 | ||||||||
 |  | |||||||||
| 244 | 245 246 | |||||||||
 |  | |||||||||
| 247 | 248 R=OH 249 R=Cl | |||||||||
 |  | |||||||||
| 250 | 251 | |||||||||
 |  | |||||||||
| 252 | 253 | |||||||||
 |  |  | ||||||||
| 254 | 255 | 256 | ||||||||
 |  | |||||||||
| 257 | 258 | |||||||||
 |  |  |
| 259 | 260 R=βOH 261 R=αOH | 262 R=H 263 R=COCH3 |
 |  |  |
| 264 | 265 | 266 |
 |  |  |
| 267 | 268 | 269 |
 |  |  |
| 270 | 271 | 272 |
 |  |  |
| 273 R=Cl 274 R=Br | 275 R=Cl 276 R=Br | 277 |
 |  |  |
| 278 | 279 | 280 |
 |  |  |
| 281 | 282 | 283 |
 |  |  |
| 284 | 285 | 286 |
 |  | |
| 287 | 288 | |
 |  | |
| 289 | 290 R1=OH, R2=CH2OH 291 R1=H, R2=CH3 | |
 |  |  |
| 292 R1=βCH3, R2=H 293 R1=αCH3, R2=H 294 R1=βCH3, R2=OH 295 R1=αCH3, R2=OH | 296 | 297 |
 |  |  |
| 298 R1=R2=H, R3=H 299 R1=R2=H, R3=OH 300 R1=OH, R2=H, R3=H 301 R1=H, R2=OH, R3=H | 302 | 303 |
 |  | |
| 304 R=H 305 R=Cl | 306 | |
 | ||
| 307 | ||
 |  |  | |
| 308 R=H 309 R=CH3 | 310 R1=R2=R3=H 311 R1=R2=H, R3=CH3 312 R1=H, R2=Cl, R3=H 313 R1=OH, R2=R3=H | 314 R=H 315 R=CH2OH 316 R=CH(OH)2 317 R=CHO 318 R=COOH | |
 |  |  | |
| 319 | 320 | 321 R=CH3 322 R=H | |
 |  | ||
| 323 R1=H, R2=αOH, R3=H 324 R1=H, R2=αOH, R3=CH3 325 R1=H, R2=OCH3, R3=H | 326 R=CH3 327 R=CH2OH | ||
 |  | ||
| 328 R=H 329 R=CH3 | 330 dixiamycin B | ||
 |  | ||
| 331 | 332 | ||
 |  | ||
| 333 | 334 | ||
 |  | ||
| 335 | 336 | ||
 |  | ||
| 337 | 338 | ||
 |  | ||
| 339 | 340 | ||
 |  | ||
| 341 R1=R2=H, R3=CH3, R4=H 342 R1=R2=H, R3=CH2OH, R4=H 343 R1=R2=H, R3=R4=CH3 344 R1=H, R2=OH, R3=CH3, R4=H 345 R1=OH, R2=H, R3=CH3, R4=H | 346 | ||
 |  |  |  |
| 347 R= | 348 R= | 349 R= | |
 | 351 R=NH2 |  |  |
| 350 R= | 352 R= | 353 R= |
 | ||||||
| R1 | R2 | R3 | R4 | |||
| 354 | OH | -CO-CH3 |  | CH3 | ||
| 355 | OH | -CO-CH3 |  | H | ||
| 356 | OH | -CO-CH3 |  | -CH2-O-CH3 | ||
| 357 | H | -CO-CH3 |  | CH3 | ||
| 358 | H | -CO-CH3 | OH | CH3 | ||
| 359 | H | -CO-CH3 |  | CH3 | ||
| 360 | H | H | H | CH3 | ||
| 361 | H | -CO-CH3 | H | CH3 | ||
| 362 | H | H | OH | CH3 | ||
| 363 | H | H |  | CH3 | ||
| 364 | H | -CO-CH3 |  | -CH2-O-CH3 | ||
 |  | 367 R=αOH | ||||
| 368 R=O | ||||||
 | ||||||
| 365 R=αOH 366 R=O | 369 R | |||||
 |  | |||||
| 370 | 371 R1=Cl, R2=CH3, R3=OCH3 372 R1=H, R2=CH3, R3=OCH3 373 R1=Cl, R2=CH3, R3=OH 374 R1=Cl, R2=CH3, R3=oxo 375 R1=Cl, R2=H, R3=OCH3 | |||||
 |  | |||||
| 376 | 377 | |||||
 |  | |||||
| 378 | 379 R=H 380 R=CH3 | |||||
 |  | |||||
| 381 R=H 382 R=OH | 383 | |||||
 |  |  |  | ||
| 384 R= | 385 R= | 386 R= | |||
 |  |  | |||
| 387 | 388 | 389 | |||
 |  |  | ||
| 390 | 391 | 392 | ||
 |  | |||
| 393 | 394 R1=OH, R2=H 395 R1=H, R2=OH | |||
 |  | |||
| 396 R1= R2=H 397 R1=OH, R2=H 398 R1=H, R2=OH | 399 R=OH | |||
 |  | |||
| 400 | 401 R=SCH3 402 R=OCH3 | |||
 |  |  | ||
| 403 R=NH2 404 R=OH | 405 | |||
 |  | |||
| 406 | ||||
| 407 | ||||
 | ||||
| 408 | ||||
2.2. Terpene Derivatives Produced by Others Actinomycetes and Their Enzymes
2.2.1. Mono- and Sesquiterpenes and Their Derivatives
 |  |  |  |
| 409 | 410 | 411 | 412 |
 |  |  |
| 413 | 414 | 415 |
 |  |  |
| 416 | 417 | 418 |
 |  |  |
| 419 | 420 | 421 |
2.2.2. Di- and Triterpenes and Their Derivatives
 |  |  |  | |
| 422 | 423 | 424 | 425 | |
 |  |  |  | |
| 426 | 427 | 428 | 429 | |
 |  |  |  | |
| 430 R=αH 431 R=βH | 432 | 433 | 434 | |
 |  |  | ||
| 435 | 436 | 437 | ||
 |  |  |  | |
| 438 | 439 | 440 | 441 | |
 |  |  |  | |
| 442 | 443 | 444 | 445 | |
 |  |  |  | |
| 446 | 447 | 448 | 449 | |
 |  |  |  | |
| 450 | 451 | 452 | 453 | |
 |  |  |  |
| 454 | 455 | 456 | 457 |
 |  |  |  |
| 458 | 459 | 460 | 461 |
 |  |  | |
| 462 | 463 | 464 |
 |  | |||||||||
 |  |  |  |  |  | |||||
| 465 R1 | 466 R1 | 467 R1 | 472 R1 | 473 R1 | 474 R1 | |||||
468 R1 | 475 R1 | |||||||||
R1 | 469 R2=H 470 R2=-COCH2CH3 471 R2=-COCH2C6H5 | R1 | 476 R2=H 477 R2=-COCH2CH3 478 R2=-COCH2C6H5 | |||||||
 |  |  | ||||||||
| 479 | 480 | 481 | ||||||||
 |  | |||||||||
| 482 | 483 | |||||||||
2.2.3. Hybrid Metabolites (Meroterpenoids)
 |  |  |  |
| 484 | 485 | 486 | 487 |
 |  | ||
| 488 | 489 | ||
 | 490 R=OCH3 | ||
| 491 R=H | |||
 |  | ||
| 492 R=OCH3 493 R=H | 494 R=OCH3 495 R=H | ||
 | |||
| 496 | |||
 |  |  499 | |
| 497 R | |||
 | |||
| 498 R | |||
3. Discussion
| Compound | Previously Isolated from Other Sources | Strain/Enzyme | Patent | Biological Activity | ||
|---|---|---|---|---|---|---|
| Mono- and sesquiterpenes | ||||||
| 1,8-Cineole (1) | Yes | Streptomyces clavuligerus ATCC 27064 | [53,54,55] | WO2018142109 | anti-inflammatory antioxidant | [333] |
| Linalool (2) | Yes | Streptomyces clavuligerus ATCC 27064 | [53,54,55] | WO2020234307 WO2018142109 | anticancer antimicrobial neuroprotective anxiolytic antidepressant anti-stress hepatoprotective | [334] |
| Streptomyces sp. GWS-BW-H5 | [53] | |||||
| Nerolidol (3) | Yes | Streptomyces clavuligerus ATCC 27064 | [53,54,55] | WO2018142109 WO2020234307 | antimicrobial anti-biofilm antioxidant antiparasitic skin-penetration enhancer skin-repellent antinociceptive anti-inflammatory anticancer | [335] |
| α-Pinene (7) β-Pinene (8) | Yes | Streptomyces coelicolor A3(2) | [63] | antimicrobial | [336] | |
| Limonene (9) | Yes | Streptomyces coelicolor A3(2) | [63] | antimicrobial antioxidant anti-inflammatory antidiabetic | [337] | |
| γ-Terpinene (10) δ-Terpinene (11) | Yes | Streptomyces coelicolor A3(2) | [63] | antioxidant | [338] | |
| (1R)-(+)-Camphor (12) | Yes | Streptomyces coelicolor A3(2) | [65] | insecticidal | [339] | |
| (-)-epi-α-Bisabolol (18) | Yes | Streptomyces citricolor NBRC 13005 | [67] | anti-inflammatory analgesic antibiotic anticancer | [340] | |
| Germacrene B (26) Germacrene D (24) | Yes | TS from Streptomyces pristinaespiralis ATCC 25486 | [82] | antileishmanial antiproliferative | [341] | |
| SAV76 from Streptomyces avermitilis | [83] | |||||
| SpS from Streptomyces xinghaiensis S187 | [84] | |||||
| Streptomyces hygroscopicus NRRL 15879 | [66] | |||||
| Bicyclogermacrene (28) | Yes | SpS from Streptomyces xinghaiensis S187 | [84] | antibacterial antifungal | [342] | |
| Isopterchiayione (415) | No | Isoptericola chiayiensis BCRC 16888 | [262] | anti-inflammatory (IC50 24.72 ± 1.25 µM) | [262] | |
| Cyperusol C (417) | Yes | Verrucosispora gifhornensis YM28-088 | [264] | antiviral (against hepatitis B virus, IC50 14.1 ± 1.1 µM) | [343] | |
| epi-Cubenol (31) | Yes | Streptomyces sp. GWS-BW-H5 | [53] | antifungal | [344] | |
| Transf. Streptomyces lividans TK21 gecA from Streptomyces griseus IFO13350 | [87] | |||||
| Streptomyces albolongus YIM 101047 | [73] | |||||
| Streptomyces griseus NBRC102592 | [88] | |||||
| Streptomyces roseosporus NRRL 11379 | [5] | |||||
| Streptomyces sp. SirexAA-E | [5] | |||||
| Streptomyces roseosporus NRRL15998 | [237] | |||||
| Streptomyces flavogriseus ATCC33331 | [237] | |||||
| Kandenol A (36) Kandenol B (37) Kandenol C (38) Kandenol D (39) Kandenol E (40) | No | Streptomyces sp. HKI0595 | [90] | antimicrobial (against Bacillus subtilis, Mycobacterium vaccae, MIC 12.5–50 µM) | [90] | |
| (2R,4S,8αR)-8,8α,1,2,3,4-Hexahydro-2-hydroxy-4,8α-dimethyl-2(2H)-naphthalenone (52) | No | Streptomyces sp. XM17 | [96] | antiviral (against influenza A virus, IC50 5–49 nM) | [96] | |
| (1S,3S,4S,4αS,8αR)-4,8α-Dimethyloctahydronaphthalene-1,3,4α(3H)-triol (53) | ||||||
| (4S,4αS,8αS)-Octahydro-4α-hydroxy-4,8α-dimethyl-1(2H)-naphthalenone (54) | ||||||
| (1β,4β,4aβ,8aα)-4,8α-Dimethyloctahydronaphthalene-1,4a(2H)-diol (55) | No | Streptomyces albolongus YIM 101047 | [73] | antifungal (against Candida parapsilosis, MIC 3.13 µg/mL) | [73] | |
| (-)-δ-Cadinene (58) | Yes | SSCG_02150 from Streptomyces clavuligerus ATCC 27074 | [97] | antimicrobial | [345] | |
| T-Muurolol (59) | Yes | SSCG_03688 from Streptomyces clavuligerus ATCC 27074 | [97] | antifungal | [346] | |
| Streptomyces sp. M491 | [98] | |||||
| 15-Hydroxy-T-muurolol (61) | No | Streptomyces sp. M491 | [98] | antitumor (IC50 6.7 µg/mL) | [98] | |
| 10-epi-δ-Eudesmol (86) | Yes | Streptomyces chartreusis NRRL 3882 | [5] | repellent (against Aedes aegypti and ticks) | [102,347] | |
| β-Eudesmol (72) | Yes | Streptomyces exfoliatus SMF19 | [66] | potential antitumor potential antiangiogenic antimicrobial | [348,349] | |
| Streptomyces hygroscopicus NRRL 15879 | [66] | |||||
| Aromadendrene oxide-(2) (79) | Yes | Streptomyces hygroscopicus NRRL 15879 | [66] | antibacterial antitumor | [350] | |
| (-)-β-Cedrene (126) (+)-β-Cedrene (127) | Yes | Streptomyces hygroscopicus NRRL 15879 | [66] | WO2015120431 | antibacterial | [351] |
| epi-isozizaene synthase from Streptomyces coelicolor A3(2) | [110,122] | |||||
| β-Patchoulene (77) | Yes | Streptomyces hygroscopicus NRRL 15879 | [66] | anti-inflammatory | [352] | |
| α-Elemol (80) | Yes | Streptomyces parvulus B1682 | [66] | insecticidal (against Ixodes scapularis, Amblyomma americanum) | [353] | |
| Streptomyces chartreusis NRRL 3882 | [102] | |||||
| Caryophyllene (93) | Yes | Streptomyces yanglinensis 3-10 | [62] | anticancer antioxidant antimicrobial | [354,355] | |
| Saccharothrix espanaensis DSM 44229 | [103] | |||||
| Caryolan-1-ol (94) | Yes | Streptomyces griseus | [105] | antifungal (against Botrytis cinerea, IC50 0.026 µM/mL) | [107] | |
| Transf. Streptomyces lividans with gcoA from S. griseus | ||||||
| Streptomyces globisporus TFH56 | [106] | |||||
| Streptomyces griseus S4–7 | [107] | WO2018062668 | ||||
| Streptomyces albolongus YIM 101047 | [73] | |||||
| Albaflavenone (109) | No | Streptomyces coelicolor A3 (2) | [112] | antibacterial (against Bacillus subtilis, MIC 8–10 µg/mL) | [356] | |
| Transf. Streptomyces avermitilis SUKA16 with sav3032 and sav4925 from S. avermitilis | [119] | |||||
| Streptomyces cyaneogriseus subsp. noncyanogenus | [5] | |||||
| Streptomyces spectabilis NRRL-2792 | [118] | |||||
| Streptomyces viridochromogenes DSM 40736 | [116] | |||||
| Streptomyces griseoflavus Tu4000 | [116] | |||||
| Streptomyces ghanaensis ATCC 14672 | [116] | |||||
| Streptomyces albus ATCC 2396 | [116] | |||||
| Streptomyces sp. CRB46 | [115] | |||||
| Streptomyces coelicolor M145 | [117] | |||||
| Streptomyces albidoflavus DSM 5415 | WO1995007878 | |||||
| (Z)-α-Bisabolene (115) (Z)-γ-Bisabolene (117) | Yes | epi-isozizaene synthase Streptomyces coelicolor A3(2) | [110,122] | WO2015120431 | antioxidant | [357] |
| Curcumene (116) | Yes | epi-isozizaene synthase Streptomyces coelicolor A3(2) | [110,122] | antifungal | [358] | |
| Sesquiphellandrene (118) | Yes | epi-isozizaene synthase Streptomyces coelicolor A3(2) | [110,122] | antiproliferative | [359] | |
| Strepsesquitriol (136) | No | Streptomyces sp. SCSIO 10355 | [123] | anti-inflammatory | [123] | |
| Pentalenolactone (132) | No | Streptomyces exfoliatus UC5319 Streptomyces avermitilis Streptomyces arenae TÜ469 | [130] | antimicrobial antiviral | [125] | |
| Streptomyces albus JA 3453-10 | DD261608 | |||||
| 1-Deoxy-8α-hydroxypentalenic acid (150) | No | Streptomyces sp. NRRL S-4 | [134] | antimicrobial (against Staphylococcus aureus, MIC 16 μg/mL; Escherichia coli, MIC 16–32 μg/mL) | [134] | |
| 1-Deoxy-9β-hydroxy-11-oxopentalenic acid (151) | ||||||
| Dihydro-β-agarofuran (78) | Yes | Streptomyces hygroscopicus NRRL 15879 | [66] | insecticidal | [360] | |
| Caryolan-1,9β-diol (96) | Yes | Streptomyces sp. AH25 | [108] | anti-inflammatory (ED50 0.34 mg/ear) | [361] | |
| Streptomyces albolongus YIM 101047 | [73] | |||||
| Viridiflorol (91) | Yes | SAV_76 from Streptomyces avermitilis | [83] | anti-inflammatory antioxidant (against DPPH, IC50 74.7 µg/mL) | [362] | |
| Di- and triterpenes and their derivatives | ||||||
| Lobocompactol (166) | No | Streptomyces cinnabarinus PK209 | [140] | antifouling (against macroalga Ulva pertusa, EC50 0.18 µg/mL; diatom Navicula annexa; EC50 0.43 µg/mL) | [140] | |
| Microeunicellol A (168) | No | Streptomyces albogriseolus SY67903 | [142] | antitumor (against MCF-7, IC50 5.3 μM; MDA-MB-231, IC50 8.6 μM) | [142] | |
| Terpentecin (427) | No | Kitasatosporia griseola MF730-N6 | [202] | antibacterial (against Staphylococcus aureus, Bacillus subtilis, Corynebacterium bovis, Shigella dysenteriae, Aeromonas salmonicida, Vibrio anguillarum, MIC 0.05 µg/mL) | [274] | |
| Isopimara-8(9),15-diene (180) | Yes | Streptomyces sp. PKU-TA00600 | [150] | anti-inflammatory | [363] | |
| Sat1646 from Salinispora sp. PKU-MA00418 | ||||||
| Isopimara-7(8),15-diene (445) Isopimara-8(14),15-diene (446) Syn-isopimara-7(8),15-diene (440) 8β-Isopimara-9(11),15-diene (441) 8β-Pimara-9(11),15-diene (442) Syn-stemod-13(17)-ene (443) Syn-pimara-7(8),15-diene (444) | No | |||||
| 2α-Hydroxy-8(14),15-pimaradien-17,18-dioic acid (450) | No | Microbispora hainanensis CSR-4 | [281] | anti-Alzheimer neuroprotective (1 ng/mL) antitumor antioxidant | [281] | |
| Gifhornenolone A (447) | No | Verrucosispora gifhornensis YM28-088 | [264] | antiandrogenic (IC50 2.8 µg/mL) | [264] | |
| Actinomadurol (452) | No | Actinomadura sp. KC 191 | [283] | antibacterial (against Staphylococcus aureus, Kocuria rhizophila, Proteus hauseri, MIC 0.39–0.78 μg/mL) | [283] | |
| k4610422 (453) | No | Actinomadura sp. AMW41E2 | [284] | cytotoxic (against P388, IC50 30 μM) | [284] | |
| Cyclooctatin (184) | No | Streptomyces melanosporofaciens MI614-43F2 | anti-inflammatory | [364] | ||
| Transf. E. coli with CotB3 or CotB4 from Streptomyces afghaniensis | ||||||
| Streptomyces sp. KCB17JA11 | ||||||
| 3,7,18-Dolabellatriene (188) | Yes | Mutant W288G of CotB2 from Streptomyces melanosporofaciens MI614-43F2 | [158] | antimicrobial (against methicillin-resistant Staphylococcus aureus, MIC 16.0 µg/mL) | [365] | |
| 2,7,18-Dolabellatriene (459) | Saccharopolyspora spinosa NRRL 18395 | [286] | ||||
| Thunbergol (464) | Yes | Allokutzneria albata DSM 44149 | [287] | antimicrobial | [366] | |
| Meroterpenoids | ||||||
| Furaquinocin A (226) Furaquinocin B (227) | No | Streptomyces sp. KO-3988 Streptomyces sp. CLl90 | [185] | WO2006081537 | antitumor (against HeLa S3, IC50 1.6–3.1 μg/mL) | [185] |
| Furaquinocin C (228) Furaquinocin D (226) Furaquinocin E (234) Furaquinocin G (235) Furaquinocin H (231) | No | Streptomyces sp. KO-3988 | cytotoxic (against B16, IC50 0.08–6.87 μg/mL; HeLa S3, IC50 0.22–5.05 μg/mL) | |||
| Furaquinocin L (238) | No | Streptomyces sp. Je 1-369 | [191] | antibacterial (against Staphylococcus aureus, MIC 2.0 μg/mL) | [191] | |
| Murayaquinone (240) | No | Streptomyces sp. TBRC7642 | [188] | antitubercular (MIC 3.13 μg/mL) | [188] | |
| cytotoxic (against MCF-7 IC50 6.0 μM; NCI–H187, IC500.85 μM; Vero, IC502.05 μM) | ||||||
| Merochlorin A (241) | No | Streptomyces sp. CNH-189 | [192] | antibacterial (against MRSA, MIC 2.0–4.0 μg/mL; Clostridium difficile 0.3–0.15 μg/mL) | [192] | |
| Merochlorin I (249) | No | Streptomyces sp. CNH-189 | [194] | antibacterial (against Bacillus subtilis, MIC 1.0 μg/mL; Kocuria rhizophila, MIC 2.0 μg/mL; Staphylococcus aureus, MIC 2.0 μg/mL) | [194] | |
| Merochlorin E (245) Merochlorin F (246) | No | Streptomyces sp. CNH-189 | [193] | antibacterial (against Bacillus subtilis, MIC 1.0 µg/mL, Kocuria rhizophila MIC 2.0 μg/mL, Staphylococcus aureus MIC 1.0–2.0 μg/mL) | [193] | |
| Flaviogeranin D (256) Flaviogeranin C2 (258) | No | Streptomyces sp. B9173 | [196] | antibacterial (against Mycobacterium smegmatis, MIC 5.2 μg/mL) | [196] | |
| cytotoxic (against A549, IC50 0.6–0.9 μM; Hela, IC50 0.4–1.1 μM) | ||||||
| Flaviogeranin A (252) | Streptomyces sp. RAC226 | [195] | neuroprotective (EC50 8.6 nM) | [195] | ||
| Naphterpin (259) | No | Streptomyces sp. CL190 Streptomyces sp. strain CLl90 | [197] | WO2006081537 | antioxidant (suppressed lipid peroxidation in rat homogenate system, IC50 5.3 μg/mL) | [197] |
| Naphterpin B (260) Naphterpin C (261) | No | Streptomyces sp. CL190 | [199] | antioxidant (suppressed lipid peroxidation in rat homogenate system, IC50 6.0–6.5 μg/mL) | [199] | |
| Napyradiomycin CNQ-525.1 (226) | No | Streptomyces sp. CNQ-525 | [208] | antibacterial (against MRSA, MIC 1.95 μg/mL; Enterococcus faecium (VREF) MIC 1.9–3.9 μg/mL) | [208] | |
| Napyradiomycin CNQ-525.2 (281) | ||||||
| Napyradiomycin CNQ-525.3 (282) | cytotoxic (against HCT, IC50 1.0–2.4 μg/mL) | |||||
| Napyradiomycin CNQ-525.4 (283) | ||||||
| Napyradiomycin D1 (287) | No | Streptomyces sp. CA-271078 | [203] | antibacterial (against MRSA, MIC 12.0–24.0 μg/mL; Mycobacterium tuberculosis, MIC 12.0–48.0 μg/mL) | [203] | |
| cytotoxic (HepG2, IC50 14.9 μM) | ||||||
| 3-Dechloro-3-bromonapyradiomycin A1 (266) | No | Streptomyces sp. SCSIO 10428 Streptomyces kebangsaanensis WS-68302 Streptomyces sp. CA-271078 | [201,204] | CN105399721 | antibacterial (against Staphylococcus aureus, MIC 0.5–1.0 μg/mL; MRSA, MIC 4.0–8.0 μg/mL; Bacillus subtilis, MIC 1.0–2.0 μg/mL; Bacillus thuringiensis, MIC 0.5–2.0 μg/mL) cytotoxic (against HCT-116, IC50 2.0–3.0 μM) | [201,204,209] |
| Napyradiomycin B1 (273) | ||||||
| Naphthomevalin (289) | ||||||
| Napyradiomycin A1 (264) | No | Streptomyces sp. CA-271078 | [201] | antibacterial (against MRSA, MIC 0.5–1.0 μg/mL) | [201] | |
| Streptomyces sp. YP127 | [200] | antiangiogenic | [200] | |||
| Streptomyces kebangsaanensis WS-68302 | CN105399721 | antibacterial (against Staphylococcus aureus, MIC 0.078 µg/mL) antiviral (against Pseudorabies virus, IC50 2.2 μg/mL) | ||||
| Napyradiomycin B2 (275) | No | Streptomyces sp. CNQ-329 Streptomyces sp. CNH-070 | [206] | cytotoxic (against HCT-116, IC50 3.18 μg/mL) antibacterial (against MRSA, MIC 3.0–6.0 μg/mL) | [206] | |
| Streptomyces sp. CA-271078 | [203] | |||||
| Napyradiomycin B3 (274) | No | Streptomyces sp. CNQ-329 Streptomyces sp. CNH-070 | [206] | cytotoxic (against HCT-116, IC50 0.2 μg/mL) antibacterial (against MRSA, MIC 2.0 μg/mL; against Staphylococcus aureus, MIC 0.5 μg/mL; Bacillus subtilis, MIC 0.2 μg/mL; Bacillus thuringiensis, MIC 0.5 μg/mL) | [203,206] | |
| Streptomyces sp. SCSIO 10428 | [203] | |||||
| Napyradiomycin B4 (284) | Streptomyces strains CNQ-329 and CNH-070 | [206] | cytotoxic (against HCT-116, IC50 1.41 μg/mL) | [206] | ||
| NPM 1 (288) | Streptomyces strains CNQ-329 and CNH-070 | [206] | cytotoxic (against HCT-116, IC50 4.2–4.8 μg/mL) | [206] | ||
| Napyradiomycin CNQ525.538 (271) | No | Streptomyces sp. CNQ-525 | [209] | cytotoxic (against HCT-116, IC50 6.0 μg/mL) | [209] | |
| A80915A (277) A80915B (278) A80915D (279) A80915G (291) | No | Streptomyces aculeolatus A80915 | - | EP0376609 | antibacterial (against Staphylococcus aureus, MIC 0.03–4.0 μg/mL; S. epidermidis, MIC 0.15–2.0 μg/mL; Streptococcus pyogenes, MIC 0.03–2.0 μg/mL; S. pneumonia, MIC 0.125–2.0 μg/mL; Enterococcus faecium, MIC 1.0–4.0 μg/mL; E. faecalis, MIC 1.0 μg/mL; Haemophilus influenzae, MIC 0.008 μg/mL; Clostridium difficile, MIC 2.0–4.0 μg/mL; C. perfringers, MIC 2.0–4.0 μg/mL; C. septicum, MIC 1.0–2.0 μg/mL; Eubacterium aerofaciens, MIC 0.5–2.0 μg/mL; Peptococcus asaccharolyticus, MIC 0.5–4.0 μg/mL; P. prevotii, MIC 1.0–2.0 μg/mL; P. intermediatus, MIC 1.0–2.0 μg/mL; Propionibacterium acnes, MIC 0.5–1.0 μg/mL; Bacteroides fragilis, MIC 2.0–4.0; B. melaninogenicus, MIC 0.5–2.0 μg/mL; B. corrodens, MIC 2.0–4.0 μg/mL; Fusobacterium symbiosum, MIC 0.5–4.0 μg/mL) | - |
| A80915A (277) A80915B (278) A80915D (279) | No | Streptomyces sp. CNQ-525 | [209] | cytotoxic (against HCT-116, IC50 1.0–3.0 μg/mL) | [209] | |
| 7-Demethyl SF2415A3 (272) 7-Demethyl A80915B (285) | No | Streptomyces antimycoticus NT17 | [202] | antibacterial (against Staphylococcus aureus, MIC 2.0–3.7 nM/mL; Bacillus subtilis, MIC 1.0–3.7 nM/mL) | [202] | |
| Napyradiomycin A4 (267) | No | Streptomyces kebangsaanensis WS-68302 | CN114805278 | antiviral (against Pseudorabies virus (PRV), IC50 2.056 μM) | ||
| 16Z-19-Hydroxynapyradiomycin A1 (265) | No | Streptomyces sp. YP127 | [205] | anti-inflammatory antioxidant | [205] | |
| (R)-3-Chloro-6-hydroxy-8-methoxy-alpha-lapachone (286) | No | Streptomyces sp. YP127 Streptomyces antimycoticus NT17 | [202,205] | anti-inflammatory | [205] | |
| Marfuraquinocin A (292) Marfuraquinocin C (294) Marfuraquinocin D (295) | No | Streptomyces niveus SCSIO 3406 | [210] | cytotoxic (against NCI-H460, IC50 3.7; 4.4; 8.8 μM) antibacterial (against Staphylococcus aureus ATCC 29213, methicillin-resistant Staphylococcus epidermidis, MIC 8.0 μg/mL) | [210] | |
| FW03105 (484) | No | Verrucosispora sp. FIM06031 | CN101921721 | antitumor (against HepG2, IC50 16.99 µM; EC109, IC50 25.33 µM; HeLA, IC50 34.64 µM) | ||
| Saccharoquinoline (492) | No | Saccharomonospora sp. CNQ-490 | [293] | cytotoxic (against HCT-116, IC50 1.0 μM) | [293] | |
| Teleocidin B (314) | No | Streptomyces mediocidicus | [211] | tumor promoter | [211] | |
| Streptomyces sp. 680560 | [367] | nematicidal | [367] | |||
| Streptomyces blastmyceticus | [214] | |||||
| Lavanducyanin (304) | No | Streptomyces sp. CNS-284 and CNY-960 Streptomyces sp. CLl90 | [216] | WO2006081537 | cytotoxic (against HCT-116, IC50 2.41 μM) | [216] |
| antimicrobial (against Staphylococcus aureus, MIC 2.92 μM; Candida albicans, MIC 5.96 μM) | ||||||
| Marinocyanin A (298) Marinocyanin B (299) Marinocyanin C (300) | No | Streptomyces sp. CNS-284 и CNY-960 | [216] | - | cytotoxic (against HCT-116, IC50 0.029–0.049 μM) | [216] |
| antimicrobial (against Staphylococcus aureus, MIC 2.37 μM; Candida albicans, MIC 0.95–3.90 μM) | ||||||
| Farneside A (306) | No | Streptomyces sp. CNT-372 | [217] | antimalarial (against Plasmodium falciparum) | [217] | |
| Xiamycin A (310) | Streptomyces sp. SCSIO 02999 | [220] | CN102757908 CN102732534 | antiviral anti-HIV cytotoxic | [220] | |
| Streptomyces sp. GT2002/1503 | [221] | antiviral (against SARS-CoV-2) | [368] | |||
| Streptomyces sp. HKI0595 | [226] | antiviral (against HSV-1) | [329] | |||
| Xiamycin methyl ester (311) | No | Streptomyces sp. SCSIO 02999 | [220] | CN102757908 | antitumor (IC50 10.13 μM) | |
| antiviral (against SARS-CoV-2) | [368] | |||||
| Dixiamycin A (328) Dixiamycin B (330) | No | Streptomyces sp. GT2002/1503 | [221] | antibacterial (against E. coli, S. aureus, MIC 8–16 µg/mL; B. thuringiensis, MIC 4–8 µg/mL) | [221] | |
| Streptomyces xinghaiensis NRRL B-24674T | [228] | |||||
| Streptomyces sp. SCSIO 02999 | CN102757908 | |||||
| Dixiamycin 6a/6b (333/334) | No | Transf. S. albus with xia from Streptomyces sp. SCSIO 02999 | [230] | WO2014029498 | antibacterial (against MRSA, MIC 0.2 µg/mL) | [230] |
| Dixiamycin 8 (337) | antibacterial (against S. aureus, MRSA, MIC 1.56 µg/mL) | |||||
| Dixiamycin 7a/7b (335/336) | No | Streptomyces olivaceus OUCLQ19-3 | [229] | antibacterial (S. aureus, E. faecalis, E. faecium, M. luteus, P. aeruginosa, MIC 6.25–12.5 µg/mL) | [229] | |
| Dixiamycin 12a/12b (331/332) | antibacterial (S. aureus, MIC 0.78–3.12 µg/mL; E. faecalis, E. faecium, M. luteus, MIC 3.12–6.25 µg/mL; P. aeruginosa, MIC 1.56 µg/mL) | |||||
| Xiamycin B (313) Indosespene (318) | No | Streptomyces sp. HKI0595 Streptomyces sp. SCSIO 02999 | [226] | CN102732534 | antimicrobial (against MRSA; vancomycin-resistant Enterococcus faecalis) | [226] |
| Sespenine (319) | antiviral (against SARS-CoV-2) | [368] | ||||
| Xiamycin D (324) | No | Streptomyces sp. HK18 | [225] | antiviral (against PEDV) | [225] | |
| Xiamycin C (323) | antiviral (against SARS-CoV-2) | [368] | ||||
| Oridamycin A (326) | No | Streptomyces sp. KS84 | [227] | antifungal (against Saprolegnia parasitica, MIC 3.0 µg/mL) | [227] | |
| Sulfonylbixiamycin A (338) | No | Transf. S. albus with xiamycin BGC from Streptomyces sp. | [231] | WO2014029498 | antibacterial (against Bacillus subtilis, MIC 6.25 µg/mL; Staphylococcus aureus, MIC 3.12 µg/mL; MRSA, MIC 6.25 µg/mL) | [231] |
| Cyslabdan A (341) | No | Streptomyces cyslabdanicus K04-0144 | [233] | enhance (1000-fold) the antibiotic imipenem action (against MRSA) | [369] | |
| Oxaloterpin A (347) | No | Streptomyces sp. KO-3988 | [151] | antibacterial (against Bacillus subtilis ATCC 43223, IC50 1.9 µM/mL; Staphylococcus aureus ATCC29213; EC50 3.7) | [151] | |
| Streptomyces griseus CB00830 | [235] | |||||
| Streptomyces sp. SN194 | [152] | |||||
| Chloroxaloterpin A (352) Chloroxaloterpin B (353) | No | Streptomyces sp. SN194 | [152] | antifungal (against Botrytis cinerea, EC50 4.40–4.96 µg/mL) | [152] | |
| Fusicomycin A (384) Fusicomycin (385) Fusicomycin B (386) | No | Streptomyces violascens YIM 100212 | [164] | cytotoxicity (against BGC-823 H460, HCT116, HeLa, SMMC7721 8.9, IC50 from 3.5 ± 0.7 to 14.1 ± 0.8 µM) | [164] | |
| Streptooctatin A (387) Streptooctatin B (388) | No | Streptomyces sp. KCB17JA11 | [243] | autophagic (against HeLa) | [243] | |
| Actinoranone (389) | Streptomyces sp. CNQ-027 | [244] | cytotoxic (against HCT-116, LD50 2.0 μg/mL) | [244] | ||
| Brasilicardin A (490) | No | Nocardia brasiliensis IFM 0406 (now N. terpenica) | [299] | immunosuppressive | [300] | |
| antiproliferative (against LN229, IC50 0.13 μM) | [306] | |||||
| Platensimycin (390) Platencin (391) | Streptomyces platensis MA7327 Streptomyces platensis MA7339 Streptomyces platensis MA7237 | [245,246] | US20090081673 | antibacterial (against S. aureus (MRSA), Enterococcus faecalis, Enterococcus faecium, MIC 0.1–1.0 μg/mL) | [245,246] | |
| Atolypene A (497) Atolypene B (498) | No | Transf. Streptomyces albus with ato gene cluster from Amycolatopsis tolypomycina NRRL B-24205 | [307] | cytotoxic (against HL-60, Jurkat, HEK293, HeLa, A549, IC50 12.0–36.7 μM) | [307] | |
| Terretonin N (499) | No | Nocardiopsis sp. LGO5 | [308] | antibacterial (against Staphylococcus warneri) | [308] | |
| Soyasaponin I (407) | Yes | Streptomyces sp. YIM 56130 | [94] | anti-inflammatory antimutagenic anticarcinogenic antimicrobial | [256] | |
| Longestin (408) | No | Streptomyces argenteolus A-2 | [257] | antiamnesic (IC50 0.065 µM) | [370] | |
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Caputi, L.; Aprea, E. Use of terpenoids as natural flavouring compounds in food industry. Recent Pat. Food Nutr. Agric. 2011, 3, 9–16. [Google Scholar] [CrossRef]
- Duttaroy, A.K. Health effects of terpenoids. In Evidence-Based Nutrition and Clinical Evidence of Bioactive Foods in Human Health and Disease; Ball, M.R., Ed.; Academic Press: London, UK, 2021; pp. 413–424. ISBN 978-0-12-822405-2. [Google Scholar] [CrossRef]
- Fordjour, E.; Mensah, E.O.; Hao, Y.; Yang, Y.; Liu, X.; Li, Y.; Liu, C.-L.; Bai, Z. Toward improved terpenoids biosynthesis: Strategies to enhance the capabilities of cell factories. Bioresour. Bioprocess. 2022, 9, 6. [Google Scholar] [CrossRef]
- Zhu, K.; Kong, J.; Zhao, B.; Rong, L.; Liu, S.; Lu, Z.; Zhang, C.; Xiao, D.; Pushpanathan, K.; Foo, J.L.; et al. Metabolic engineering of microbes for monoterpenoid production. Biotechnol. Adv. 2021, 53, 107837. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Kuzuyama, T.; Komatsu, M.; Shin-ya, K.; Omura, S.; Cane, D.E.; Ikeda, H. Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 857–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickschat, J.S. Bacterial terpene cyclases. Nat. Prod. Rep. 2016, 33, 87–110. [Google Scholar] [CrossRef]
- Smanski, M.J.; Peterson, R.M.; Huang, S.X.; Shen, B. Bacterial diterpene synthases: New opportunities for mechanistic enzymology and engineered biosynthesis. Curr. Opin. Chem. Biol. 2012, 16, 132–141. [Google Scholar] [CrossRef] [Green Version]
- Lewin, G.R.; Carlos, C.; Chevrette, M.G.; Horn, H.A.; McDonald, B.R.; Stankey, R.J.; Fox, B.G.; Currie, C.R. Evolution and ecology of Actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 2016, 70, 235–254. [Google Scholar] [CrossRef] [Green Version]
- Salwan, R.; Sharma, V. Bioactive compounds of Streptomyces: Biosynthesis to applications. Stud. Nat. Prod. Chem. 2020, 64, 467–491. [Google Scholar] [CrossRef]
- Jose, P.A.; Maharshi, A.; Jha, B. Actinobacteria in natural products research: Progress and prospects. Microbiol. Res. 2021, 246, 126708. [Google Scholar] [CrossRef]
- Gong, R.; Yu, L.; Qin, Y.; Price, N.P.J.; He, X.; Deng, Z.; Chen, W. Harnessing synthetic biology-based strategies for engineered biosynthesis of nucleoside natural products in actinobacteria. Biotechnol. Adv. 2021, 46, 107673. [Google Scholar] [CrossRef]
- Patzer, S.I.; Braun, V. Gene cluster involved in the biosynthesis of griseobactin, a catechol-peptide siderophore of Streptomyces sp. ATCC 700974. J. Bacteriol. 2010, 192, 426–435. [Google Scholar] [CrossRef] [Green Version]
- Anandan, R.; Dharumadurai, D.; Manogaran, G.P. An Introduction to Actinobacteria. In Basics and Biotechnological Applications; Dhanasekaran, D., Jiang, Y., Eds.; OpenIntech: London, UK, 2016; pp. 3–38. [Google Scholar]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salwan, R.; Sharma, V. The role of Actinobacteria in the production of industrial enzymes. In New and Future Developments in Microbial Biotechnology and Bioengineering: Actinobacteria: Diversity and Biotechnological Applications; Singh, B.P., Gupta, V.K., Passari, A.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 165–177. ISBN 9780444639950. [Google Scholar]
- Jagannathan, S.V.; Manemann, E.M.; Rowe, S.E.; Callender, M.C.; Soto, W. Marine Actinomycetes, new sources of biotechnological products. Mar. Drugs 2021, 19, 365. [Google Scholar] [CrossRef] [PubMed]
- Arulprakasam, K.R.; Dharumadurai, D. Genome mining of biosynthetic gene clusters intended for secondary metabolites conservation in actinobacteria. Microb. Pathog. 2021, 161, 105252. [Google Scholar] [CrossRef]
- Kuyukina, M.S.; Ivshina, I.B. Rhodococcus biosurfactants: Biosynthesis, properties, and potential applications. In Biology of Rhodococcus; Alvarez, H.M., Ed.; Springer: Berlin, Germany, 2010; pp. 291–313. ISBN 9783642129377. [Google Scholar]
- Kuyukina, M.S.; Ivshina, I.B. Production of trehalolipid biosurfactants by Rhodococcus. In Biology of Rhodococcus; Alvarez, H., Ed.; Springer: Cham, Switzerland, 2019; pp. 271–298. [Google Scholar]
- Shi, L.; Wu, Z.; Zhang, Y.; Zhang, Z.; Fang, W.; Wang, Y.; Wan, Z.; Wang, K.; Ke, S. Herbicidal secondary metabolites from Actinomycetes: Structure diversity, modes of action, and their roles in the development of herbicides. J. Agric. Food Chem. 2020, 68, 17–32. [Google Scholar] [CrossRef]
- Silva, L.J.; Crevelin, E.J.; Souza, D.T.; Lacerda-Júnior, G.V.; de Oliveira, V.M.; Ruiz, A.L.T.G.; Rosa, L.H.; Moraes, L.A.B.; Melo, I.S. Actinobacteria from Antarctica as a source for anticancer discovery. Sci. Rep. 2020, 10, 13870. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, Z.; Zhao, J.; Zhuang, X.; Cao, P.; Guo, X.; Liu, C.; Xiang, W. Community composition, antifungal activity and chemical analyses of ant-derived Actinobacteria. Front. Microbiol. 2020, 11, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuyukina, M.S.; Ivshina, I.B.; Gein, S.V.; Baeva, T.A.; Chereshnev, V.A. In vitro immunomodulating activity of biosurfactant glycolipid complex from Rhodococcus ruber. Bull. Exp. Biol. Med. 2007, 144, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Gein, S.V.; Kuyukina, M.S.; Ivshina, I.B.; Baeva, T.A.; Chereshnev, V.A. In vitro cytokine stimulation assay for glycolipid biosurfactant from Rhodococcus ruber: Role of monocyte adhesion. Cytotechnology 2011, 63, 559–566. [Google Scholar] [CrossRef] [Green Version]
- Gein, S.V.; Kochina, O.A.; Kuyukina, M.S.; Ivshina, I.B. Effects of glycolipid Rhodococcus biosurfactant on innate and adaptive immunity parameters in vivo. Bull. Exp. Biol. Med. 2018, 165, 368–372. [Google Scholar] [CrossRef]
- Mast, Y.; Stegmann, E. Actinomycetes: The antibiotics producers. Antibiotics 2019, 8, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamedi, J.; Mohammadipanah, F. Biotechnological application and taxonomical distribution of plant growth promoting actinobacteria. J. Ind. Microbiol. Biotechnol. 2015, 42, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Betancur, L.A.; Naranjo-Gaybor, S.J.; Vinchira-Villarraga, D.M.; Moreno-Sarmiento, N.C.; Maldonado, L.A.; Suarez-Moreno, Z.R.; Acosta-González, A.; Padilla-Gonzalez, G.F.; Puyana, M.; Castellanos, L.; et al. Marine Actinobacteria as a source of compounds for phytopathogen control: An integrative metabolic-profiling/bioactivity and taxonomical approach. PLoS ONE 2017, 12, e0170148. [Google Scholar] [CrossRef] [Green Version]
- Hariprasad, K.V. Recent advancement in the development of biopesticides by Actinomycetes for the control of insect pests. In Plant Growth Promoting Actinobacteria; Subramaniam, G., Arumugam, S., Rajendran, V., Eds.; Springer: Singapore; Gateway: Singapore, 2016; pp. 47–62. [Google Scholar]
- Paulraj, M.G.; Kumar, P.S.; Ignacimuthu, S.; Sukumaran, D. Natural insecticides from Actinomycetes and other microbes for vector mosquito control. In Herbal Insecticides, Repellents and Biomedicines: Effectiveness and Commercialization; Veer, V., Gopalakrishnan, R., Eds.; Springer: New Delhi, India, 2016; pp. 85–99. [Google Scholar]
- Jose, P.A.; Jha, B. New dimensions of research on Actinomycetes: Quest for next generation antibiotics. Front. Microbiol. 2016, 7, 1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Simeis, D.; Serra, S. Actinomycetes: A never-ending source of bioactive compounds—An overview on antibiotics production. Antibiotics 2021, 10, 483. [Google Scholar] [CrossRef]
- Raja, A.; Prabakaran, P. Actinomycetes and drug–An overview. Am. J. Drug Discov. Dev. 2011, 1, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Ding, T.; Yang, L.-J.; Zhang, W.-D.; Shen, Y.-H. The secondary metabolites of rare Actinomycetes: Chemistry and bioactivity. RSC Adv. 2019, 9, 21964–21988. [Google Scholar] [CrossRef] [Green Version]
- El-Shahidy, S.; Mansour, S.R.; Al-Bassiony, A.D. Exploring the Bioactive Compounds from Actinobacteria: Inhabiting Different Habitats (Natural Products, the Future Approach for Drug Discovery); LAP LAMBERT Academic Publishing: London, UK, 2014; ISBN 3659627976. [Google Scholar]
- Selim, M.S.M.; Abdelhamid, S.A.; Mohamed, S.S. Secondary metabolites and biodiversity of Actinomycetes. J. Genet. Eng. Biotechnol. 2021, 19, 72. [Google Scholar] [CrossRef]
- Al-shaibani, M.M.; Radin Mohamed, R.M.S.; Sidik, N.M.; El Enshasy, H.A.; Al-Gheethi, A.; Noman, E.; Al-Mekhlafi, N.A.; Zin, N.M. Biodiversity of secondary metabolites compounds isolated from phylum Actinobacteria and its therapeutic applications. Molecules 2021, 26, 4504. [Google Scholar] [CrossRef]
- Binda, C.; Lopetuso, L.R.; Rizzatti, G.; Gibiino, G.; Cennamo, V.; Gasbarrini, A. Actinobacteria: A relevant minority for the maintenance of gut homeostasis. Dig. Liver Dis. 2018, 50, 421–428. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Ho, C.L. Recent development of probiotic Bifidobacteria for treating human diseases. Front. Bioeng. Biotechnol. 2021, 9, 770248. [Google Scholar] [CrossRef] [PubMed]
- Kügler, J.H.; Le Roes-Hill, M.; Syldatk, C.; Hausmann, R. Surfactants tailored by the class Actinobacteria. Front. Microbiol. 2015, 6, 212. [Google Scholar] [CrossRef] [Green Version]
- Gong, K.; Yong, D.; Fu, J.; Li, A.; Zhang, Y.; Li, R. Diterpenoids from Streptomyces: Structures, biosyntheses and bioactivities. ChemBioChem 2022, 23, e202200231. [Google Scholar] [CrossRef]
- Dickschat, J.S. Bacterial diterpene biosynthesis. Angew. Chem. Int. Ed. 2019, 58, 15964–15976. [Google Scholar] [CrossRef] [PubMed]
- Rudolf, J.D.; Alsup, T.A.; Xu, B.; Li, Z. Bacterial terpenome. Nat. Prod. Rep. 2021, 38, 905–980. [Google Scholar] [CrossRef]
- Avalos, M.; Garbeva, P.; Vader, L.; Van Wezel, G.P.; Dickschat, J.S.; Ulanova, D. Biosynthesis, evolution and ecology of microbial terpenoids. Nat. Prod. Rep. 2022, 39, 249–272. [Google Scholar] [CrossRef]
- Christianson, D.W. Structural and chemical biology of terpenoid cyclases. Chem. Rev. 2017, 117, 11570–11648. [Google Scholar] [CrossRef] [Green Version]
- Helfrich, E.J.N.; Lin, G.-M.; Voigt, C.A.; Clardy, J. Bacterial terpene biosynthesis: Challenges and opportunities for pathway engineering. Beilstein J. Org. Chem. 2019, 15, 2889–2906. [Google Scholar] [CrossRef] [PubMed]
- Cane, D.E.; Ikeda, H. Exploration and mining of the bacterial terpenome. Acc. Chem. Res. 2012, 45, 463–472. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, M.; Uchiyama, T.; Ōmura, S.; Cane, D.E.; Ikeda, H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. USA 2010, 107, 2646–2651. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, Y.; Rebets, Y.; Estévez, M.R.; Zapp, J.; Myronovskyi, M.; Luzhetskyy, A. Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters. Microb. Cell Fact. 2020, 19, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nah, H.-J.; Pyeon, H.-R.; Kang, S.-H.; Choi, S.-S.; Kim, E.-S. Cloning and heterologous expression of a large-sized natural product biosynthetic gene cluster in Streptomyces species. Front. Microbiol. 2017, 8, 394. [Google Scholar] [CrossRef] [Green Version]
- Baltz, R.H. Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J. Ind. Microbiol. Biotechnol. 2010, 37, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Palazzotto, E.; Weber, T. Omics and multi-omics approaches to study the biosynthesis of secondary metabolites in microorganisms. Curr. Opin. Microbiol. 2018, 45, 109–116. [Google Scholar] [CrossRef]
- Dickschat, J.S.; Martens, T.; Brinkhoff, T.; Simon, M.; Schulz, S. Volatiles released by a Streptomyces species isolated from the North sea. Chem. Biodivers. 2005, 2, 837–865. [Google Scholar] [CrossRef] [PubMed]
- Nakano, C.; Kim, H.-K.; Ohnishi, Y. Identification of the first bacterial monoterpene cyclase, a 1,8-cineole synthase, that catalyzes the direct conversion of geranyl diphosphate. ChemBioChem 2011, 12, 1988–1991. [Google Scholar] [CrossRef] [PubMed]
- Karuppiah, V.; Ranaghan, K.E.; Leferink, N.G.H.; Johannissen, L.O.; Shanmugam, M.; Ní Cheallaigh, A.; Bennett, N.J.; Kearsey, L.J.; Takano, E.; Gardiner, J.M.; et al. Structural basis of catalysis in the bacterial monoterpene synthases linalool synthase and 1,8-cineole synthase. ACS Catal. 2017, 7, 6268–6282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferraz, C.A.; Leferink, N.G.H.; Kosov, I.; Scrutton, N.S. Isopentenol utilization pathway for the production of linalool in Escherichia coli using an improved bacterial linalool/nerolidol synthase. ChemBioChem 2021, 22, 2325–2334. [Google Scholar] [CrossRef]
- Li, L.; Liu, R.; Han, L.; Jiang, Y.; Liu, J.; Li, Y.; Yuan, C.; Huang, X. Structure determination of two new nerolidol-type sesquiterpenoids from the soil actinomycete Streptomyces scopuliridis. Magn. Reson. Chem. 2016, 54, 606–609. [Google Scholar] [CrossRef]
- Komatsu, M.; Tsuda, M.; Omura, S.; Oikawa, H.; Ikeda, H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc. Natl. Acad. Sci. USA 2008, 105, 7422–7427. [Google Scholar] [CrossRef] [Green Version]
- Schrader, K.K.; Harries, M.D.; Page, P.N. Temperature effects on biomass, geosmin, and 2-methylisoborneol production and cellular activity by Nocardia spp. and Streptomyces spp. isolated from rainbow trout recirculating aquaculture systems. J. Ind. Microbiol. Biotechnol. 2015, 42, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, Z.; Qiao, X.; Li, Z.; Li, F.; Chen, M.; Wang, Y.; Huang, Y.; Cui, H. Antifungal activity of volatile organic compounds from Streptomyces alboflavus TD-1. FEMS Microbiol. Lett. 2013, 341, 45–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saadoun, I. Production of 2-methylisoborneol by Streptomyces violaceusniger and its transformation by selected species ofPseudomonas. J. Basic Microbiol. 2005, 45, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Lyu, A.; Yang, L.; Wu, M.; Zhang, J.; Li, G. High efficacy of the volatile organic compounds of Streptomyces yanglinensis 3-10 in suppression of Aspergillus contamination on peanut kernels. Front. Microbiol. 2020, 11, 142. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-M.; Cane, D.E. Biochemistry and molecular genetics of the biosynthesis of the earthy odorant methylisoborneol in Streptomyces coelicolor biosynthesis of methylisoborneol scheme 2. Cyclization of GPP by SCO7700. J. Am. Chem. Soc 2008, 130, 8908–8909. [Google Scholar] [CrossRef] [PubMed]
- Köksal, M.; Chou, W.K.W.; Cane, D.E.; Christianson, D.W. Structure of 2-methylisoborneol synthase from Streptomyces coelicolor and implications for the cyclization of a noncanonical C -methylated monoterpenoid substrate. Biochemistry 2012, 51, 3011–3020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köksal, M.; Chou, W.K.W.; Cane, D.E.; Christianson, D.W. Unexpected reactivity of 2-fluorolinalyl diphosphate in the active site of crystalline 2-methylisoborneol synthase. Biochemistry 2013, 52, 5247–5255. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; McCann, S.; Faraone, N.; Clarke, J.-A.; Hudson, E.A.; Cloonan, K.; Hillier, N.K.; Tahlan, K. Production of plant-associated volatiles by select model and industrially important Streptomyces spp. Microorganisms 2020, 8, 1767. [Google Scholar] [CrossRef] [PubMed]
- Nakano, C.; Kudo, F.; Eguchi, T.; Ohnishi, Y. Genome mining reveals two novel bacterial sesquiterpene cyclases: (−)-germacradien-4-ol and (−)-epi-α-bisabolol synthases from Streptomyces citricolor. ChemBioChem 2011, 12, 2271–2275. [Google Scholar] [CrossRef] [PubMed]
- Grundy, D.J.; Chen, M.; González, V.; Leoni, S.; Miller, D.J.; Christianson, D.W.; Allemann, R.K. Mechanism of germacradien-4-ol synthase-controlled water capture. Biochemistry 2016, 55, 2112–2121. [Google Scholar] [CrossRef] [PubMed]
- Rabe, P.; Barra, L.; Rinkel, J.; Riclea, R.; Citron, C.A.; Klapschinski, T.A.; Janusko, A.; Dickschat, J.S. Conformational analysis, thermal rearrangement, and EI-MS fragmentation mechanism of (1(10)e,4e,6s,7r)-germacradien-6-ol by 13C-labeling experiments. Angew. Chem. Int. Ed. 2015, 54, 13448–13451. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.L.; Escorcia, A.M.; Huynh, F.; Miller, D.J.; Allemann, R.K.; Van Der Kamp, M.W. Redesigning the molecular choreography to prevent hydroxylation in germacradien-11-ol synthase catalysis. ACS Catal. 2021, 11, 1033–1041. [Google Scholar] [CrossRef]
- Guan, S.; Grabley, S.; Groth, I.; Lin, W.; Christner, A.; Guo, D.; Sattler, I. Structure determination of germacrane-type sesquiterpene alcohols from an endophyte Streptomyces griseus subsp. Magn. Reson. Chem. 2005, 43, 1028–1031. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Wang, R.; Wang, G.; Liu, M.; Liao, Z.; Liao, G.; Chen, M. Roseosporol A, the first isolation of a novel sesquiterpenoid from Streptomyces roseosporus. Nat. Prod. Res. 2018, 33, 2038–2043. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Jiang, Y.; Han, L.; Chen, X.; Ma, J.; Qu, X.; Mu, Y.; Liu, J.; Li, L.; Jiang, C.; et al. Bafilomycins and odoriferous sesquiterpenoids from Streptomyces albolongus isolated from Elephas maximus feces. J. Nat. Prod. 2016, 79, 799–805. [Google Scholar] [CrossRef]
- Cane, D.E.; He, X.; Kobayashi, S.; Ōmura, S.; Ikeda, H. Geosmin biosynthesis in Streptomyces avermitilis. Molecular cloning, expression, and mechanistic study of the germacradienol/geosmin synthase. J. Antibiot. 2006, 59, 471–479. [Google Scholar] [CrossRef]
- Cane, D.E.; Watt, R.M. Expression and mechanistic analysis of a germacradienol synthase from Streptomyces coelicolor implicated in geosmin biosynthesis. Proc. Natl. Acad. Sci. USA 2003, 100, 1547–1551. [Google Scholar] [CrossRef] [Green Version]
- Schrader, K.K.; Blevins, W.T. Effects of carbon source, phosphorus concentration, and several micronutrients on biomass and geosmin production by Streptomyces halstedii. J. Ind. Microbiol. Biotechnol. 2001, 26, 241–247. [Google Scholar] [CrossRef]
- Jiang, J.; He, X.; Cane, D.E. Geosmin biosynthesis. Streptomyces coelicolor germacradienol/germacrene d synthase converts farnesyl diphosphate to geosmin. J. Am. Chem. Soc. 2006, 128, 8128–8129. [Google Scholar] [CrossRef]
- Jiang, J.; He, X.; Cane, D.E. Biosynthesis of the earthy odorant geosmin by a bifunctional Streptomyces coelicolor enzyme. Nat. Chem. Biol. 2007, 3, 711–715. [Google Scholar] [CrossRef] [Green Version]
- Nawrath, T.; Dickschat, J.S.; Müller, R.; Jiang, J.; Cane, D.E.; Schulz, S. Identification of (8S,9S,10S)-8,10-dimethyl-1-octalin, a key intermediate in the biosynthesis of geosmin in bacteria. J. Am. Chem. Soc. 2008, 130, 430–431. [Google Scholar] [CrossRef] [Green Version]
- Harris, G.G.; Lombardi, P.M.; Pemberton, T.A.; Matsui, T.; Weiss, T.M.; Cole, K.E.; Köksal, M.; Murphy, F.V.; Vedula, L.S.; Chou, W.K.W.; et al. Structural studies of geosmin synthase, a bifunctional sesquiterpene synthase with αα domain architecture that catalyzes a unique cyclization-fragmentation reaction sequence. Biochemistry 2015, 54, 7142–7155. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.; Tae, O.J.; Sohng, K.J. Exploration of geosmin synthase from Streptomyces peucetius ATCC 27952 by deletion of doxorubicin biosynthetic gene cluster. J. Ind. Microbiol. Biotechnol. 2009, 36, 1257–1265. [Google Scholar] [CrossRef]
- Baer, P.; Rabe, P.; Fischer, K.; Citron, C.A.; Klapschinski, T.A.; Groll, M.; Dickschat, J.S. Induced-fit mechanism in class i terpene cyclases. Angew. Chem. Int. Ed. 2014, 53, 7652–7656. [Google Scholar] [CrossRef]
- Chou, W.K.W.; Fanizza, I.; Uchiyama, T.; Komatsu, M.; Ikeda, H.; Cane, D.E. Genome mining in Streptomyces avermitilis: Cloning and characterization of sav-76, the synthase for a new sesquiterpene, avermitilol. J. Am. Chem. Soc. 2010, 132, 8850–8851. [Google Scholar] [CrossRef] [Green Version]
- Rinkel, J.; Lauterbach, L.; Dickschat, J.S. Spata-13,17-diene synthase-an enzyme with sesqui-, di-, and sesterterpene synthase activity from Streptomyces xinghaiensis. Angew. Chem. Int. Ed. 2017, 56, 16385–16389. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.J.; Fotso, S.; Li, F.; Qin, S.; Kelter, G.; Fiebig, H.H.; Laatsch, H. N-Carboxamido-staurosporine and selina-4(14),7(11)-diene-8,9-diol, new metabolites from a marine Streptomyces sp. J. Antibiot. 2006, 59, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Xu, H.; Zou, J.; Chen, X.-B.; Zhuang, Y.-Q.; Liu, W.-L.; Celik, E.; Chen, G.-D.; Hu, D.; Gao, H.; et al. Catalytic role of carbonyl oxygens and water in selinadiene synthase. Nat. Catal. 2022, 5, 128–135. [Google Scholar] [CrossRef]
- Nakano, C.; Tezuka, T.; Horinouchi, S.; Ohnishi, Y. Identification of the SGR6065 gene product as a sesquiterpene cyclase involved in (+)-epicubenol biosynthesis in Streptomyces griseus. J. Antibiot. 2012, 65, 551–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citron, C.A.; Dickschat, J.S. [2H26]-1-epi-Cubenol, a completely deuterated natural product from Streptomyces griseus. Beilstein J. Org. Chem. 2013, 9, 2841–2845. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Görls, H.; Hertweck, C. Plant-like cadinane sesquiterpenes from an actinobacterial mangrove endophyte. Magn. Reson. Chem. 2021, 59, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Maier, A.; Fiebig, H.-H.; Lin, W.-H.; Peschel, G.; Hertweck, C. Kandenols A–E, Eudesmenes from an endophytic streptomyces sp. of the mangrove tree Kandelia candel. J. Nat. Prod. 2012, 75, 2223–2227. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Jiang, Y.; Liu, J.; Li, Q.; Wang, X.; Mu, Y.; Han, L.; Huang, X. Structure determination of two new sesquiterpenoids from Streptomyces sanglieri. Magn. Reson. Chem. 2016, 54, 930–932. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Han, L.; Jiang, Y.; Li, G.; Liu, J.; Mu, Y.; Huang, X. Sesquiterpenoids from Streptomyces anulatus isolated from Giraffa camelopardalis feces. Magn. Reson. Chem. 2018, 56, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, K.A.; Singh, S.; Elshahawi, S.I.; Wang, X.; Ponomareva, L.V.; Sunkara, M.; Copley, G.C.; Hower, J.C.; Morris, A.J.; Kharel, M.K.; et al. The native production of the sesquiterpene isopterocarpolone by Streptomyces sp. RM-14-6. Nat. Prod. Res. 2014, 28, 337–339. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Yang, Y.; Yang, X.; Zhang, Y.; Zhao, L.; Xu, L.; Ding, Z. Sesquiterpenes from the secondary metabolites of Streptomyces sp. (YIM 56130). Chem. Pharm. Bull. 2011, 59, 1430–1433. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Mei, W.; Zhao, Y.-X.; Hong, K.; Dai, H. A new degraded sesquiterpene from marine actinomycete Streptomyces sp. 0616208. Chinese Chem. Lett. 2006, 17, 1463–1465. [Google Scholar]
- Lu, S.; Xie, X.; Hu, J.; Lin, H.; Li, F.; Zhou, R.; Guo, J.; Wu, S.; He, J. New anti-influenza A viral norsesquiterpenoids isolated from feces-residing Streptomyces sp. Fitoterapia 2022, 157, 105107. [Google Scholar] [CrossRef]
- Hu, Y.; Chou, W.K.W.; Hopson, R.; Cane, D.E. Genome mining in Streptomyces clavuligerus: Expression and biochemical characterization of two new cryptic sesquiterpene synthases. Chem. Biol. 2011, 18, 32–37. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Pfoh, R.; Rühl, S.; Qin, S.; Laatsch, H. T-Muurolol sesquiterpenes from the marine Streptomyces sp. M491 and revision of the configuration of previously reported amorphanes. J. Nat. Prod. 2009, 72, 99–101. [Google Scholar] [CrossRef]
- Lauterbach, L.; Hou, A.; Dickschat, J.S. Rerouting and improving dauc-8-en-11-ol synthase from Streptomyces venezuelae to a high yielding biocatalyst. Chem. Eur. J. 2021, 27, 7923–7929. [Google Scholar] [CrossRef] [PubMed]
- Klapschinski, T.A.; Rabe, P.; Dickschat, J.S. Pristinol, a sesquiterpene alcohol with an unusual skeleton from Streptomyces pristinaespiralis. Angew. Chem. Int. Ed. 2016, 55, 10141–10144. [Google Scholar] [CrossRef] [PubMed]
- Rabe, P.; Rinkel, J.; Klapschinski, T.A.; Barra, L.; Dickschat, J.S. A method for investigating the stereochemical course of terpene cyclisations. Org. Biomol. Chem. 2016, 14, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Kracht, O.N.; Correia Cordeiro, R.S.; Håkansson, M.; Stockmann, J.; Sander, D.; Bandow, J.; Senges, C.H.R.; Logan, D.T.; Kourist, R. Discovery of three novel sesquiterpene synthases from Streptomyces chartreusis NRRL 3882 and crystal structure of an α-eudesmol synthase. J. Biotechnol. 2019, 297, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Rabe, P.; Schmitz, T.; Dickschat, J.S. Mechanistic investigations on six bacterial terpene cyclases. Beilstein J. Org. Chem. 2016, 12, 1839–1850. [Google Scholar] [CrossRef] [Green Version]
- Rabe, P.; Samborskyy, M.; Leadlay, P.F.; Dickschat, J.S. Isoafricanol synthase from Streptomyces malaysiensis. Org. Biomol. Chem. 2017, 15, 2353–2358. [Google Scholar] [CrossRef] [PubMed]
- Nakano, C.; Horinouchi, S.; Ohnishi, Y. Characterization of a novel sesquiterpene cyclase involved in (+)-caryolan-1-ol biosynthesis in Streptomyces griseus. J. Biol. Chem. 2011, 286, 27980–27987. [Google Scholar] [CrossRef] [Green Version]
- Cho, G.; Kwak, Y.S. Evolution of antibiotic synthesis gene clusters in the Streptomyces globisporus TFH56, isolated from tomato flower. G3 Genes Genomes Genet. 2019, 9, 1807–1813. [Google Scholar] [CrossRef] [Green Version]
- Cho, G.; Kim, J.; Park, C.G.; Nislow, C.; Weller, D.M.; Kwak, Y.-S. Caryolan-1-ol, an antifungal volatile produced by Streptomyces spp., inhibits the endomembrane system of fungi. Open Biol. 2017, 7, 170075. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Zhu, Y.; Zhang, M.; Li, H.; Sun, P. Micaryolanes A and B, two new caryolane-type sesquiterpenoids from marine Streptomyces sp. AH25. Chem. Biodivers. 2020, 17, e2000769. [Google Scholar] [CrossRef]
- Ding, L.; Goerls, H.; Dornblut, K.; Lin, W.; Maier, A.; Fiebig, H.-H.; Hertweck, C. Bacaryolanes A–C, rare bacterial caryolanes from a mangrove endophyte. J. Nat. Prod. 2015, 78, 2963–2967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aaron, J.A.; Lin, X.; Cane, D.E.; Christianson, D.W. Structure of epi-isozizaene synthase from Streptomyces coelicolor A3(2), a platform for new terpenoid cyclization templates. Biochemistry 2010, 49, 1787–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.L.; Tian, T.; Alonso-Gutierrez, J.; Garabedian, B.; Wang, S.; Baidoo, E.E.K.; Benites, V.; Chen, Y.; Petzold, C.J.; Adams, P.D.; et al. Renewable production of high density jet fuel precursor sesquiterpenes from Escherichia coli. Biotechnol. Biofuels 2018, 11, 285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Hopson, R.; Cane, D.E. Genome mining in Streptomyces coelicolor: Molecular cloning and characterization of a new sesquiterpene synthase. J. Am. Chem. Soc. 2006, 128, 6022–6023. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Lin, X.; Lei, L.; Lamb, D.C.; Kelly, S.L.; Waterman, M.R.; Cane, D.E. Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor A3(2). J. Biol. Chem. 2008, 283, 8183–8189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Cane, D.E. Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor. Mechanism and stereochemistry of the enzymatic formation of epi-isozizaene. J. Am. Chem. Soc. 2009, 131, 6332–6333. [Google Scholar] [CrossRef] [Green Version]
- Ambarwati, A.; Wahyuono, S.; Moeljopawiro, S.; Yuwono, T. Antimicrobial activity of ethyl acetate extracts of Streptomyces sp. CRB46 and the prediction of their bioactive compounds chemical structure. Biodiversitas 2020, 21, 3380–3390. [Google Scholar] [CrossRef]
- Moody, S.C.; Zhao, B.; Lei, L.; Nelson, D.R.; Mullins, J.G.L.; Waterman, M.R.; Kelly, S.L.; Lamb, D.C. Investigating conservation of the albaflavenone biosynthetic pathway and CYP170 bifunctionality in Streptomycetes. FEBS J. 2012, 279, 1640–1649. [Google Scholar] [CrossRef]
- Čihák, M.; Kameník, Z.; Šmídová, K.; Bergman, N.; Benada, O.; Kofroňová, O.; Petříčková, K.; Bobek, J. Secondary metabolites produced during the germination of Streptomyces coelicolor. Front. Microbiol. 2017, 8, 2495. [Google Scholar] [CrossRef] [Green Version]
- Sinha, A.; Phillips-Salemka, S.; Niraula, T.-A.; Short, K.A.; Niraula, N.P. The complete genomic sequence of Streptomyces spectabilis NRRL-2792 and identification of secondary metabolite biosynthetic gene clusters. J. Ind. Microbiol. Biotechnol. 2019, 46, 1217–1223. [Google Scholar] [CrossRef]
- Takamatsu, S.; Lin, X.; Nara, A.; Komatsu, M.; Cane, D.E.; Ikeda, H. Characterization of a silent sesquiterpenoid biosynthetic pathway in Streptomyces avermitilis controlling epi-isozizaene albaflavenone biosynthesis and isolation of a new oxidized epi-isozizaene metabolite. Microb. Biotechnol. 2011, 4, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Raju, R.; Gromyko, O.; Fedorenko, V.; Luzketskyy, A.; Müller, R. Albaflavenol B, a new sesquiterpene isolated from the terrestrial actinomycete, Streptomyces sp. J. Antibiot. 2015, 68, 286–288. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Ding, N.; Jiang, Y.; Zhang, J.; Ma, J.; Chen, X.; Liu, J.; Han, L.; Huang, X. Albaflavenoid, a new tricyclic sesquiterpenoid from Streptomyces violascens. J. Antibiot. 2016, 69, 773–775. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chou, W.K.W.; Himmelberger, J.A.; Litwin, K.M.; Harris, G.G.; Cane, D.E.; Christianson, D.W. Reprogramming the chemodiversity of terpenoid cyclization by remolding the active site contour of epi -isozizaene synthase. Biochemistry 2014, 53, 1155–1168. [Google Scholar] [CrossRef]
- Yang, X.-W.; Peng, K.; Liu, Z.; Zhang, G.-Y.; Li, J.; Wang, N.; Steinmetz, A.; Liu, Y. Strepsesquitriol, a rearranged zizaane-type sesquiterpenoid from the deep-sea-derived actinomycete Streptomyces sp. SCSIO 10355. J. Nat. Prod. 2013, 76, 2360–2363. [Google Scholar] [CrossRef] [PubMed]
- Lauterbach, L.; Dickschat, J.S. Sesquiterpene synthases for bungoene, pentalenene and epi-isozizaene from Streptomyces bungoensis. Org. Biomol. Chem. 2020, 18, 4547–4550. [Google Scholar] [CrossRef]
- Tetzlaff, C.N.; You, Z.; Cane, D.E.; Takamatsu, S.; Omura, S.; Ikeda, H. A gene cluster for biosynthesis of the sesquiterpenoid antibiotic pentalenolactone in Streptomyces avermitilis. Biochemistry 2006, 45, 6179–6186. [Google Scholar] [CrossRef] [Green Version]
- Quaderer, R.; Omura, S.; Ikeda, H.; Cane, D.E. Pentalenolactone biosynthesis. Molecular cloning and assignment of biochemical function to PtlI, a cytochrome P450 of Streptomyces avermitilis. J. Am. Chem. Soc. 2006, 128, 13036–13037. [Google Scholar] [CrossRef] [Green Version]
- You, Z.; Omura, S.; Ikeda, H.; Cane, D.E.; Jogl, G. Crystal structure of the non-heme iron dioxygenase PtlH in pentalenolactone biosynthesis. J. Biol. Chem. 2007, 282, 36552–36560. [Google Scholar] [CrossRef] [Green Version]
- You, Z.; Omura, S.; Ikeda, H.; Cane, D.E. Pentalenolactone biosynthesis: Molecular cloning and assignment of biochemical function to PtlF, a short-chain dehydrogenase from Streptomyces avermitilis, and identification of a new biosynthetic intermediate. Arch. Biochem. Biophys. 2007, 459, 233–240. [Google Scholar] [CrossRef] [Green Version]
- Seo, M.J.; Zhu, D.; Endo, S.; Ikeda, H.; Cane, D.E. Genome mining in streptomyces. elucidation of the role of Baeyer-Villiger monooxygenases and non-heme iron-dependent dehydrogenase/oxygenases in the final steps of the biosynthesis of pentalenolactone and neopentalenolactone. Biochemistry 2011, 50, 1739–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Seo, M.J.; Ikeda, H.; Cane, D.E. Genome mining in Streptomyces. Discovery of an unprecedented P450-catalyzed oxidative rearrangement that is the final step in the biosynthesis of pentalenolactone. J. Am. Chem. Soc. 2011, 133, 2128–2131. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Wang, Y.; Zhang, M.; Ikeda, H.; Deng, Z.; Cane, D.E. Product-mediated regulation of pentalenolactone biosynthesis in Streptomyces species by the MarR/SlyA aamily Activators PenR and PntR. J. Bacteriol. 2013, 195, 1255–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Tetzlaff, C.N.; Takamatsu, S.; Iwatsuki, M.; Komatsu, M.; Ikeda, H.; Cane, D.E. Genome mining in Streptomyces avermitilis: A biochemical Baeyer-Villiger reaction and discovery of a new branch of the pentalenolactone family tree. Biochemistry 2009, 48, 6431–6440. [Google Scholar] [CrossRef] [Green Version]
- Takamatsu, S.; Xu, L.H.; Fushinobu, S.; Shoun, H.; Komatsu, M.; Cane, D.E.; Ikeda, H. Pentalenic acid is a shunt metabolite in the biosynthesis of the pentalenolactone family of metabolites: Hydroxylation of 1-deoxypentalenic acid mediated by CYP105D7 (SAV-7469) of Streptomyces avermitilis. J. Antibiot. 2011, 64, 65–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Li, H.; Chen, S.; Wu, W.; Sun, P. Isolation and identification of pentalenolactone analogs from Streptomyces sp. NRRL S-4. Molecules 2021, 26, 7377. [Google Scholar] [CrossRef]
- Yamada, Y.; Arima, S.; Nagamitsu, T.; Johmoto, K.; Uekusa, H.; Eguchi, T.; Shin-Ya, K.; Cane, D.E.; Ikeda, H. Novel terpenes generated by heterologous expression of bacterial terpene synthase genes in an engineered Streptomyces host. J. Antibiot. 2015, 68, 385–394. [Google Scholar] [CrossRef] [Green Version]
- Chow, J.-Y.; Tian, B.-X.; Ramamoorthy, G.; Hillerich, B.S.; Seidel, R.D.; Almo, S.C.; Jacobson, M.P.; Poulter, C.D. Computational-guided discovery and characterization of a sesquiterpene synthase from Streptomyces clavuligerus. Proc. Natl. Acad. Sci. USA 2015, 112, 5661–5666. [Google Scholar] [CrossRef] [Green Version]
- Blank, P.N.; Pemberton, T.A.; Chow, J.-Y.; Poulter, C.D.; Christianson, D.W. Crystal structure of cucumene synthase, a terpenoid cyclase that generates a linear triquinane sesquiterpene. Biochemistry 2018, 57, 6326–6335. [Google Scholar] [CrossRef]
- Xu, H.; Rinkel, J.; Dickschat, J.S. Isoishwarane synthase from Streptomyces lincolnensis. Org. Chem. Front. 2021, 8, 1177–1184. [Google Scholar] [CrossRef]
- Meguro, A.; Tomita, T.; Nishiyama, M.; Kuzuyama, T. Identification and characterization of bacterial diterpene cyclases that synthesize the cembrane skeleton. ChemBioChem 2013, 14, 316–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CHO, J.Y.; KIM, M.S. Induction of antifouling diterpene production by Streptomyces cinnabarinus PK209 in co-culture with marine-derived Alteromonas sp. KNS-16. Biosci. Biotechnol. Biochem. 2012, 76, 1849–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Xu, B.; Adpressa, D.A.; Rudolf, J.D.; Loesgen, S. Discovery and biosynthesis of a structurally dynamic antibacterial diterpenoid. Angew. Chem. Int. Ed. 2021, 60, 14163–14170. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-F.; Chen, M.-J.; Liang, D.-E.; Shi, L.-M.; Ying, Y.-M.; Shan, W.-G.; Li, G.-Q.; Zhan, Z.-J. Streptomyces albogriseolus SY67903 produces eunicellin diterpenoids structurally similar to terpenes of the gorgonian Muricella sibogae, the bacterial source. J. Nat. Prod. 2020, 83, 1641–1645. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Tantillo, D.J.; Rudolf, J.D. Mechanistic insights into the formation of the 6,10-bicyclic eunicellane skeleton by the bacterial diterpene synthase Bnd4. Angew. Chem. Int. Ed. 2021, 60, 23159–23163. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Komatsu, M.; Ikeda, H. Chemical diversity of labdane-type bicyclic diterpene biosynthesis in Actinomycetales microorganisms. J. Antibiot. 2016, 69, 515–523. [Google Scholar] [CrossRef]
- Serrano-Posada, H.; Centeno-Leija, S.; Rojas-Trejo, S.; Stojanoff, V.; Rodríguez-Sanoja, R.; Rudiño-Piñera, E.; Sánchez, S. Crystallization and X-ray diffraction analysis of a putative bacterial class I labdane-related diterpene synthase. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 1194–1199. [Google Scholar] [CrossRef] [Green Version]
- Centeno-Leija, S.; Tapia-Cabrera, S.; Guzmán-Trampe, S.; Esquivel, B.; Esturau-Escofet, N.; Tierrafría, V.H.; Rodríguez-Sanoja, R.; Zárate-Romero, A.; Stojanoff, V.; Rudiño-Piñera, E.; et al. The structure of (E)-biformene synthase provides insights into the biosynthesis of bacterial bicyclic labdane-related diterpenoids. J. Struct. Biol. 2019, 207, 29–39. [Google Scholar] [CrossRef]
- Guzmán-Trampe, S.M.; Ikeda, H.; Vinuesa, P.; Macías-Rubalcava, M.L.; Esquivel, B.; Centeno-Leija, S.; Tapia-Cabrera, S.M.; Mora-Herrera, S.I.; Ruiz-Villafán, B.; Rodríguez-Sanoja, R.; et al. Production of distinct labdane-type diterpenoids using a novel cryptic labdane-like cluster from Streptomyces thermocarboxydus K155. Appl. Microbiol. Biotechnol. 2020, 104, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.-J.; Huang, J.; Yan, Y.; Wang, L.; Wang, Z.; Yang, J.; Luo, J.; Li, J.; Huang, S.-X. Isolation and biosynthesis of labdanmycins: Four new labdane diterpenes from endophytic Streptomyces. Org. Chem. Front. 2018, 5, 1272–1279. [Google Scholar] [CrossRef]
- Ikeda, H.; Shin-ya, K.; Nagamitsu, T.; Tomoda, H. Biosynthesis of mercapturic acid derivative of the labdane-type diterpene, cyslabdan that potentiates imipenem activity against methicillin-resistant Staphylococcus aureus: Cyslabdan is generated by mycothiol-mediated xenobiotic detoxification. J. Ind. Microbiol. Biotechnol. 2016, 43, 325–342. [Google Scholar] [CrossRef]
- Xing, B.; Yu, J.; Chi, C.; Ma, X.; Xu, Q.; Li, A.; Ge, Y.; Wang, Z.; Liu, T.; Jia, H.; et al. Functional characterization and structural bases of two class I diterpene synthases in pimarane-type diterpene biosynthesis. Commun. Chem. 2021, 4, 140. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, C.; Hayashi, Y.; Itoh, N.; Seto, H.; Dairi, T. Functional analysis of eubacterial ent-copalyl diphosphate synthase and pimara-9(11),15-diene synthase with unique primary sequences. J. Biochem. 2006, 141, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Yu, Z. Diterpenoids from Streptomyces sp. SN194 and their antifungal activity against Botrytis cinerea. J. Agric. Food Chem. 2016, 64, 8525–8529. [Google Scholar] [CrossRef]
- Kim, S.Y.; Zhao, P.; Igarashi, M.; Sawa, R.; Tomita, T.; Nishiyama, M.; Kuzuyama, T. Cloning and heterologous expression of the cyclooctatin biosynthetic gene cluster afforda diterpene cyclase and two P450 hydroxylases. Chem. Biol. 2009, 16, 736–743. [Google Scholar] [CrossRef] [Green Version]
- Janke, R.; Görner, C.; Hirte, M.; Brück, T.; Loll, B. The first structure of a bacterial diterpene cyclase: CotB2. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1528–1537. [Google Scholar] [CrossRef] [PubMed]
- Driller, R.; Janke, S.; Fuchs, M.; Warner, E.; Mhashal, A.R.; Major, D.T.; Christmann, M.; Brück, T.; Loll, B. Towards a comprehensive understanding of the structural dynamics of a bacterial diterpene synthase during catalysis. Nat. Commun. 2018, 9, 3971. [Google Scholar] [CrossRef] [Green Version]
- Driller, R.; Garbe, D.; Mehlmer, N.; Fuchs, M.; Raz, K.; Major, D.T.; Brück, T.; Loll, B. Current understanding and biotechnological application of the bacterial diterpene synthase CotB2. Beilstein J. Org. Chem. 2019, 15, 2355–2368. [Google Scholar] [CrossRef]
- Tomita, T.; Kim, S.-Y.; Teramoto, K.; Meguro, A.; Ozaki, T.; Yoshida, A.; Motoyoshi, Y.; Mori, N.; Ishigami, K.; Watanabe, H.; et al. Structural insights into the CotB2-catalyzed cyclization of geranylgeranyl diphosphate to the diterpene cyclooctat-9-en-7-ol. ACS Chem. Biol. 2017, 12, 1621–1628. [Google Scholar] [CrossRef] [Green Version]
- Görner, C.; Hirte, M.; Huber, S.; Schrepfer, P.; Brück, T. Stereoselective chemo-enzymatic oxidation routes for (1R,3E,7E,11S,12S)-3,7,18-dolabellatriene. Front. Microbiol. 2015, 6, 1115. [Google Scholar] [CrossRef]
- Görner, C.; Schrepfer, P.; Redai, V.; Wallrapp, F.; Loll, B.; Eisenreich, W.; Haslbeck, M.; Brück, T. Identification, characterization and molecular adaptation of class I redox systems for the production of hydroxylated diterpenoids. Microb. Cell Fact. 2016, 15, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, G.S.; Li, S.R.; Wang, Y.Y.; Hao, H.L.; Shen, Y.M.; Lu, C.H. 16,17-dihydroxycyclooctatin, a new diterpene from Streptomyces sp. LZ35. Drug Discov. Ther. 2013, 7, 185–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawamura, A.; Iacovidou, M.; Hirokawa, E.; Soll, C.E.; Trujillo, M. 17-Hydroxycyclooctatin, a fused 5−8−5 ring diterpene, from Streptomyces sp. MTE4a. J. Nat. Prod. 2011, 74, 492–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.R.; Lee, D.; Park, M.; Lee, J.C.; Park, H.-J.; Kang, K.S.; Kim, C.-E.; Beemelmanns, C.; Kim, K.H. Absolute configuration and corrected nmr assignment of 17-hydroxycyclooctatin, a fused 5–8–5 tricyclic diterpene. J. Nat. Prod. 2020, 83, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.; Li, Q.; Song, T.; Chen, L.; Li, X.-C.; Zhang, Z.; Lian, X.-Y. Isolation, structure elucidation, and antibacterial evaluation of the metabolites produced by the marine-sourced Streptomyces sp. ZZ820. Tetrahedron 2019, 75, 1186–1193. [Google Scholar] [CrossRef]
- Zheng, D.; Han, L.; Qu, X.; Chen, X.; Zhong, J.; Bi, X.; Liu, J.; Jiang, Y.; Jiang, C.; Huang, X. Cytotoxic fusicoccane-type diterpenoids from Streptomyces violascens isolated from Ailuropoda melanoleuca feces. J. Nat. Prod. 2017, 80, 837–844. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, S.; Ma, K.; Xu, Y.; Tao, Q.; Chen, Y.; Chen, J.; Guo, S.; Ren, J.; Wang, W.; et al. Discovery and characterization of a new family of diterpene cyclases in bacteria and fungi. Angew. Chem. Int. Ed. 2017, 56, 4749–4752. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, Y.; Zhang, X.; Chang, Y.; Li, S.; Zhang, X.; Zheng, S.; Geng, C.; Men, P.; Ma, L.; et al. Fragrant venezuelaenes a and b with a 5–5–6–7 tetracyclic skeleton: Discovery, biosynthesis, and mechanisms of central catalysts. ACS Catal. 2020, 10, 5846–5851. [Google Scholar] [CrossRef]
- Rabe, P.; Rinkel, J.; Dolja, E.; Schmitz, T.; Nubbemeyer, B.; Luu, T.H.; Dickschat, J.S. Mechanistic investigations of two bacterial diterpene cyclases: Spiroviolene synthase and tsukubadiene synthase. Angew. Chem. Int. Ed. 2017, 56, 2776–2779. [Google Scholar] [CrossRef]
- Xu, H.; Dickschat, J.S. Revision of the cyclisation mechanism for the diterpene spiroviolene and investigations of its mass spectrometric fragmentation. ChemBioChem 2021, 22, 850–854. [Google Scholar] [CrossRef]
- Rinkel, J.; Steiner, S.T.; Dickschat, J.S. Diterpene biosynthesis in Actinomycetes: Studies on cattleyene synthase and phomopsene synthase. Angew. Chem. Int. Ed. 2019, 58, 9230–9233. [Google Scholar] [CrossRef] [PubMed]
- Ringel, M.; Reinbold, M.; Hirte, M.; Haack, M.; Huber, C.; Eisenreich, W.; Masri, M.A.; Schenk, G.; Guddat, L.W.; Loll, B.; et al. Towards a sustainable generation of pseudopterosin-type bioactives. Green Chem. 2020, 22, 6033–6046. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Y.; Zhang, S.; Chen, Q.; Ma, K.; Bao, L.; Tao, Y.; Yin, W.; Wang, G.; Liu, H. Identification and characterization of a membrane-bound sesterterpene cyclase from Streptomyces somaliensis. J. Nat. Prod. 2018, 81, 1089–1092. [Google Scholar] [CrossRef] [PubMed]
- Hou, A.; Dickschat, J.S. The biosynthetic gene cluster for sestermobaraenes—Discovery of a geranylfarnesyl diphosphate synthase and a multiproduct sesterterpene synthase from Streptomyces mobaraensis. Angew. Chem. Int. Ed. 2020, 59, 19961–19965. [Google Scholar] [CrossRef]
- Ghimire, G.P.; Lee, H.C.; Sohng, J.K. Improved squalene production via modulation of the methylerythritol 4-phosphate pathway and heterologous expression of genes from Streptomyces peucetius ATCC 27952 in Escherichia coli. Appl. Environ. Microbiol. 2009, 75, 7291–7293. [Google Scholar] [CrossRef] [Green Version]
- Khalid, A.; Takagi, H.; Panthee, S.; Muroi, M.; Chappell, J.; Osada, H.; Takahashi, S. Development of a terpenoid-production platform in Streptomyces reveromyceticus SN-593. ACS Synth. Biol. 2017, 6, 2339–2349. [Google Scholar] [CrossRef]
- Poralla, K.; Muth, G.; Hartner, T. Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 2000, 189, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Sakr, E.; Schaeffer, P.; Talbot, H.M.; Donisi, J.; Härtner, T.; Kannenberg, E.; Takano, E.; Rohmer, M. Ribosylhopane, a novel bacterial hopanoid, as precursor of C35 bacteriohopanepolyols in Streptomyces coelicolor A3(2). ChemBioChem 2015, 16, 2156–2161. [Google Scholar] [CrossRef] [Green Version]
- Ghimire, G.P.; Koirala, N.; Sohng, J.K. Activation of cryptic hop genes from Streptomyces peucetius ATCC 27952 involved in hopanoid biosynthesis. J. Microbiol. Biotechnol. 2015, 25, 658–661. [Google Scholar] [CrossRef]
- Seipke, R.F.; Loria, R. Hopanoids are not essential for growth of Streptomyces scabies 87-22. J. Bacteriol. 2009, 191, 5216–5223. [Google Scholar] [CrossRef] [Green Version]
- Ghimire, G.P.; Oh, T.-J.; Lee, H.C.; Sohng, J.K. Squalene-hopene cyclase (Spterp25) from Streptomyces peucetius: Sequence analysis, expression and functional characterization. Biotechnol. Lett. 2009, 31, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Russo, D.; Milella, L. Analysis of meroterpenoids. In Recent Advances in Natural Products Analysis; Silva, A.S., Nabavi, S.F., Saeedi, M., Nabavi, S.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 477–501. [Google Scholar] [CrossRef]
- Murray, L.A.M.; McKinnie, S.M.K.; Moore, B.S.; George, J.H. Meroterpenoid natural products from Streptomyces bacteria—The evolution of chemoenzymatic syntheses. Nat. Prod. Rep. 2020, 37, 1334–1366. [Google Scholar] [CrossRef] [PubMed]
- Kaysser, L.; Bernhardt, P.; Nam, S.-J.; Loesgen, S.; Ruby, J.G.; Skewes-Cox, P.; Jensen, P.R.; Fenical, W.; Moore, B.S. Merochlorins A–D, cyclic meroterpenoid antibiotics biosynthesized in divergent pathways with vanadium-dependent chloroperoxidases. J. Am. Chem. Soc. 2012, 134, 11988–11991. [Google Scholar] [CrossRef] [Green Version]
- Winter, J.M.; Moffitt, M.C.; Zazopoulos, E.; McAlpine, J.B.; Dorrestein, P.C.; Moore, B.S. Molecular Basis for Chloronium-mediated meroterpene cyclization. J. Biol. Chem. 2007, 282, 16362–16368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.Y.-T.; Adak, S.; Chekan, J.R.; Liscombe, D.K.; Miyanaga, A.; Bernhardt, P.; Diethelm, S.; Fielding, E.N.; George, J.H.; Miles, Z.D.; et al. Structural basis of stereospecific vanadium-dependent haloperoxidase family enzymes in napyradiomycin biosynthesis. Biochemistry 2022, 61, 1844–1852. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, K.; Funayama, S.; Anraku, Y.; Ishibashi, M.; Takahashi, Y.; Omura, S. Novel antibiotics, furaquinocins A and B. Taxonomy, fermentation, isolation and physico-chemical and biological characteristics. J. Antibiot. 1990, 43, 247–252. [Google Scholar] [CrossRef] [Green Version]
- Ishibashi, M.; Funayama, S.; Anraku, Y.; Komiyama, K.; Omura, S. Novel antibiotics, furaquinocins C, D, E, F, G and H. J. Antibiot. 1991, 44, 390–395. [Google Scholar] [CrossRef]
- Kawasaki, T.; Hayashi, Y.; Kuzuyama, T.; Furihata, K.; Itoh, N.; Seto, H.; Dairi, T. Biosynthesis of a natural polyketide-isoprenoid hybrid compound, furaquinocin A: Identification and heterologous expression of the gene cluster. J. Bacteriol. 2006, 188, 1236–1244. [Google Scholar] [CrossRef] [Green Version]
- Bunbamrung, N.; Intaraudom, C.; Dramae, A.; Thawai, C.; Tadtong, S.; Auncharoen, P.; Pittayakhajonwut, P. Antibacterial, antitubercular, antimalarial and cytotoxic substances from the endophytic Streptomyces sp. TBRC7642. Phytochemistry 2020, 172, 112275. [Google Scholar] [CrossRef]
- Panthee, S.; Takahashi, S.; Takagi, H.; Nogawa, T.; Oowada, E.; Uramoto, M.; Osada, H. Furaquinocins I and J: Novel polyketide isoprenoid hybrid compounds from Streptomyces reveromyceticus SN-593. J. Antibiot. 2011, 64, 509–513. [Google Scholar] [CrossRef]
- Kawahara, T.; Nagai, A.; Takagi, M.; Shin-ya, K. A new furaquinocin derivative, JBIR-136, from Streptomyces sp. 4963H2. J. Antibiot. 2012, 65, 579–581. [Google Scholar] [CrossRef] [Green Version]
- Tistechok, S.; Stierhof, M.; Myronovskyi, M.; Zapp, J.; Gromyko, O.; Luzhetskyy, A. Furaquinocins K and L: Novel naphthoquinone-based meroterpenoids from Streptomyces sp. Je 1-369. Antibiotics 2022, 11, 1587. [Google Scholar] [CrossRef]
- Sakoulas, G.; Nam, S.-J.; Loesgen, S.; Fenical, W.; Jensen, P.R.; Nizet, V.; Hensler, M. Novel bacterial metabolite merochlorin A demonstrates in vitro activity against multi-drug resistant methicillin-resistant Staphylococcus aureus. PLoS ONE 2012, 7, e29439. [Google Scholar] [CrossRef] [PubMed]
- Ryu, M.-J.; Hwang, S.; Kim, S.; Yang, I.; Oh, D.-C.; Nam, S.-J.; Fenical, W. Meroindenon and merochlorins E and F, antibacterial meroterpenoids from a marine-derived sediment bacterium of the genus Streptomyces. Org. Lett. 2019, 21, 5779–5783. [Google Scholar] [CrossRef] [PubMed]
- Ryu, M.-J.; Hillman, P.F.; Lee, J.; Hwang, S.; Lee, E.-Y.; Cha, S.-S.; Yang, I.; Oh, D.-C.; Nam, S.-J.; Fenical, W. Antibacterial meroterpenoids, merochlorins G–J from the marine bacterium Streptomyces sp. Mar. Drugs 2021, 19, 618. [Google Scholar] [CrossRef]
- Hayakawa, Y.; Yamazaki, Y.; Kurita, M.; Kawasaki, T.; Takagi, M.; Shin-ya, K. Flaviogeranin, a new neuroprotective compound from Streptomyces sp. J. Antibiot. 2010, 63, 379–380. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Wang, X.; Huang, T.; Deng, Z.; Lin, S. Naphthoquinone-based meroterpenoids from marine-derived Streptomyces sp. B9173. Biomolecules 2020, 10, 1187. [Google Scholar] [CrossRef] [PubMed]
- Shin-Ya, K.; Imai, S.; Furihata, K.; Hayakawa, Y.; Kato, Y.; Vanduyne, G.D.; Clardy, J.; Seto, H. Isolation and structural elucidation of an antioxidative agent, naphterpin. J. Antibiot. 1990, 43, 444–447. [Google Scholar] [CrossRef] [Green Version]
- Park, J.-S.; Kwon, H. New naphthoquinone terpenoids from marine actinobacterium, Streptomyces sp. CNQ-509. Mar. Drugs 2018, 16, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagi, H.; Motohashi, K.; Miyamoto, T.; Shin-ya, K.; Furihata, K.; Seto, H. Studies on terpenoids produced by Actinomycetes. J. Antibiot. 2005, 58, 275–278. [Google Scholar] [CrossRef]
- Hwang, J.S.; Kim, G.J.; Choi, H.G.; Kim, M.C.; Hahn, D.; Nam, J.-W.; Nam, S.-J.; Kwon, H.C.; Chin, J.; Cho, S.J.; et al. Identification of antiangiogenic potential and cellular mechanisms of napyradiomycin A1 isolated from the marine-derived Streptomyces sp. YP127. J. Nat. Prod. 2017, 80, 2269–2275. [Google Scholar] [CrossRef] [PubMed]
- Lacret, R.; Pérez-Victoria, I.; Oves-Costales, D.; de la Cruz, M.; Domingo, E.; Martín, J.; Díaz, C.; Vicente, F.; Genilloud, O.; Reyes, F. MDN-0170, a new napyradiomycin from Streptomyces sp. strain CA-271078. Mar. Drugs 2016, 14, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motohashi, K.; Sue, M.; Furihata, K.; Ito, S.; Seto, H. Terpenoids produced by Actinomycetes: Napyradiomycins from Streptomyces antimycoticus NT17. J. Nat. Prod. 2008, 71, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Molina, D.; Ortiz-López, F.J.; Martín, J.; Oves-Costales, D.; Díaz, C.; de la Cruz, M.; Cautain, B.; Vicente, F.; Genilloud, O.; Reyes, F. New napyradiomycin analogues from Streptomyces sp. strain CA-271078. Mar. Drugs 2019, 18, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Li, S.; Li, J.; Chen, Y.; Saurav, K.; Zhang, Q.; Zhang, H.; Zhang, W.; Zhang, W.; Zhang, S.; et al. Antibacterial and cytotoxic new napyradiomycins from the marine-derived Streptomyces sp. SCSIO 10428. Mar. Drugs 2013, 11, 2113–2125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.W.; Kim, G.J.; Kim, H.J.; Nam, J.-W.; Kim, J.; Chin, J.; Park, J.-H.; Choi, H.; Park, K.D. Identification and evaluation of a napyradiomycin as a potent Nrf2 activator: Anti-oxidative and anti-inflammatory activities. Bioorg. Chem. 2020, 105, 104434. [Google Scholar] [CrossRef]
- Cheng, Y.-B.; Jensen, P.R.; Fenical, W. Cytotoxic and antimicrobial napyradiomycins from two marine-derived Streptomyces strains. Eur. J. Org. Chem. 2013, 2013, 3751–3757. [Google Scholar] [CrossRef] [Green Version]
- Pereira, F.; Almeida, J.R.; Paulino, M.; Grilo, I.R.; Macedo, H.; Cunha, I.; Sobral, R.G.; Vasconcelos, V.; Gaudêncio, S.P. Antifouling napyradiomycins from marine-derived actinomycetes Streptomyces aculeolatus. Mar. Drugs 2020, 18, 63. [Google Scholar] [CrossRef] [Green Version]
- Soria-Mercado, I.E.; Prieto-Davo, A.; Jensen, P.R.; Fenical, W. Antibiotic terpenoid chloro-dihydroquinones from a new marine actinomycete. J. Nat. Prod. 2005, 68, 904–910. [Google Scholar] [CrossRef]
- Farnaes, L.; Coufal, N.G.; Kauffman, C.A.; Rheingold, A.L.; DiPasquale, A.G.; Jensen, P.R.; Fenical, W. Napyradiomycin derivatives, produced by a marine-derived actinomycete, illustrate cytotoxicity by induction of apoptosis. J. Nat. Prod. 2014, 77, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Huang, H.; Chen, Y.; Ding, J.; Zhang, Y.; Sun, A.; Zhang, W.; Ju, J. Cytotoxic and antibacterial marfuraquinocins from the deep South China Sea-derived Streptomyces niveus SCSIO 3406. J. Nat. Prod. 2013, 76, 2263–2268. [Google Scholar] [CrossRef]
- Takashima, M.; Sakai, H. A New toxic substance, teleocidin, produced by Streptomyces. Bull. Agric. Chem. Soc. Japan 1960, 24, 647–655. [Google Scholar] [CrossRef]
- Awakawa, T.; Abe, I. Biosynthesis of the teleocidin-type terpenoid indole alkaloids. Org. Biomol. Chem. 2018, 16, 4746–4752. [Google Scholar] [CrossRef] [PubMed]
- Awakawa, T. Enzymatic reactions in teleocidin B biosynthesis. J. Nat. Med. 2021, 75, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Abe, I. Biosynthetic studies on teleocidins in Streptomyces. J. Antibiot. 2018, 71, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Kwon, H.C.; Williams, P.G.; Jensen, P.R.; Fenical, W. Azamerone, a terpenoid phthalazinone from a marine-derived bacterium related to the genus Streptomyces (Actinomycetales). Org. Lett. 2006, 8, 2471–2474. [Google Scholar] [CrossRef] [Green Version]
- Asolkar, R.N.; Singh, A.; Jensen, P.R.; Aalbersberg, W.; Carté, B.K.; Feussner, K.-D.; Subramani, R.; DiPasquale, A.; Rheingold, A.L.; Fenical, W. Marinocyanins, cytotoxic bromo-phenazinone meroterpenoids from a marine bacterium from the streptomycete clade MAR4. Tetrahedron 2017, 73, 2234–2241. [Google Scholar] [CrossRef] [Green Version]
- Zafrir Ilan, E.; Torres, M.R.; Prudhomme, J.; Le Roch, K.; Jensen, P.R.; Fenical, W. Farnesides A and B, sesquiterpenoid nucleoside ethers from a marine-derived Streptomyces sp., strain CNT-372 from Fiji. J. Nat. Prod. 2013, 76, 1815–1818. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.; Cheng, W.; Song, Y.; He, Q.; Ju, J.; Li, Q. Complete genome sequence of the deep South China Sea-derived Streptomyces niveus SCSIO 3406, the producer of cytotoxic and antibacterial marfuraquinocins. PLoS ONE 2021, 16, e0248404. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Q.; Li, S.; Zhu, Y.; Zhang, G.; Zhang, H.; Tian, X.; Zhang, S.; Ju, J.; Zhang, C. Identification and characterization of xiamycin A and oxiamycin gene cluster reveals an oxidative cyclization strategy tailoring indolosesquiterpene biosynthesis. J. Am. Chem. Soc. 2012, 134, 8996–9005. [Google Scholar] [CrossRef]
- Zhang, Q.; Mándi, A.; Li, S.; Chen, Y.; Zhang, W.; Tian, X.; Zhang, H.; Li, H.; Zhang, W.; Zhang, S.; et al. N-N-coupled indolo-sesquiterpene atropo-diastereomers from a marine-derived actinomycete. Eur. J. Org. Chem. 2012, 2012, 5256–5262. [Google Scholar] [CrossRef]
- Ding, L.; Münch, J.; Goerls, H.; Maier, A.; Fiebig, H.-H.; Lin, W.-H.; Hertweck, C. Xiamycin, a pentacyclic indolosesquiterpene with selective anti-HIV activity from a bacterial mangrove endophyte. Bioorg. Med. Chem. Lett. 2010, 20, 6685–6687. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, Y.; Zhang, Q.; Zhu, Y.; Li, S.-M.; Li, A.; Zhang, C. Elucidating the cyclization cascades in xiamycin biosynthesis by substrate synthesis and enzyme characterizations. Org. Lett. 2015, 17, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, H.; Li, S.; Zhu, Y.; Zhang, G.; Zhang, H.; Zhang, W.; Shi, R.; Zhang, C. Carboxyl formation from methyl via triple hydroxylations by XiaM in xiamycin A biosynthesis. Org. Lett. 2012, 14, 6142–6145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, H.; Yu, L.; Sun, Y.; Zhu, Y.; Zhu, H.; Zhang, L.; Li, S.-M.; Shen, Y.; Tian, C.; et al. Characterization of the flavoenzyme XiaK as an N-hydroxylase and implications in indolosesquiterpene diversification. Chem. Sci. 2017, 8, 5067–5077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.-H.; Ha, T.-K.-Q.; Oh, W.K.; Shin, J.; Oh, D.-C. Antiviral indolosesquiterpenoid xiamycins C–E from a halophilic actinomycete. J. Nat. Prod. 2016, 79, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Maier, A.; Fiebig, H.-H.; Lin, W.-H.; Hertweck, C. A family of multicyclic indolosesquiterpenes from a bacterial endophyte. Org. Biomol. Chem. 2011, 9, 4029. [Google Scholar] [CrossRef]
- Takada, K.; Kajiwara, H.; Imamura, N. Oridamycins A and B, anti-Saprolegnia parasitica indolosesquiterpenes isolated from Streptomyces sp. KS84. J. Nat. Prod. 2010, 73, 698–701. [Google Scholar] [CrossRef]
- Chen, L.-Y.; Wang, X.-Q.; Wang, Y.-M.; Geng, X.; Xu, X.-N.; Su, C.; Yang, Y.-L.; Tang, Y.-J.; Bai, F.-W.; Zhao, X.-Q. Genome mining of Streptomyces xinghaiensis NRRL B-24674T for the discovery of the gene cluster involved in anticomplement activities and detection of novel xiamycin analogs. Appl. Microbiol. Biotechnol. 2018, 102, 9549–9562. [Google Scholar] [CrossRef]
- Jin, E.; Li, H.; Liu, Z.; Xiao, F.; Li, W. Antibiotic dixiamycins from a cold-seep-derived Streptomyces olivaceus. J. Nat. Prod. 2021, 84, 2606–2611. [Google Scholar] [CrossRef]
- Baunach, M.; Ding, L.; Bruhn, T.; Bringmann, G.; Hertweck, C. Regiodivergent N-C and N-N aryl coupling reactions of indoloterpenes and cycloether formation mediated by a single bacterial flavoenzyme. Angew. Chem. Int. Ed. 2013, 52, 9040–9043. [Google Scholar] [CrossRef]
- Baunach, M.; Ding, L.; Willing, K.; Hertweck, C. Bacterial synthesis of unusual sulfonamide and sulfone antibiotics by flavoenzyme-mediated sulfur dioxide capture. Angew. Chem. Int. Ed. 2015, 54, 13279–13283. [Google Scholar] [CrossRef] [PubMed]
- Také, A.; Matsumoto, A.; Ōmura, S.; Takahashi, Y. Streptomyces lactacystinicus sp. nov. and Streptomyces cyslabdanicus sp. nov., producing lactacystin and cyslabdan, respectively. J. Antibiot. 2015, 68, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Koyama, N.; Tokura, Y.; Takahashi, Y.; Tomoda, H. New cyslabdans B and C, potentiators of imipenem activity against methicillin-resistant Staphylococcus aureus produced by Streptomyces sp. K04-0144. Acta Pharm. Sin. B 2011, 1, 236–239. [Google Scholar] [CrossRef] [Green Version]
- Motohashi, K.; Ueno, R.; Sue, M.; Furihata, K.; Matsumoto, T.; Dairi, T.; Omura, S.; Seto, H. Studies on terpenoids produced by Actinomycetes: Oxaloterpins A, B, C, D, and E, diterpenes from Streptomyces sp. KO-3988. J. Nat. Prod. 2007, 70, 1712–1717. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Ma, M.; Rateb, M.E.; Shaaban, K.A.; Yu, Z.; Huang, S.-X.; Zhao, L.-X.; Zhu, X.; Yan, Y.; Peterson, R.M.; et al. Biosynthetic potential-based strain prioritization for natural product discovery: A showcase for diterpenoid-producing Actinomycetes. J. Nat. Prod. 2014, 77, 377–387. [Google Scholar] [CrossRef]
- Dürr, C.; Schnell, H.J.; Luzhetskyy, A.; Murillo, R.; Weber, M.; Welzel, K.; Vente, A.; Bechthold, A. Biosynthesis of the terpene phenalinolactone in Streptomyces sp. Tü6071: Analysis of the gene cluster and generation of derivatives. Chem. Biol. 2006, 13, 365–377. [Google Scholar] [CrossRef] [Green Version]
- Gebhardt, K.; Meyer, S.W.; Schinko, J.; Bringmann, G.; Zeeck, A.; Fiedler, H.-P. Phenalinolactones A-D, terpenoglycoside antibiotics from Streptomyces sp. Tü 6071. J. Antibiot. 2011, 64, 229–232. [Google Scholar] [CrossRef]
- Daum , M.; Schnell , H.-J.; Herrmann, S.; Günther, A.; Murillo, R.; Müller, R.; Bisel, P.; Müller, M.; Bechthold, A. Functions of genes and enzymes involved in phenalinolactone biosynthesis. ChemBioChem 2010, 11, 1383–1391. [Google Scholar] [CrossRef]
- Kiske, C.; Erxleben, A.; Lucas, X.; Willmann, L.; Klementz, D.; Günther, S.; Römer, W.; Kammerer, B. Metabolic pathway monitoring of phenalinolactone biosynthesis from Streptomyces sp. Tü6071 by liquid chromatography/mass spectrometry coupling. Rapid Commun. Mass Spectrom. 2014, 28, 1459–1467. [Google Scholar] [CrossRef]
- Binz, T.M.; Wenzel, S.C.; Schnell, H.-J.; Bechthold, A.; Müller, R. Heterologous expression and genetic engineering of the phenalinolactone biosynthetic gene cluster by using Red/ET recombineering. ChemBioChem 2008, 9, 447–454. [Google Scholar] [CrossRef]
- Dong, L.-B.; Rudolf, J.D.; Deng, M.-R.; Yan, X.; Shen, B. Discovery of the tiancilactone antibiotics by genome mining of atypical bacterial type II diterpene synthases. ChemBioChem 2018, 19, 1727–1733. [Google Scholar] [CrossRef]
- Wei, F.; Li, W.; Song, R.; Shen, Y. Trinulactones A-D, new dinorsesterterpenoids from Streptomyces sp. S006. Nat. Prod. Commun. 2018, 13, 1433–1436. [Google Scholar] [CrossRef] [Green Version]
- Jang, J.-P.; Jang, M.; Hwang, G.J.; Kim, M.H.; Ahn, J.S.; Ko, S.-K.; Jang, J.-H. Streptooctatins A and B, fusicoccane-type diterpenoids with autophagic activity from Streptomyces sp. KCB17JA11. Bioorg. Med. Chem. Lett. 2022, 57, 128504. [Google Scholar] [CrossRef]
- Nam, S.-J.; Kauffman, C.A.; Paul, L.A.; Jensen, P.R.; Fenical, W. Actinoranone, a cytotoxic meroterpenoid of unprecedented structure from a marine adapted Streptomyces sp. Org. Lett. 2013, 15, 5400–5403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Soisson, S.M.; Young, K.; Shoop, W.; Kodali, S.; Galgoci, A.; Painter, R.; Parthasarathy, G.; Tang, Y.S.; Cummings, R.; et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 2006, 441, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kodali, S.; Sang, H.L.; Galgoci, A.; Painter, R.; Dorso, K.; Racine, F.; Motyl, M.; Hernandez, L.; Tinney, E.; et al. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc. Natl. Acad. Sci. USA 2007, 104, 7612–7616. [Google Scholar] [CrossRef] [Green Version]
- Smanski, M.J.; Yu, Z.; Casper, J.; Lin, S.; Peterson, R.M.; Chen, Y.; Wendt-Pienkowski, E.; Rajski, S.R.; Shen, B. Dedicated ent-kaurene and ent-atiserene synthases for platensimycin and platencin biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, 13498–13503. [Google Scholar] [CrossRef] [Green Version]
- Herath, K.B.; Attygalle, A.B.; Singh, S.B. Biosynthetic studies of platensimycin. J. Am. Chem. Soc. 2007, 129, 15422–15423. [Google Scholar] [CrossRef]
- Smanski, M.J.; Peterson, R.M.; Shen, B. Platensimycin and platencin biosynthesis in Streptomyces platensis, showcasing discovery and characterization of novel bacterial diterpene synthases. Methods Enzymol. 2012, 515, 163–186. [Google Scholar] [CrossRef]
- Rudolf, J.D.; Dong, L.B.; Cao, H.; Hatzos-Skintges, C.; Osipiuk, J.; Endres, M.; Chang, C.Y.; Ma, M.; Babnigg, G.; Joachimiak, A.; et al. Structure of the ent-copalyl diphosphate synthase PtmT2 from Streptomyces platensis CB00739, a bacterial type II diterpene synthase. J. Am. Chem. Soc. 2016, 138, 10905–10915. [Google Scholar] [CrossRef] [PubMed]
- Smanski, M.J.; Peterson, R.M.; Rajski, S.R.; Shen, B.; Doctoral, M.; Program, T. Engineered Streptomyces platensis strains that overproduce antibiotics platensimycin and platencin. Antimicrob. Agents Chemother. 2009, 53, 1299–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martens, E.; Demain, A.L. Platensimycin and platencin: Promising antibiotics for future application in human medicine. J. Antibiot. 2011, 64, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Smanski, M.J.; Peterson, R.M.; Marchillo, K.; Andes, D.; Rajski, S.R.; Shen, B. Engineering of Streptomyces platensis MA7339 for overproduction of platencin and congeners. Org. Lett. 2010, 12, 1744–1747. [Google Scholar] [CrossRef] [Green Version]
- Rudolf, J.D.; Dong, L.-B.; Shen, B. Platensimycin and platencin: Inspirations for chemistry, biology, enzymology, and medicine. Biochem. Pharmacol. 2017, 133, 139–151. [Google Scholar] [CrossRef]
- Das, M.; Sakha Ghosh, P.; Manna, K. A Review on platensimycin: A selective FabF inhibitor. Int. J. Med. Chem. 2016, 2016, 9706753. [Google Scholar] [CrossRef] [Green Version]
- Guang, C.; Chen, J.; Sang, S.; Cheng, S. Biological functionality of soyasaponins and soyasapogenols. J. Agric. Food Chem. 2014, 62, 8247–8255. [Google Scholar] [CrossRef]
- Hayashi, Y.; Onaka, H.; Itoh, N.; Seto, H.; Dairi, T. Cloning of the gene cluster responsible for biosynthesis of KS-505a (longestin), a unique tetraterpenoid. Biosci. Biotechnol. Biochem. 2007, 71, 3072–3081. [Google Scholar] [CrossRef]
- Sun, M.W.; Zhang, X.M.; Bi, H.L.; Li, W.J.; Lu, C.H. Two new sesquiterpenoids produced by halophilic Nocardiopsis chromatogenes YIM 90109. Nat. Prod. Res. 2017, 31, 77–83. [Google Scholar] [CrossRef]
- Baer, P.; Rabe, P.; Citron, C.A.; de Oliveira Mann, C.C.; Kaufmann, N.; Groll, M.; Dickschat, J.S. Hedycaryol synthase in complex with nerolidol reveals terpene cyclase mechanism. ChemBioChem 2014, 15, 213–216. [Google Scholar] [CrossRef]
- Rinkel, J.; Dickschat, J.S. Addressing the chemistry of germacrene A by isotope labeling experiments. Org. Lett. 2019, 21, 2426–2429. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-M.; Li, X.-M.; Lu, C.-H. Abscisic acid-type sesquiterpenes and ansamycins from Amycolatopsis alba DSM 44262. J. Asian Nat. Prod. Res. 2017, 19, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Cheng, M.; Wu, M.; Chai, C.; Kwan, A.; Su, S.; Kuo, Y. Chemical constituents from a mangrove-derived actinobacteria Isoptericola chiayiensis BCRC 16888 and evaluation of their anti-NO activity. Chem. Biodivers. 2021, 18, e2100211. [Google Scholar] [CrossRef] [PubMed]
- Rinkel, J.; Dickschat, J.S. Mechanistic studies on trichoacorenol synthase from Amycolatopsis benzoatilytica. ChemBioChem 2020, 21, 807–810. [Google Scholar] [CrossRef] [Green Version]
- Shirai, M.; Okuda, M.; Motohashi, K.; Imoto, M.; Furihata, K.; Matsuo, Y.; Katsuta, A.; Shizuri, Y.; Seto, H. Terpenoids produced by Actinomycetes: Isolation, structural elucidation and biosynthesis of new diterpenes, gifhornenolones A and B from Verrucosispora gifhornensis YM28-088. J. Antibiot. 2010, 63, 245–250. [Google Scholar] [CrossRef]
- Rabe, P.; Dickschat, J.S. The EIMS fragmentation mechanisms of the sesquiterpenes corvol ethers A and B, epi-cubebol and isodauc-8-en-11-ol. Beilstein J. Org. Chem. 2016, 12, 1380–1394. [Google Scholar] [CrossRef] [Green Version]
- Rabe, P.; Pahirulzaman, K.A.K.; Dickschat, J.S. Structures and biosynthesis of corvol ethers-sesquiterpenes from the actinomycete Kitasatospora setae. Angew. Chem. Int. Ed. 2015, 54, 6041–6045. [Google Scholar] [CrossRef]
- Rinkel, J.; Dickschat, J.S. Characterization of micromonocyclol synthase from the marine actinomycete Micromonospora marina. Org. Lett. 2019, 21, 9442–9445. [Google Scholar] [CrossRef]
- Nakano, C.; Okamura, T.; Sato, T.; Dairi, T.; Hoshino, T. Mycobacterium tuberculosis H37Rv3377c encodes the diterpene cyclase for producing the halimane skeleton. Chem. Commun. 2005, 8, 1016–1018. [Google Scholar] [CrossRef]
- Nakano, C.; Hoshino, T. Characterization of the Rv3377c gene product, a type-B diterpene cyclase, from the Mycobacterium tuberculosis H37 genome. ChemBioChem 2009, 10, 2060–2071. [Google Scholar] [CrossRef]
- Mann, F.M.; Xu, M.; Chen, X.; Fulton, D.B.; Russell, D.G.; Peters, R.J. Edaxadiene: A new bioactive diterpene from Mycobacterium tuberculosis. J. Am. Chem. Soc. 2009, 131, 17526–17527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prach, L.; Kirby, J.; Keasling, J.D.; Alber, T. Diterpene production in Mycobacterium tuberculosis. FEBS J. 2010, 277, 3588–3595. [Google Scholar] [CrossRef] [PubMed]
- Nakano, C.; Ootsuka, T.; Takayama, K.; Mitsui, T.; Sato, T.; Hoshino, T. Characterization of the Rv3378c gene product, a new diterpene synthase for producing tuberculosinol and (13r,s)-isotuberculosinol (nosyberkol), from the Mycobacterium tuberculosis H37Rv genome. Biosci. Biotechnol. Biochem. 2011, 75, 75–81. [Google Scholar] [CrossRef]
- Zhang, Y.; Prach, L.M.; O’Brien, T.E.; DiMaio, F.; Prigozhin, D.M.; Corn, J.E.; Alber, T.; Siegel, J.B.; Tantillo, D.J. Crystal structure and mechanistic molecular modeling studies of Mycobacterium tuberculosis diterpene cyclase Rv3377c. Biochemistry 2020, 59, 4507–4515. [Google Scholar] [CrossRef]
- Tamamura, T.; Sawa, T.; Isshiki, K.; Masuda, T.; Homma, Y.; Iinuma, H.; Naganawa, H.; Hamada, M.; Takeuchi, T.; Umezawa, H. Isolation and characterization of terpentecin, a new antitumor antibiotic. J. Antibiot. 1985, 38, 1664–1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dairi, T.; Hamano, Y.; Kuzuyama, T.; Itoh, N.; Furihata, K.; Seto, H. Eubacterial diterpene cyclase genes essential for production of the isoprenoid antibiotic terpentecin. J. Bacteriol. 2001, 183, 6085–6094. [Google Scholar] [CrossRef] [Green Version]
- Hamano, Y.; Dairi, T.; Yamamoto, M.; Kawasaki, T.; Kaneda, K.; Kuzuyama, T.; Itoh, N.; Seto, H. Cloning of a gene cluster encoding enzymes responsible for the mevalonate pathway from a terpenoid-antibiotic-producing Streptomyces strain. Biosci. Biotechnol. Biochem. 2001, 65, 1627–1635. [Google Scholar] [CrossRef]
- Hamano, Y.; Kuzuyama, T.; Itoh, N.; Furihata, K.; Seto, H.; Dairi, T. Functional analysis of eubacterial diterpene cyclases responsible for biosynthesis of a diterpene antibiotic, terpentecin. J. Biol. Chem. 2002, 277, 37098–37104. [Google Scholar] [CrossRef] [Green Version]
- Nakano, C.; Hoshino, T.; Sato, T.; Toyomasu, T.; Dairi, T.; Sassa, T. Substrate specificity of the CYC2 enzyme from Kitasatospora griseola: Production of sclarene, biformene, and novel bicyclic diterpenes by the enzymatic reactions of labdane- and halimane-type diterpene diphosphates. Tetrahedron Lett. 2010, 51, 125–128. [Google Scholar] [CrossRef]
- Xu, M.; Hillwig, M.L.; Lane, A.L.; Tiernan, M.S.; Moore, B.S.; Peters, R.J. Characterization of an orphan diterpenoid biosynthetic operon from Salinispora arenicola. J. Nat. Prod. 2014, 77, 2144–2147. [Google Scholar] [CrossRef] [Green Version]
- Mullowney, M.; Ó hAinmhire, E.; Tanouye, U.; Burdette, J.; Pham, V.; Murphy, B. A Pimarane diterpene and cytotoxic angucyclines from a marine-derived Micromonospora sp. in Vietnam’s East sea. Mar. Drugs 2015, 13, 5815–5827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thawai, C.; Bunbamrung, N.; Pittayakhajonwut, P.; Chongruchiroj, S.; Pratuangdejkul, J.; He, Y.-W.; Tadtong, S.; Sareedenchai, V.; Prombutara, P.; Qian, Y. A novel diterpene agent isolated from Microbispora hainanensis strain CSR-4 and its in vitro and in silico inhibition effects on acetylcholine esterase enzyme. Sci. Rep. 2020, 10, 11058. [Google Scholar] [CrossRef] [PubMed]
- Takagi, M.; Motohashi, K.; Khan, S.T.; Hashimoto, J.; Shin-ya, K. JBIR-65, a new diterpene, isolated from a sponge-derived Actinomadura sp. SpB081030SC-15. J. Antibiot. 2010, 63, 401–403. [Google Scholar] [CrossRef] [Green Version]
- Shin, B.; Kim, B.-Y.; Cho, E.; Oh, K.-B.; Shin, J.; Goodfellow, M.; Oh, D.-C. Actinomadurol, an antibacterial norditerpenoid from a rare actinomycete, Actinomadura sp. KC 191. J. Nat. Prod. 2016, 79, 1886–1890. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Oku, N.; Harunari, E.; Panbangred, W.; Igarashi, Y. Complete NMR assignment and absolute configuration of k4610422, a norditerpenoid inhibitor of testosterone-5α-reductase originally from Streptosporangium: Rediscovery from a thermophilic Actinomadura. J. Antibiot. 2020, 73, 60–65. [Google Scholar] [CrossRef]
- Li, G.; Guo, Y.W.; Dickschat, J.S. Diterpene biosynthesis in Catenulispora acidiphila: On the mechanism of catenul-14-en-6-ol synthase. Angew. Chem. Int. Ed. 2021, 60, 1488–1492. [Google Scholar] [CrossRef]
- Rinkel, J.; Lauterbach, L.; Dickschat, J.S. A Branched Diterpene cascade: The mechanism of spinodiene synthase from Saccharopolyspora spinosa. Angew. Chem. Int. Ed. 2019, 58, 452–455. [Google Scholar] [CrossRef]
- Rinkel, J.; Lauterbach, L.; Rabe, P.; Dickschat, J.S. Two Diterpene synthases for spiroalbatene and cembrene A from Allokutzneria albata. Angew. Chem. Int. Ed. 2018, 57, 3238–3241. [Google Scholar] [CrossRef]
- Lauterbach, L.; Rinkel, J.; Dickschat, J.S. Two bacterial diterpene synthases from Allokutzneria albata produce bonnadiene, phomopsene, and allokutznerene. Angew. Chem. Int. Ed. 2018, 57, 8280–8283. [Google Scholar] [CrossRef]
- Rosa-Putra, S.; Nalin, R.; Domenach, A.M.; Rohmer, M. Novel hopanoids from Frankia spp. and related soil bacteria: Squalene cyclization and significance of geological biomarkers revisited. Eur. J. Biochem. 2001, 268, 4300–4306. [Google Scholar] [CrossRef]
- Dobritsa, S.V.; Potter, D.; Gookin, T.E.; Berry, A.M. Hopanoid lipids in Frankia: Identification of squalene-hopene cyclase gene sequences. Can. J. Microbiol. 2001, 47, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Takagi, R.; Orito, Y.; Ono, E.; Hoshino, T. Novel Compounds of octahydroheptaprenyl mycolic acyl ester and monocyclic C 35-terpene, heptaprenylcycline B, from non-pathogenic Mycobacterium species. Biosci. Biotechnol. Biochem. 2010, 74, 147–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Kigawa, A.; Takagi, R.; Adachi, T.; Hoshino, T. Biosynthesis of a novel cyclic C35-terpene via the cyclisation of a Z-type C35-polyprenyl diphosphate obtained from a nonpathogenic Mycobacterium species. Org. Biomol. Chem. 2008, 6, 3788. [Google Scholar] [CrossRef] [PubMed]
- Le, T.; Lee, E.; Lee, J.; Hong, A.; Yim, C.-Y.; Yang, I.; Choi, H.; Chin, J.; Cho, S.; Ko, J.; et al. Saccharoquinoline, a cytotoxic alkaloidal meroterpenoid from marine-derived bacterium Saccharomonospora sp. Mar. Drugs 2019, 17, 98. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Adnani, N.; Braun, D.R.; Ellis, G.A.; Barns, K.J.; Parker-Nance, S.; Guzei, I.A.; Bugni, T.S. Micromonohalimanes A and B: Antibacterial halimane-type diterpenoids from a marine Micromonospora species. J. Nat. Prod. 2016, 79, 2968–2972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Layre, E.; Lee, H.J.; Young, D.C.; Jezek Martinot, A.; Buter, J.; Minnaard, A.J.; Annand, J.W.; Fortune, S.M.; Snider, B.B.; Matsunaga, I.; et al. Molecular profiling of Mycobacterium tuberculosis identifies tuberculosinyl nucleoside products of the virulence-associated enzyme Rv3378c. Proc. Natl. Acad. Sci. USA 2014, 111, 2978–2983. [Google Scholar] [CrossRef] [Green Version]
- Pan, S.-J.; Tapley, A.; Adamson, J.; Little, T.; Urbanowski, M.; Cohen, K.; Pym, A.; Almeida, D.; Dorasamy, A.; Layre, E.; et al. Biomarkers for tuberculosis based on secreted, species-specific, bacterial small molecules. J. Infect. Dis. 2015, 212, 1827–1834. [Google Scholar] [CrossRef] [Green Version]
- Young, D.C.; Layre, E.; Pan, S.-J.; Tapley, A.; Adamson, J.; Seshadri, C.; Wu, Z.; Buter, J.; Minnaard, A.J.; Coscolla, M.; et al. In vivo biosynthesis of terpene nucleosides provides unique chemical markers of Mycobacterium tuberculosis infection. Chem. Biol. 2015, 22, 516–526. [Google Scholar] [CrossRef] [Green Version]
- Ghanem, M.; Dubé, J.-Y.; Wang, J.; McIntosh, F.; Houle, D.; Domenech, P.; Reed, M.B.; Raman, S.; Buter, J.; Minnaard, A.J.; et al. Heterologous production of 1-tuberculosinyladenosine in Mycobacterium kansasii models pathoevolution towards the transcellular lifestyle of Mycobacterium tuberculosis. mBio 2020, 11, e02645-20. [Google Scholar] [CrossRef]
- Komaki, H.; Nemoto, A.; Tanaka, Y.; Takagi, H.; Yazawa, K.; Mikami, Y.; Shigemori, H.; Kobayashi, J.; Ando, A.; Nagata, Y. Brasilicardin A, a new terpenoid antibiotic from pathogenic Nocardia brasiliensis: Fermentation, isolation and biological activity. J. Antibiot. 1999, 52, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, K.; Tsuda, M.; Tanaka, Y.; Mikami, Y.; Kobayashi, J. Absolute stereochemistry of immunosuppressive macrolide brasilinolide A and its new congener brasilinolide C. J. Org. Chem. 2004, 69, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, K.; Tsuda, M.; Shiro, M.; Tanaka, Y.; Mikami, Y.; Kobayashi, J. Brasilicardins B-D, new tricyclic terpernoids from actinomycete Nocardia brasiliensis. Bioorganic Med. Chem. 2004, 12, 5545–5551. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Matsuura, N.; Toshima, H.; Itoh, N.; Ishikawa, J.; Mikami, Y.; Dairi, T. Cloning of the gene cluster responsible for the biosynthesis of brasilicardin A, a unique diterpenoid. J. Antibiot. 2008, 61, 164–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, P.N.; Buchmann, A.; Roller, L.; Kulik, A.; Gross, H.; Wohlleben, W.; Stegmann, E. The immunosuppressant brasilicardin: Determination of the biosynthetic gene cluster in the heterologous host Amycolatopsis japonicum. Biotechnol. J. 2018, 13, 1700527. [Google Scholar] [CrossRef]
- Wolański, M.; Krawiec, M.; Schwarz, P.N.; Stegmann, E.; Wohlleben, W.; Buchmann, A.; Gross, H.; Eitel, M.; Koch, P.; Botas, A.; et al. A novel LysR-type regulator negatively affects biosynthesis of the immunosuppressant brasilicardin. Eng. Life Sci. 2021, 21, 4–18. [Google Scholar] [CrossRef]
- Schwarz, P.N.; Roller, L.; Kulik, A.; Wohlleben, W.; Stegmann, E. Engineering metabolic pathways in Amycolatopsis japonicum for the optimization of the precursor supply for heterologous brasilicardin congeners production. Synth. Syst. Biotechnol. 2018, 3, 56–63. [Google Scholar] [CrossRef]
- Botas, A.; Eitel, M.; Schwarz, P.N.; Buchmann, A.; Costales, P.; Núñez, L.E.; Cortés, J.; Morís, F.; Krawiec, M.; Wolański, M.; et al. Genetic engineering in combination with semi-synthesis leads to a new route for gram-scale production of the immunosuppressive natural product brasilicardin A. Angew. Chem. Int. Ed. 2021, 60, 13536–13541. [Google Scholar] [CrossRef]
- Kim, S.-H.; Lu, W.; Ahmadi, M.K.; Montiel, D.; Ternei, M.A.; Brady, S.F. Atolypenes, tricyclic bacterial sesterterpenes discovered using a multiplexed in vitro Cas9-TAR gene cluster refactoring approach. ACS Synth. Biol. 2019, 8, 109–118. [Google Scholar] [CrossRef]
- Hamed, A.; Abdel-Razek, A.; Frese, M.; Stammler, H.; El-Haddad, A.; Ibrahim, T.; Sewald, N.; Shaaban, M. Terretonin N: A new meroterpenoid from Nocardiopsis sp. Molecules 2018, 23, 299. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Kim, W.; Hwang, S.; Lee, Y.; Cho, S.; Palsson, B.; Cho, B.-K. Thirty complete Streptomyces genome sequences for mining novel secondary metabolite biosynthetic gene clusters. Sci. Data 2020, 7, 55. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Hwang, S.; Lee, Y.; Cho, S.; Palsson, B.; Cho, B.-K. Synthetic biology tools for novel secondary metabolite discovery in Streptomyces. J. Microbiol. Biotechnol. 2019, 29, 667–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayed, A.M.; Abdel-Wahab, N.M.; Hassan, H.M.; Abdelmohsen, U.R. Saccharopolyspora: An underexplored source for bioactive natural products. J. Appl. Microbiol. 2020, 128, 314–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennur, T.; Ravi Kumar, A.; Zinjarde, S.S.; Javdekar, V. Nocardiopsis species: A potential source of bioactive compounds. J. Appl. Microbiol. 2016, 120, 1–16. [Google Scholar] [CrossRef]
- Thompson, D.; Cognat, V.; Goodfellow, M.; Koechler, S.; Heintz, D.; Carapito, C.; Van Dorsselaer, A.; Mahmoud, H.; Sangal, V.; Ismail, W. Phylogenomic classification and biosynthetic potential of the fossil fuel-biodesulfurizing Rhodococcus strain IGTS8. Front. Microbiol. 2020, 11, 1417. [Google Scholar] [CrossRef] [PubMed]
- Ivshina, I.; Bazhutin, G.; Tyumina, E. Rhodococcus strains as a good biotool for neutralizing pharmaceutical pollutants and obtaining therapeutically valuable products: Through the past into the future. Front. Microbiol. 2022, 13, 967127. [Google Scholar] [CrossRef]
- Jensen, P.R.; Moore, B.S.; Fenical, W. The marine actinomycete genus Salinispora: A model organism for secondary metabolite discovery. Nat. Prod. Rep. 2015, 32, 738–751. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, S.J.; Cella, E.; Jubair, M.; Azarian, T.; Baker, B.J.; Shaw, L.N. Draft genome sequence of Verrucosispora sp. strain CWR15, isolated from a Gulf of Mexico sponge. Microbiol. Resour. Announc. 2020, 9, e00176-20. [Google Scholar] [CrossRef] [Green Version]
- Kusserow, K.; Gulder, T.A.M. Complete genome sequence of Actinomadura parvosata subsp. kistnae, a rich source of novel natural product (bio-)chemistry. J. Genomics 2017, 5, 75–76. [Google Scholar] [CrossRef] [Green Version]
- Arens, J.C.; Haltli, B.; Kerr, R.G. Draft genome sequence of Kitasatospora griseola strain MF730-N6, a bafilomycin, terpentecin, and satosporin producer. Genome Announc. 2016, 3, e00208-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchmann, A.; Eitel, M.; Koch, P.; Schwarz, P.N.; Stegmann, E.; Wohlleben, W.; Wolański, M.; Krawiec, M.; Zakrzewska-Czerwińska, J.; Méndez, C.; et al. High-quality draft genome sequence of the actinobacterium Nocardia terpenica IFM 0406, producer of the immunosuppressant brasilicardins, using illumina and PacBio technologies. Genome Announc. 2016, 4, e01391-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Zhao, Y.; Huang, C.; Luo, Y. Recent advances in silent gene cluster activation in Streptomyces. Front. Bioeng. Biotechnol. 2021, 9, 632230. [Google Scholar] [CrossRef]
- Wu, M.C.; Law, B.; Wilkinson, B.; Micklefield, J. Bioengineering natural product biosynthetic pathways for therapeutic applications. Curr. Opin. Biotechnol. 2012, 23, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Ivshina, I.B.; Kuyukina, M.S. Turning Russian specialized microbial culture collections into resource centers for biotechnology. Trends Biotechnol. 2013, 31, 609–611. [Google Scholar] [CrossRef]
- Grishko, V.V.; Tarasova, E.V.; Ivshina, I.B. Biotransformation of betulin to betulone by growing and resting cells of the actinobacterium Rhodococcus rhodochrous IEGM 66. Process Biochem. 2013, 48, 1640–1644. [Google Scholar] [CrossRef]
- Cheremnykh, K.M.; Luchnikova, N.A.; Grishko, V.V.; Ivshina, I.B. Bioconversion of ecotoxic dehydroabietic acid using Rhodococcus actinobacteria. J. Hazard. Mater. 2018, 346, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Ivshina, I.B.; Luchnikova, N.A.; Maltseva, P.Y.; Ilyina, I.V.; Volcho, K.P.; Gatilov, Y.V.; Korchagina, D.V.; Kostrikina, N.A.; Sorokin, V.V.; Mulyukin, A.L.; et al. Biotransformation of (–)-Isopulegol by Rhodococcus rhodochrous. Pharmaceuticals 2022, 15, 964. [Google Scholar] [CrossRef] [PubMed]
- Luchnikova, N.A.; Grishko, V.V.; Kostrikina, N.A.; Sorokin, V.V.; Mulyukin, A.L.; Ivshina, I.B. Biotransformation of oleanolic acid using Rhodococcus rhodochrous IEGM 757. Catalysts 2022, 12, 1352. [Google Scholar] [CrossRef]
- Citron, C.A.; Gleitzmann, J.; Laurenzano, G.; Pukall, R.; Dickschat, J.S. Terpenoids are widespread in Actinomycetes: A correlation of secondary metabolism and genome data. ChemBioChem 2012, 13, 202–214. [Google Scholar] [CrossRef]
- Snyder, S.A.; Tang, Z.-Y.; Gupta, R. Enantioselective total synthesis of (−)-napyradiomycin A1 via asymmetric chlorination of an isolated olefin. J. Am. Chem. Soc. 2009, 131, 5744–5745. [Google Scholar] [CrossRef]
- Meng, Z.; Yu, H.; Li, L.; Tao, W.; Chen, H.; Wan, M.; Yang, P.; Edmonds, D.J.; Zhong, J.; Li, A. Total synthesis and antiviral activity of indolosesquiterpenoids from the xiamycin and oridamycin families. Nat. Commun. 2015, 6, 6096. [Google Scholar] [CrossRef] [Green Version]
- Brandstätter, M.; Freis, M.; Huwyler, N.; Carreira, E.M. Total synthesis of (−)-merochlorin A. Angew. Chem. Int. Ed. 2019, 58, 2490–2494. [Google Scholar] [CrossRef] [PubMed]
- Becher, P.G.; Verschut, V.; Bibb, M.J.; Bush, M.J.; Molnár, B.P.; Barane, E.; Al-Bassam, M.M.; Chandra, G.; Song, L.; Challis, G.L.; et al. Developmentally regulated volatiles geosmin and 2-methylisoborneol attract a soil arthropod to Streptomyces bacteria promoting spore dispersal. Nat. Microbiol. 2020, 5, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Zaroubi, L.; Ozugergin, I.; Mastronardi, K.; Imfeld, A.; Law, C.; Gélinas, Y.; Piekny, A.; Findlay, B.L. The ubiquitous soil terpene geosmin acts as a warning chemical. Appl. Environ. Microbiol. 2022, 88, e00093-22. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.-M.; Peng, J.-Q.; Chen, Y.; Tao, L.; Zhang, Y.-Y.; Fu, L.-Y.; Long, Q.-D.; Shen, X.-C. 1,8-Cineole: A review of source, biological activities, and application. J. Asian Nat. Prod. Res. 2021, 23, 938–954. [Google Scholar] [CrossRef]
- An, Q.; Ren, J.-N.; Li, X.; Fan, G.; Qu, S.-S.; Song, Y.; Li, Y.; Pan, S.-Y. Recent updates on bioactive properties of linalool. Food Funct. 2021, 12, 10370–10389. [Google Scholar] [CrossRef]
- Chan, W.-K.; Tan, L.; Chan, K.-G.; Lee, L.-H.; Goh, B.-H. Nerolidol: A sesquiterpene alcohol with multi-faceted pharmacological and biological activities. Molecules 2016, 21, 529. [Google Scholar] [CrossRef] [Green Version]
- da Silva, A.C.R.; Lopes, P.M.; de Azevedo, M.M.B.; Costa, D.C.M.; Alviano, C.S.; Alviano, D.S. Biological activities of a-pinene and β-pinene enantiomers. Molecules 2012, 17, 6305–6316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anandakumar, P.; Kamaraj, S.; Vanitha, M.K. D-limonene: A multifunctional compound with potent therapeutic effects. J. Food Biochem. 2021, 45, e13566. [Google Scholar] [CrossRef]
- Li, G.-X.; Liu, Z.-Q. Unusual antioxidant behavior of α- and γ-terpinene in protecting methyl linoleate, DNA, and erythrocyte. J. Agric. Food Chem. 2009, 57, 3943–3948. [Google Scholar] [CrossRef]
- El-Minshawy, A.M.; Abdelgaleil, S.A.M.; Gadelhak, G.G.; AL-Eryan, M.A.; Rabab, R.A. Effects of monoterpenes on mortality, growth, fecundity, and ovarian development of Bactrocera zonata (Saunders) (Diptera: Tephritidae). Environ. Sci. Pollut. Res. 2018, 25, 15671–15679. [Google Scholar] [CrossRef]
- Kamatou, G.P.P.; Viljoen, A.M. A review of the application and pharmacological properties of α-bisabolol and α-bisabolol-rich oils. J. Am. Oil Chem. Soc. 2010, 87, 1–7. [Google Scholar] [CrossRef]
- Siqueira, C.A.T.; Serain, A.F.; Pascoal, A.C.R.F.; Andreazza, N.L.; de Lourenço, C.C.; Góis Ruiz, A.L.T.; de Carvalho, J.E.; de Souza, A.C.O.; Tonini Mesquita, J.; Tempone, A.G.; et al. Bioactivity and chemical composition of the essential oil from the leaves of Guatteria australis A.St.-Hil. Nat. Prod. Res. 2015, 29, 1966–1969. [Google Scholar] [CrossRef]
- Cascaes, M.M.; dos Santos Carneiro, O.; do Nascimento, L.D.; de Moraes, Â.A.B.; de Oliveira, M.S.; Cruz, J.N.; Guilhon, G.M.S.P.; de Aguiar Andrade, E.H. Essential oils from Annonaceae Species from Brazil: A systematic review of their phytochemistry, and biological activities. Int. J. Mol. Sci. 2021, 22, 12140. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.B.; Ma, Y.B.; Huang, X.Y.; Geng, C.A.; Wang, H.; Zhao, Y.; Yang, T.H.; Chen, X.L.; Yang, C.Y.; Zhang, X.M.; et al. Bioactivity-guided isolation of anti-hepatitis B virus active sesquiterpenoids from the traditional Chinese medicine: Rhizomes of Cyperus rotundus. J. Ethnopharmacol. 2015, 171, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Takao, Y.; Kuriyama, I.; Yamada, T.; Mizoguchi, H.; Yoshida, H.; Mizushina, Y. Antifungal properties of Japanese cedar essential oil from waste wood chips made from used sake barrels. Mol. Med. Rep. 2012, 5, 1163–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulyaningsih, S.; Youns, M.; El-Readi, M.Z.; Ashour, M.L.; Nibret, E.; Sporer, F.; Herrmann, F.; Reichling, J.; Wink, M. Biological activity of the essential oil of Kadsura longipedunculata (Schisandraceae) and its major components. J. Pharm. Pharmacol. 2010, 62, 1037–1044. [Google Scholar] [CrossRef]
- Cheng, S.-S.; Wu, C.-L.; Chang, H.-T.; Kao, Y.-T.; Chang, S.-T. Antitermitic and antifungal activities of essential oil of Calocedrus formosana leaf and its composition. J. Chem. Ecol. 2004, 30, 1957–1967. [Google Scholar] [CrossRef]
- Tabanca, N.; Wang, M.; Avonto, C.; Chittiboyina, A.G.; Parcher, J.F.; Carroll, J.F.; Kramer, M.; Khan, I.A. Bioactivity-guided investigation of geranium essential oils as natural tick repellents. J. Agric. Food Chem. 2013, 61, 4101–4107. [Google Scholar] [CrossRef]
- Tshering, G.; Pimtong, W.; Plengsuriyakarn, T.; Na-Bangchang, K. Anti-angiogenic effects of β-eudesmol and atractylodin in developing zebrafish embryos. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2021, 243, 108980. [Google Scholar] [CrossRef]
- Acharya, B.; Chaijaroenkul, W.; Na-Bangchang, K. Therapeutic potential and pharmacological activities of β-eudesmol. Chem. Biol. Drug Des. 2021, 97, 984–996. [Google Scholar] [CrossRef]
- Pavithra, P.S.; Mehta, A.; Verma, R.S. Aromadendrene oxide 2, induces apoptosis in skin epidermoid cancer cells through ROS mediated mitochondrial pathway. Life Sci. 2018, 197, 19–29. [Google Scholar] [CrossRef]
- Kim, M.-G.; Lee, H.-S. Growth-inhibiting effects and chemical composition of essential oils extracted from Platycladus orientalis leaves and stems toward human intestinal bacteria. Food Sci. Biotechnol. 2015, 24, 427–431. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, X.; Chen, H.; Wang, L.; Liang, J.; Luo, D.; Liu, Y.; Yang, H.; Li, Y.; Xie, J.; et al. Anti-inflammatory activity of β-patchoulene isolated from patchouli oil in mice. Eur. J. Pharmacol. 2016, 781, 229–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, J.F.; Paluch, G.; Coats, J.; Kramer, M. Elemol and amyris oil repel the ticks Ixodes scapularis and Amblyomma americanum (Acari: Ixodidae) in laboratory bioassays. Exp. Appl. Acarol. 2010, 51, 383–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahham, S.; Tabana, Y.; Iqbal, M.; Ahamed, M.; Ezzat, M.; Majid, A.; Majid, A. The anticancer, antioxidant and antimicrobial properties of the sesquiterpene β-caryophyllene from the essential oil of Aquilaria crassna. Molecules 2015, 20, 11808–11829. [Google Scholar] [CrossRef] [Green Version]
- Larionov, O.V.; Corey, E.J. An unconventional approach to the enantioselective synthesis of caryophylloids. J. Am. Chem. Soc. 2008, 130, 2954–2955. [Google Scholar] [CrossRef]
- Gūrtler, H.; Pedersen, R.; Anthoni, U.; Christophersen, C.; Nielsen, P.H.; Wellington, E.M.H.; Pedersen, C.; Bock, K. Albaflavenone, a sesquiterpene ketone with a zizaene skeleton produced by a streptomycete with a new rope morphology. J. Antibiot. 1994, 47, 434–439. [Google Scholar] [CrossRef] [Green Version]
- Kazemi, M.; Rostami, H. Chemical composition, antimicrobial and antioxidant activities of the essential oil of Psammogeton canescens. Nat. Prod. Res. 2015, 29, 277–280. [Google Scholar] [CrossRef]
- Valarezo, E.; del Carmen Guamán, M.; Paguay, M.; Meneses, M.A. Chemical composition and biological activity of the essential oil from Gnaphalium elegans kunth from Loja, Ecuador. J. Essent. Oil Bear. Plants 2019, 22, 1372–1378. [Google Scholar] [CrossRef]
- Tyagi, A.K.; Prasad, S.; Yuan, W.; Li, S.; Aggarwal, B.B. Identification of a novel compound (β-sesquiphellandrene) from turmeric (Curcuma longa) with anticancer potential: Comparison with curcumin. Investig. New Drugs 2015, 33, 1175–1186. [Google Scholar] [CrossRef]
- Gao, J.-M.; Wu, W.-J.; Zhang, J.-W.; Konishi, Y. The dihydro-β-agarofuran sesquiterpenoids. Nat. Prod. Rep. 2007, 24, 1153. [Google Scholar] [CrossRef] [PubMed]
- Delgado, G.; Del Socorro Olivares, M.; Chávez, M.I.; Ramírez-Apan, T.; Linares, E.; Bye, R.; Espinosa-García, F.J. Antiinflammatory constituents from Heterotheca inuloides. J. Nat. Prod. 2001, 64, 861–864. [Google Scholar] [CrossRef] [PubMed]
- Trevizan, L.N.F.; do Nascimento, K.F.; Santos, J.A.; Kassuya, C.A.L.; Cardoso, C.A.L.; do Carmo Vieira, M.; Moreira, F.M.F.; Croda, J.; Formagio, A.S.N. Anti-inflammatory, antioxidant and anti-Mycobacterium tuberculosis activity of viridiflorol: The major constituent of Allophylus edulis (A. St.-Hil., A. Juss. & Cambess.) Radlk. J. Ethnopharmacol. 2016, 192, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Win, N.N.; Hardianti, B.; Ngwe, H.; Hayakawa, Y.; Morita, H. Anti-inflammatory activities of isopimara-8(9),15-diene diterpenoids and mode of action of kaempulchraols B–D from Kaempferia pulchra rhizomes. J. Nat. Med. 2020, 74, 487–494. [Google Scholar] [CrossRef]
- Aoyagi, T.; Aoyama, T.; Kojima, F.; Hattori, S.; Honma, Y.; Hamada, M.; Takeuchi, T. Cyclooctatin, a new inhibitor of lysophospholipase, produced by Streptomyces melanosporofaciens Mi614-43F2 taxonomy, production, isolation, physico-chemical properties and biological activities. J. Antibiot. 1992, 45, 1587–1591. [Google Scholar] [CrossRef] [Green Version]
- Ioannou, E.; Quesada, A.; Rahman, M.M.; Gibbons, S.; Vagias, C.; Roussis, V. Dolabellanes with antibacterial activity from the brown alga Dilophus spiralis. J. Nat. Prod. 2011, 74, 213–222. [Google Scholar] [CrossRef]
- Mitić, Z.S.; Jovanović, B.; Jovanović, S.Č.; Stojanović-Radić, Z.Z.; Mihajilov-Krstev, T.; Jovanović, N.M.; Nikolić, B.M.; Marin, P.D.; Zlatković, B.K.; Stojanović, G.S. Essential oils of Pinus halepensis and P. heldreichii: Chemical composition, antimicrobial and insect larvicidal activity. Ind. Crop. Prod. 2019, 140, 111702. [Google Scholar] [CrossRef]
- Kang, M.; Kim, M.; Liu, M.; Jin, C.Z.; Park, S.H.; Lee, J.M.; Kim, J.; Park, D.; Park, H.; Kim, Y.H.; et al. Nematicidal activity of teleocidin B4 isolated from Streptomyces sp. against pine wood nematode, Bursaphelenchus xylophilus. Pest Manag. Sci. 2021, 77, 1607–1615. [Google Scholar] [CrossRef]
- Muhammad, S.; Qaisar, M.; Iqbal, J.; Khera, R.A.; Al-Sehemi, A.G.; Alarfaji, S.S.; Adnan, M. Exploring the inhibitory potential of novel bioactive compounds from mangrove Actinomycetes against nsp10 the major activator of SARS-CoV-2 replication. Chem. Pap. 2022, 76, 3051–3064. [Google Scholar] [CrossRef]
- Fukumoto, A.; Kim, Y.-P.; Hanaki, H.; Shiomi, K.; Tomoda, H.; Ōmura, S. Cyslabdan, a new potentiator of imipenem activity against methicillin-resistant Staphylococcus aureus, produced by Streptomyces sp. K04-0144. J. Antibiot. 2008, 61, 7–10. [Google Scholar] [CrossRef] [Green Version]
- Nakanishi, S.; Osawa, K.; Saito, Y.; Kawamoto, I.; Kuroda, K.; Kase, H. Ks-505A, A novel inhibitor of bovine brain ca2+ and calmodulin-dependent cyclic-nucleotide phosphodiesterase from Streptomyces argenteolus. J. Antibiot. 1992, 45, 341–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Application Area | Review, Book Chapter | |
|---|---|---|
| Agriculture | Plant growth promoting | [27] |
| Phytopathogen control | [28] | |
| Bioherbicides | [20] | |
| Biopesticides | [29] | |
| Bioinsecticides | Against insects, mites | [30] |
| Medicine | Antibiotics | [26,31,32] |
| Pharmaceuticals (antitumor, anti-inflammatory, antifungals, antihelminthics, etc.) | [33,34,35,36,37] | |
| Probiotics | [38,39] | |
| Industry | Detergents (Surfactants) | [40] |
| Biofuel | [8] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarasova, E.V.; Luchnikova, N.A.; Grishko, V.V.; Ivshina, I.B. Actinomycetes as Producers of Biologically Active Terpenoids: Current Trends and Patents. Pharmaceuticals 2023, 16, 872. https://doi.org/10.3390/ph16060872
Tarasova EV, Luchnikova NA, Grishko VV, Ivshina IB. Actinomycetes as Producers of Biologically Active Terpenoids: Current Trends and Patents. Pharmaceuticals. 2023; 16(6):872. https://doi.org/10.3390/ph16060872
Chicago/Turabian StyleTarasova, Ekaterina V., Natalia A. Luchnikova, Victoria V. Grishko, and Irina B. Ivshina. 2023. "Actinomycetes as Producers of Biologically Active Terpenoids: Current Trends and Patents" Pharmaceuticals 16, no. 6: 872. https://doi.org/10.3390/ph16060872
APA StyleTarasova, E. V., Luchnikova, N. A., Grishko, V. V., & Ivshina, I. B. (2023). Actinomycetes as Producers of Biologically Active Terpenoids: Current Trends and Patents. Pharmaceuticals, 16(6), 872. https://doi.org/10.3390/ph16060872