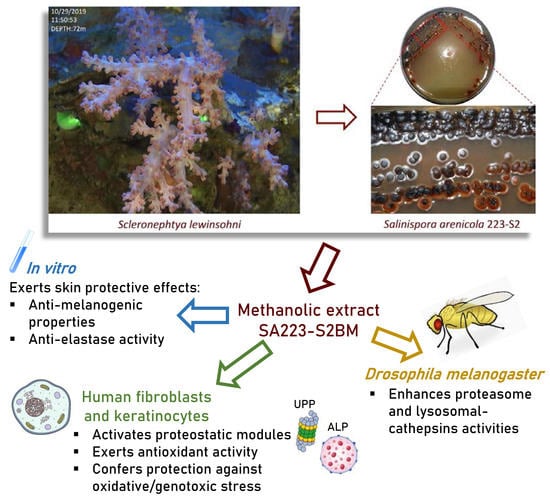
Isolation of an Extract from the Soft Coral Symbiotic Microorganism Salinispora arenicola Exerting Cytoprotective and Anti-Aging Effects
Abstract
:1. Introduction
2. Materials and Methods
2.1. SA223-S2BM Extract’s Origin and Isolation
2.1.1. Coral Collection
2.1.2. Strain Isolation and Identification
2.1.3. Phylogeny Investigation
2.1.4. Cultivation Conditions
2.2. Elastase and Tyrosinase Inhibitory Activities
2.3. Cell Lines, Cell Culture Conditions and Cell Viability Assay
2.4. Drosophila Lines and Longevity Assay
2.5. Measurement of Reactive Oxygen Species (ROS)
2.6. RNA Extraction, cDNA Synthesis and Quantitative Real-Time PCR (Q-RT-PCR) Analysis
2.7. Immunoblotting Analysis and Measurement of Proteasome and Cathepsins Activity
2.8. Antibodies Used
2.9. Statistical Analyses
3. Results
3.1. Identification of the Actinomycete Associated with the Soft Coral S. lewinsohni
3.2. The Extract SA223-S2BM Shows no Toxicity in Human Cells and Activates Key Components of the PN
3.3. Treatment of Flies with the Extract SA223-S2BM Induces Cell Proteostatic Modules Activity
3.4. The Extract SA223-S2BM Exhibits Anti-Melanogenic and Anti-Elastase Properties
3.5. Incubation of Human Cells with the Extract SA223-S2BM Activates Antioxidant Responses and Protects Cells from Oxidative and Genotoxic Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Argyropoulou, A.; Aligiannis, N.; Trougakos, I.P.; Skaltsounis, A.-L. Natural compounds with anti-ageing activity. Nat. Prod. Rep. 2013, 30, 1412–1437. [Google Scholar] [CrossRef]
- Riera, C.E.; Merkwirth, C.; De Magalhaes-Filho, C.D.; Dillin, A. Signaling Networks Determining Life Span. Annu. Rev. Biochem. 2016, 85, 35–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, H.; Finkel, T. Key proteins and pathways that regulate lifespan. J. Biol. Chem. 2017, 292, 6452–6460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trougakos, I.P.; Sesti, F.; Tsakiri, E.; Gorgoulis, V.G. Non-enzymatic post-translational protein modifications and proteostasis network deregulation in carcinogenesis. J. Proteom. 2013, 92, 274–298. [Google Scholar] [CrossRef]
- Niforou, K.; Cheimonidou, C.; Trougakos, I.P. Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2014, 2, 323–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsakiri, E.N.; Trougakos, I.P. The Amazing Ubiquitin-Proteasome System: Structural Components and Implication in Aging. Int. Rev. Cell Mol. Biol. 2015, 314, 171–237. [Google Scholar] [CrossRef] [PubMed]
- Sala, A.J.; Bott, L.C.; Morimoto, R.I. Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol. 2017, 216, 1231–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaushik, S.; Cuervo, A.M. Proteostasis and aging. Nat. Med. 2015, 21, 1406–1415. [Google Scholar] [CrossRef]
- Klaips, C.L.; Jayaraj, G.G.; Hartl, F.U. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 2017, 217, 51–63. [Google Scholar] [CrossRef]
- Labbadia, J.; Morimoto, R.I. The Biology of Proteostasis in Aging and Disease. Annu. Rev. Biochem. 2015, 84, 435–464. [Google Scholar] [CrossRef] [Green Version]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; Kasturi, P.; Hartl, F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Botbol, Y.; Macian, F.; Cuervo, A.M. Autophagy and disease: Always two sides to a problem. J. Pathol. 2011, 226, 255–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balch, W.E.; Morimoto, R.I.; Dillin, A.; Kelly, J.W. Adapting Proteostasis for Disease Intervention. Science 2008, 319, 916–919. [Google Scholar] [CrossRef] [Green Version]
- Powers, E.T.; Morimoto, R.I.; Dillin, A.; Kelly, J.W.; Balch, W.E. Biological and Chemical Approaches to Diseases of Proteostasis Deficiency. Annu. Rev. Biochem. 2009, 78, 959–991. [Google Scholar] [CrossRef] [Green Version]
- Calamini, B.; Morimoto, R.I. Protein Homeostasis as a Therapeutic Target for Diseases of Protein Conformation. Curr. Top. Med. Chem. 2013, 12, 2623–2640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sklirou, A.D.; Papanagnou, E.-D.; Fokialakis, N.; Trougakos, I.P. Cancer chemoprevention via activation of proteostatic modules. Cancer Lett. 2018, 413, 110–121. [Google Scholar] [CrossRef]
- Zhang, S.; Duan, E. Fighting against Skin Aging: The Way from Bench to Bedside. Cell Transplant. 2018, 27, 729–738. [Google Scholar] [CrossRef]
- Yasin, Z.A.M.; Ibrahim, F.; Rashid, N.N.; Razif, M.F.M.; Yusof, R.; Yasin, F.I.Z.A.M. The Importance of Some Plant Extracts as Skin Anti-aging Resources: A Review. Curr. Pharm. Biotechnol. 2018, 18, 864–876. [Google Scholar] [CrossRef]
- Bae-Harboe, Y.S.C.; Park, H.Y. Tyrosinase: A central regulatory protein for cutaneous pigmentation. J. Investig. Dermatol. 2012, 132, 2678–2680. [Google Scholar] [CrossRef] [Green Version]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V. Skin whitening agents: Medicinal chemistry perspective of tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2017, 32, 403–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, Y.; Zhan, Q.; Du, S.; Ding, Y.; Fang, B.; Du, W.; Wu, Q.; Yu, H.; Li, L.; Huang, W. Catalysis-based specific detection and inhibition of tyrosinase and their application. J. Pharm. Anal. 2020, 10, 414–425. [Google Scholar] [CrossRef]
- Decorte, B.L. Underexplored Opportunities for Natural Products in Drug Discovery. J. Med. Chem. 2016, 59, 9295–9304. [Google Scholar] [CrossRef] [PubMed]
- Beutler, J.A. Natural Products as a Foundation for Drug Discovery. Curr. Protoc. Pharmacol. 2019, 86, 9.11.1–9.11.21. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, W.; Dong, X.; Fu, C.; Yuan, J.; Xu, M.; Liang, Z.; Qiu, C.; Xu, C. A natural product solution to aging and aging-associated diseases. Pharmacol. Ther. 2020, 216, 107673. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.; Reker, D.; Schneider, P.; Schneider, G. Counting on natural products for drug design. Nat. Chem. 2016, 8, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [Green Version]
- Christensen, S. Natural Products That Changed Society. Biomedicines 2021, 9, 472. [Google Scholar] [CrossRef]
- Montaser, R.; Luesch, H. Marine natural products: A new wave of drugs? Future Med. Chem. 2011, 3, 1475–1489. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Luo, D.; Luesch, H. Advances in exploring the therapeutic potential of marine natural products. Pharmacol. Res. 2019, 147, 104373. [Google Scholar] [CrossRef]
- Sang, V.T.; Dat, T.T.H.; Vinh, L.B.; Cuong, L.C.V.; Oanh, P.T.T.; Ha, H.; Kim, Y.H.; Anh, H.L.T.; Yang, S.Y. Coral and coral-associated microorganisms: A prolific source of potential bioactive natural products. Mar. Drugs 2019, 17, 468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Le Goff, G.; Adelin, E.; Cortial, S.; Servy, C.; Ouazzani, J. Application of solid-phase extraction to agar-supported fermentation. Bioprocess Biosyst. Eng. 2012, 36, 1285–1290. [Google Scholar] [CrossRef]
- Hassane, C.S.; Fouillaud, M.; Le Goff, G.; Sklirou, A.D.; Boyer, J.B.; Trougakos, I.P.; Jerabek, M.; Bignon, J.; De Voogd, N.J.; Ouazzani, J.; et al. Microorganisms Associated with the Marine Sponge Scopalina hapalia: A Reservoir of Bioactive Molecules to Slow Down the Aging Process. Microorganisms 2020, 8, 1262. [Google Scholar] [CrossRef] [PubMed]
- Campos, P.E.; Herbette, G.; Chendo, C.; Clerc, P.; Tintillier, F.; de Voogd, N.J.; Papanagnou, E.-D.; Trougakos, I.P.; Jerabek, M.; Bignon, J.; et al. New Long-Chain Highly Oxygenated Polyacetylenes from the Mayotte Marine Sponge Haliclona sp. Mar. Drugs 2020, 18, 350. [Google Scholar] [CrossRef] [PubMed]
- Nikou, T.; Liaki, V.; Stathopoulos, P.; Sklirou, A.D.; Tsakiri, E.N.; Jakschitz, T.; Bonn, G.; Trougakos, I.P.; Halabalaki, M.; Skaltsounis, L.A. Comparison survey of EVOO polyphenols and exploration of healthy aging-promoting properties of oleocanthal and oleacein. Food Chem. Toxicol. 2019, 125, 403–412. [Google Scholar] [CrossRef]
- Tsakiri, E.N.; Sykiotis, G.P.; Papassideri, I.S.; Gorgoulis, V.G.; Bohmann, D.; Trougakos, I.P. Differential regulation of pro-teasome functionality in reproductive vs. somatic tissues of Drosophila during aging or oxidative stress. FASEB J. 2013, 27, 2407–2420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gumeni, S.; Evangelakou, Z.; Tsakiri, E.; Scorrano, L.; Trougakos, I.P. Functional wiring of proteostatic and mitostatic modules ensures transient organismal survival during imbalanced mitochondrial dynamics. Redox Biol. 2019, 24, 101219. [Google Scholar] [CrossRef] [PubMed]
- Ransy, C.; Vaz, C.; Lombès, A.; Bouillaud, F. Use of H2O2 to Cause Oxidative Stress, the Catalase Issue. Int. J. Mol. Sci. 2020, 21, 9149. [Google Scholar] [CrossRef]
- Sklirou, A.D.; Gaboriaud-Kolar, N.; Papassideri, I.; Skaltsounis, A.L.; Trougakos, I.P. 6-bromo-indirubin-3′-oxime (6BIO), a Glycogen synthase kinase-3β inhibitor, activates cytoprotective cellular modules and suppresses cellular senescence-mediated bio-molecular damage in human fibroblasts. Sci. Rep. 2017, 7, 11713. [Google Scholar] [CrossRef] [Green Version]
- Sklirou, A.D.; Ralli, M.; Dominguez, M.; Papassideri, I.; Skaltsounis, A.-L.; Trougakos, I.P. Hexapeptide-11 is a novel modulator of the proteostasis network in human diploid fibroblasts. Redox Biol. 2015, 5, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Rasband, W.S. ImageJ U.S.; 1.48v/Java 1.6.0-20 (32-bit); National Institutes of Health: Bethesda, MD, USA, 2014. [Google Scholar]
- Papanagnou, E.-D.; Terpos, E.; Kastritis, E.; Papassideri, I.S.; Tsitsilonis, O.E.; Dimopoulos, M.A.; Trougakos, I.P. Molecular responses to therapeutic proteasome inhibitors in multiple myeloma patients are donor-, cell type- and drug-dependent. Oncotarget 2018, 9, 17797–17809. [Google Scholar] [CrossRef] [Green Version]
- Tsakiri, E.N.; Gumeni, S.; Vougas, K.; Pendin, D.; Papassideri, I.; Daga, A.; Gorgoulis, V.; Juhász, G.; Scorrano, L.; Trougakos, I.P. Proteasome dysfunction induces excessive proteome instability and loss of mitostasis that can be mitigated by enhancing mitochondrial fusion or autophagy. Autophagy 2019, 15, 1757–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsakiri, E.N.; Gaboriaud-Kolar, N.; Iliaki, K.K.; Tchoumtchoua, J.; Papanagnou, E.-D.; Chatzigeorgiou, S.; Tallas, K.D.; Mikros, E.; Halabalaki, M.; Skaltsounis, A.L.; et al. The Indirubin Derivative 6-Bromoindirubin-3′-Oxime Activates Proteostatic Modules, Reprograms Cellular Bioenergetic Pathways, and Exerts Antiaging Effects. Antioxid. Redox Signal. 2017, 27, 1027–1047. [Google Scholar] [CrossRef]
- Sarkar, S.; Ravikumar, B.; Floto, R.A.; Rubinsztein, D.C. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009, 16, 46–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benayahu, Y. Octocorals of the Indo-Pacific. In Mesophotic Coral Ecosystems; Loya, Y., Puglise, K., Bridge, T., Eds.; Coral Reefs of the World; Springer: Cham, Switzerland, 2019; Volume 12, pp. 709–728. [Google Scholar]
- Millán-Aguiñaga, N.; Chavarria, K.L.; Ugalde, J.A.; Letzel, A.-C.; Rouse, G.; Jensen, P.R. Phylogenomic Insight into Salinispora (Bacteria, Actinobacteria) Species Designations. Sci. Rep. 2017, 7, 3564. [Google Scholar] [CrossRef] [Green Version]
- Kruszewski, M.; Szumiel, I. Cytotoxic and mutagenic effects of hydrogen peroxide in murine L5178Y sublines. Acta Biochim. Pol. 1993, 40, 42–45. [Google Scholar] [CrossRef]
- Campisi, J.; d’Adda di Fagagna, F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007, 8, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, R.; Hamidullah, H.M.; Pandey, P.; Maheshwari, M.; Singh, L.R.; Siddiqui, M.Q.; Konwar, R.; Sashidhara, K.V.; Sarkar, J. Coumarin-chalcone hybrid instigates DNA damage by minor groove binding and stabilizes p53 through post translational modifications. Sci. Rep. 2017, 7, 45287. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef]
- Bielak-Zmijewska, A.; Mosieniak, G. Cellular Senescence in Ageing, Age-Related Disease and Longevity. Curr. Vasc. Pharmacol. 2013, 12, 698–706. [Google Scholar] [CrossRef]
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; D’Adda Di Fagagna, F. Cellular senescence in ageing: From mechanisms to ther-apeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021, 22, 75–95. [Google Scholar] [CrossRef]
- Taylor, R.; Dillin, A. Aging as an Event of Proteostasis Collapse. Cold Spring Harb. Perspect. Biol. 2011, 3, a004440. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, R.I.; Cuervo, A.M. Proteostasis and the Aging Proteome in Health and Disease. J. Gerontol. Ser. A Boil. Sci. Med. Sci. 2014, 69, S33–S38. [Google Scholar] [CrossRef] [Green Version]
- Maldonado, L.A.; Fenical, W.; Jensen, P.R.; Kauffman, C.A.; Mincer, T.J.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromono-sporaceae. Int. J. Syst. Evol. Microbiol. 2005, 55, 1759–1766. [Google Scholar] [CrossRef] [Green Version]
- Fontana, L.; Partridge, L.; Longo, V.D. Dietary restriction, growth factors and aging: From yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef] [Green Version]
- Wedel, S.; Manola, M.; Cavinato, M.; Trougakos, I.P.; Jansen-Dürr, P. Targeting protein quality control mechanisms by natural products to promote healthy ageing. Molecules 2018, 23, 1219. [Google Scholar] [CrossRef] [Green Version]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2008, 8, 69–85. [Google Scholar] [CrossRef]
- Vasilopoulou, M.; Ioannou, E.; Roussis, V.; Chondrogianni, N. Modulation of the ubiquitin-proteasome system by marine natural products. Redox Biol. 2021, 41, 101897. [Google Scholar] [CrossRef] [PubMed]
- Royer, M.; Prado, M.; García-Pérez, M.E.; Diouf, P.N.; Stevanovic, T. Study of nutraceutical, nutricosmetics and cosmeceutical potentials of polyphenolic bark extracts from Canadian forest species. PharmaNutrition 2013, 1, 158–167. [Google Scholar] [CrossRef]
- Tundis, R.; Loizzo, M.R.; Bonesi, M.; Menichini, F. Potential Role of Natural Compounds Against Skin Aging. Curr. Med. Chem. 2015, 22, 1515–1538. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.; Vieira, H.; Gaspar, H.; Santos, S. Marketed Marine Natural Products in the Pharmaceutical and Cosmeceutical Industries: Tips for Success. Mar. Drugs 2014, 12, 1066–1101. [Google Scholar] [CrossRef] [Green Version]
- Corinaldesi, C.; Barone, G.; Marcellini, F.; Dell’Anno, A.; Danovaro, R. Marine Microbial-Derived Molecules and Their Potential Use in Cosmeceutical and Cosmetic Products. Mar. Drugs 2017, 15, 118. [Google Scholar] [CrossRef]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A Highly Cytotoxic Proteasome Inhibitor from a Novel Microbial Source, a Marine Bacterium of the New Genus Salinospora. Angew. Chem. Int. Ed. 2003, 42, 355–357. [Google Scholar] [CrossRef]
- Niewerth, D.; Jansen, G.; Riethoff, L.F.V.; van Meerloo, J.; Kale, A.J.; Moore, B.S.; Assaraf, Y.G.; Anderl, J.L.; Zweegman, S.; Kaspers, G.J.L.; et al. Antileukemic Activity and Mechanism of Drug Resistance to the Marine Salinispora tropica Proteasome Inhibitor Salinosporamide A (Marizomib). Mol. Pharmacol. 2014, 86, 12–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asolkar, R.N.; Kirkland, T.N.; Jensen, P.R.; Fenical, W. Arenimycin, an antibiotic effective against rifampin- and methicillin-resistant Staphylococcus aureus from the marine actinomycete Salinispora arenicola. J. Antibiot. 2010, 63, 37–39. [Google Scholar] [CrossRef] [Green Version]
- Williams, P.G.; Asolkar, R.N.; Kondratyuk, T.; Pezzuto, J.M.; Jensen, P.R.; Fenical, W. Saliniketals A and B, Bicyclic Polyketides from the Marine Actinomycete Salinispora arenicola. J. Nat. Prod. 2006, 70, 83–88. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Louka, X.P.; Sklirou, A.D.; Le Goff, G.; Lopes, P.; Papanagnou, E.-D.; Manola, M.S.; Benayahu, Y.; Ouazzani, J.; Trougakos, I.P. Isolation of an Extract from the Soft Coral Symbiotic Microorganism Salinispora arenicola Exerting Cytoprotective and Anti-Aging Effects. Curr. Issues Mol. Biol. 2022, 44, 14-30. https://doi.org/10.3390/cimb44010002
Louka XP, Sklirou AD, Le Goff G, Lopes P, Papanagnou E-D, Manola MS, Benayahu Y, Ouazzani J, Trougakos IP. Isolation of an Extract from the Soft Coral Symbiotic Microorganism Salinispora arenicola Exerting Cytoprotective and Anti-Aging Effects. Current Issues in Molecular Biology. 2022; 44(1):14-30. https://doi.org/10.3390/cimb44010002
Chicago/Turabian StyleLouka, Xanthippi P., Aimilia D. Sklirou, Géraldine Le Goff, Philippe Lopes, Eleni-Dimitra Papanagnou, Maria S. Manola, Yehuda Benayahu, Jamal Ouazzani, and Ioannis P. Trougakos. 2022. "Isolation of an Extract from the Soft Coral Symbiotic Microorganism Salinispora arenicola Exerting Cytoprotective and Anti-Aging Effects" Current Issues in Molecular Biology 44, no. 1: 14-30. https://doi.org/10.3390/cimb44010002
APA StyleLouka, X. P., Sklirou, A. D., Le Goff, G., Lopes, P., Papanagnou, E. -D., Manola, M. S., Benayahu, Y., Ouazzani, J., & Trougakos, I. P. (2022). Isolation of an Extract from the Soft Coral Symbiotic Microorganism Salinispora arenicola Exerting Cytoprotective and Anti-Aging Effects. Current Issues in Molecular Biology, 44(1), 14-30. https://doi.org/10.3390/cimb44010002