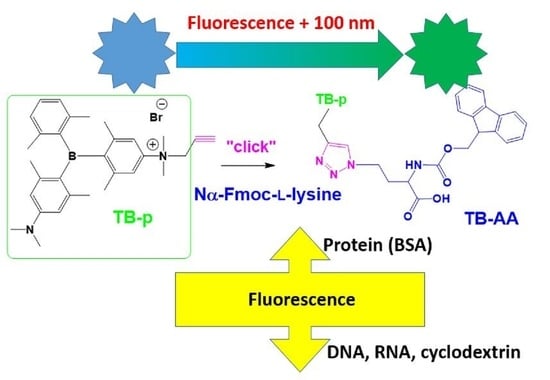
Triarylborane-“Click” Fluorescent Tag for Orthogonal Amino Acid Labelling, Interactions with DNA, Protein, and Cyclodextrins
Abstract
:1. Introduction
2. Results and Discussion
2.1. Spectrophotometric Properties of TB-p and TB-AA
2.2. Non-Covalent Interactions of TB-p and TB-AA with ds-DNA, ds-RNA, and BSA
| Compound | a ct-DNA | a poly A–poly U | b BSA |
|---|---|---|---|
| TB-p | 5.0(c 0.87) | 7.1(c 0.51) | 4.5(c 47.83) |
| TB-AA | 5.9(c 0.13) | 6.0(c 0.16) | 4.7(c 3.78) |
2.3. Interactions of TB-AA with Cyclodextrins
2.4. Biological Activity of TB-p and TB-AA
2.4.1. TB-AA Exhibits No Cytotoxic Effect Regardless of UV Light Irradiation
2.4.2. Intracellular Localisation of Compounds
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, G.; Zheng, S.Q.; Liu, H.P.; Chen, P.R. Illuminating biological processes through site-specific protein labeling. Chem. Soc. Rev. 2015, 44, 3405–3417. [Google Scholar] [CrossRef] [PubMed]
- Vrhovac Madunic, I.; Skelin Klemen, M.; Madunic, J.; Kah Yun Wee, N.; Sampaio, P. Imaging Modalities for Biological and Preclinical Research: A Compendium; Walter, A., Mannheim, J., Caruana, C., Eds.; IOP Publishing Ltd.: Bristol, UK, 2021; Volume 1, pp. I.1.b-1–I.1.b-10. [Google Scholar] [CrossRef]
- Specht, E.A.; Braselmann, E.; Palmer, A.E. A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging. Annu. Rev. Physiol. 2017, 79, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Smidlehner, T.; Badovinac, M.; Piantanida, I. Pyrene-cyanine conjugates as multipurpose fluorescent probes for non-covalent recognition of ds-DNA, RNA and proteins. N. J. Chem. 2018, 42, 6655–6663. [Google Scholar] [CrossRef]
- Amini, H.; Ban, Z.; Ferger, M.; Lorenzen, S.; Rauch, F.; Friedrich, A.; Crnolatac, I.; Kendel, A.; Miljanic, S.; Piantanida, I.; et al. Tetracationic bis-triarylborane 1,3-butadiyne as a combined fluorimetric and Raman probe for simultaneous and selective sensing of various DNA, RNA and proteins. Chem. Eur. J. 2020, 26, 6017–6028. [Google Scholar] [CrossRef]
- Ferger, M.; Ban, Z.; Krosl, I.; Tomic, S.; Dietrich, L.; Lorenzen, S.; Rauch, F.; Sieh, D.; Friedrich, A.; Griesbeck, S.; et al. Bis(phenylethynyl)arene Linkers in Tetracationic Bis-triarylborane Chromophores Control Fluorimetric and Raman Sensing of Various DNAs and RNAs. Eur. J. Chem. 2021, 27, 5142–5159. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.M.; Ruhe, J.; Schwarzmann, J.; Phillipps, A.; Richard, A.K.; Ferger, M.; Krummenacher, I.; Tumir, L.M.; Ban, Z.; Crnolatac, I.; et al. Bithiophene-Cored, mono-, bis-, and tris-(Trimethylammonium)-Substituted, bis-Triarylborane Chromophores: Effect of the Number and Position of Charges on Cell Imaging and DNA/RNA Sensing. Eur. J. Chem. 2021, 27, 14057–14072. [Google Scholar] [CrossRef]
- Ban, Z.; Griesbeck, S.; Tomic, S.; Nitsch, J.; Marder, T.B.; Piantanida, I. A Quadrupolar Bis-Triarylborane Chromophore as a Fluorimetric and Chirooptic Probe for Simultaneous and Selective Sensing of DNA, RNA and Proteins. Eur. J. Chem. 2020, 26, 2195–2203. [Google Scholar] [CrossRef]
- Ban, Z.; Karačić, Z.; Tomić, S.; Amini, H.; Marder, T.B.; Piantanida, I. Triarylborane Dyes as a Novel Non-Covalent and Non-Inhibitive Fluorimetric Markers for DPP III Enzyme. Molecules 2021, 26, 4816. [Google Scholar] [CrossRef]
- Janshoff, A.; Steinem, C.; Michalke, A.; Henke, C.; Galla, H.J. Monofunctionalized β-cyclodextrins as sensor elements for the detection of small molecules. Sens. Actuators B Chem. 2000, 70, 243–253. [Google Scholar] [CrossRef]
- Fenyvesi, É.; Vikmon, M.; Szente, L. Cyclodextrins in Food Technology and Human Nutrition: Benefits and Limitations. Crit. Rev. Food Sci. Nutr. 2016, 56, 1981–2004. [Google Scholar] [CrossRef]
- Bom, A.; Bradley, M.; Cameron, K.; Clark, J.K.; van Egmond, J.; Feilden, H.; MacLean, E.J.; Muir, A.W.; Palin, R.; Rees, D.C.; et al. A novel concept of reversing neuromuscular block: Chemical encapsulation of rocuronium bromide by a cyclodextrin-based synthetic host. Angew. Chem. Int. Ed. 2002, 41, 266–269. [Google Scholar] [CrossRef]
- Yang, L.P.H.; Keam, S.J. Sugammadex: A review of its use in anaesthetic practice. Drugs 2009, 69, 919–942. [Google Scholar] [CrossRef] [PubMed]
- Zwiers, A.; Van Den Heuvel, M.; Smeets, J.; Rutherford, S. Assessment of the potential for displacement interactions with sugammadex: A pharmacokinetic-pharmacodynamic modelling approach. Clin. Drug Investig. 2011, 31, 101–111. [Google Scholar] [CrossRef] [PubMed]
- McGhee, J.D.; Von Hippel, P.H. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef] [PubMed]
- Scatchard, G. The attractions of proteins for small molecules and ions. Ann. N. Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- Sharma, S.; Takkella, D.; Kumar, P.; Gavvala, K. Spectroscopic analysis to identify the binding site for Rifampicin on Bovine Serum Albumin. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 283, 121721. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Lacroix, L. Analysis of Thermal Melting Curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef]
- Demeunynck, M.; Bailly, C.; Wilson, W.D. Small Molecule DNA and RNA Binders: From Synthesis to Nucleic Acid Complexes; Wiley- VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2004. [Google Scholar] [CrossRef]
- Eriksson, M.; Nordén, B. Linear and Circular Dichroism of Drug-Nucleic Acid Complexes. Meth. Enzymol. 2001, 340, 68–98. [Google Scholar]
- Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization spectroscopy methods in the determination of interactions of small molecules with nucleic acids—Tutorial. Beil. J. Org. Chem. 2018, 14, 84–105. [Google Scholar] [CrossRef]
- Becer, C.R.; Hoogenboom, R.; Schubert, U.S. Click Chemistry beyond Metal-Catalyzed Cycloaddition. Angew. Chem. Int. Ed. 2009, 48, 4900–4908. [Google Scholar] [CrossRef]
- Sun, Y.; Ma, X.W.; Cheng, K.; Wu, B.Y.; Duan, J.L.; Chen, H.; Bu, L.H.; Zhang, R.P.; Hu, X.M.; Deng, Z.X.; et al. Strained Cyclooctyne as a Molecular Platform for Construction of Multimodal Imaging Probes. Angew. Chem. Int. Ed. 2015, 54, 5981–5984. [Google Scholar] [CrossRef] [PubMed]
- Bolte, S.; Cordelieres, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Griesbeck, S.; Zhang, Z.; Gutmann, M.; Lühmann, T.; Edkins, R.M.; Clermont, G.; Lazar, A.N.; Haehnel, M.; Edkins, K.; Eichhorn, A.; et al. Water-Soluble Triarylborane Chromophores for One- and Two-Photon Excited Fluorescence Imaging of Mitochondria in Cells. Chem. Eur. J. 2016, 22, 14701–14706. [Google Scholar] [CrossRef]
- Lau, Y.H.; Spring, D.R. Efficient synthesis of Fmoc-protected azido amino acids. Synlett 2011, 13, 1917–1919. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Xu, W.; Liu, S.; Sun, H.; Zhao, X.; Zhao, Q.Q.; Sun, S.; Cheng, S.; Ma, T.; Zhoua, L.; Huang, W. FRET-based probe for fluoride based on a phosphorescent iridium(iii) complex containing triarylboron groups. J. Mater. Chem. 2011, 21, 7572–7581. [Google Scholar] [CrossRef]
- Jewett, J.C.; Sletten, E.M.; Bertozzi, C.R. Rapid Cu-Free Click Chemistry with Readily Synthesized Biarylazacyclooctynones. J. Am. Chem. Soc. 2010, 132, 3688–3690. [Google Scholar] [CrossRef]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jurković, M.; Ferger, M.; Drašković, I.; Marder, T.B.; Piantanida, I. Triarylborane-“Click” Fluorescent Tag for Orthogonal Amino Acid Labelling, Interactions with DNA, Protein, and Cyclodextrins. Pharmaceuticals 2023, 16, 1208. https://doi.org/10.3390/ph16091208
Jurković M, Ferger M, Drašković I, Marder TB, Piantanida I. Triarylborane-“Click” Fluorescent Tag for Orthogonal Amino Acid Labelling, Interactions with DNA, Protein, and Cyclodextrins. Pharmaceuticals. 2023; 16(9):1208. https://doi.org/10.3390/ph16091208
Chicago/Turabian StyleJurković, Marta, Matthias Ferger, Isabela Drašković, Todd B. Marder, and Ivo Piantanida. 2023. "Triarylborane-“Click” Fluorescent Tag for Orthogonal Amino Acid Labelling, Interactions with DNA, Protein, and Cyclodextrins" Pharmaceuticals 16, no. 9: 1208. https://doi.org/10.3390/ph16091208
APA StyleJurković, M., Ferger, M., Drašković, I., Marder, T. B., & Piantanida, I. (2023). Triarylborane-“Click” Fluorescent Tag for Orthogonal Amino Acid Labelling, Interactions with DNA, Protein, and Cyclodextrins. Pharmaceuticals, 16(9), 1208. https://doi.org/10.3390/ph16091208