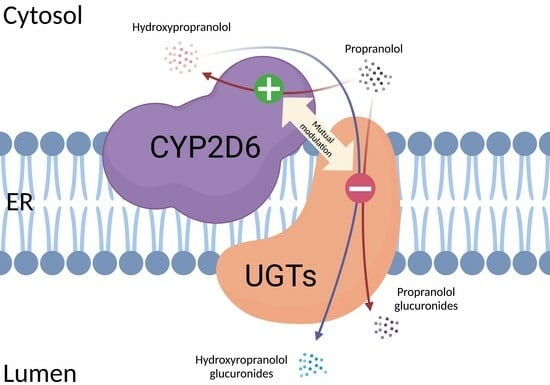
Mutual Modulation of the Activities of Human CYP2D6 and Four UGTs during the Metabolism of Propranolol
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. pH Extraction Assay
2.3. Metabolite Quantitation
2.4. qPCR Analysis of CYP2D6 in Diploid Yeast Strains
2.5. Yeast Strains, Preparation, and Long-Term Storage of Enzyme Bags
2.6. Metabolization of Propranolol in Diploid Yeast Enzyme Bags
2.7. LC-MS Analysis
3. Results
3.1. Optimization of the Biotransformation Protocol
3.1.1. Liquid–Liquid Extraction with Ethyl Acetate
3.1.2. Long-Term Storage for Enzyme Bags
3.1.3. Optimization of the CYP Reaction Time for Diploid Yeast
3.2. Influence of Four UGTs on CYP2D6 Activity
3.3. UGT Activities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meyer, U.A. Overview of enzymes of drug metabolism. J. Pharmacokinet. Biopharm. 1996, 24, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Josephy, P.D.; Guengerich, F.P.; Miners, J.O. “Phase I and Phase II” drug metabolism: Terminology that we should phase out? Drug Metab. Rev. 2005, 37, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Intersection of the Roles of Cytochrome P450 Enzymes with Xenobiotic and Endogenous Substrates: Relevance to Toxicity and Drug Interactions. Chem. Res. Toxicol. 2017, 30, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Hannemann, F.; Bichet, A.; Ewen, K.M.; Bernhardt, R. Cytochrome P450 systems-biological variations of electron transport chains. Biochim. Biophys. Acta 2007, 1770, 330–344. [Google Scholar] [CrossRef]
- Di, L.; Kerns, E.H. Drug-Like Properties: Concepts, Structure Design and Methods from ADME to Toxicity Optimization; Academic Press: Waltham, MA, USA, 2016. [Google Scholar]
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Guillemette, C.; Levesque, E.; Rouleau, M. Pharmacogenomics of human uridine diphospho-glucuronosyltransferases and clinical implications. Clin. Pharmacol. Ther. 2014, 96, 324–339. [Google Scholar] [CrossRef]
- Miners, J.O.; Rowland, A.; Novak, J.J.; Lapham, K.; Goosen, T.C. Evidence-based strategies for the characterisation of human drug and chemical glucuronidation in vitro and UDP-glucuronosyltransferase reaction phenotyping. Pharmacol. Ther. 2021, 218, 107689. [Google Scholar] [CrossRef]
- Taura, K.I.; Yamada, H.; Hagino, Y.; Ishii, Y.; Mori, M.A.; Oguri, K. Interaction between cytochrome P450 and other drug-metabolizing enzymes: Evidence for an association of CYP1A1 with microsomal epoxide hydrolase and UDP-glucuronosyltransferase. Biochem. Biophys. Res. Commun. 2000, 273, 1048–1052. [Google Scholar] [CrossRef]
- Taura, K.; Naito, E.; Ishii, Y.; Mori, M.A.; Oguri, K.; Yamada, H. Cytochrome P450 1A1 (CYP1A1) inhibitor alpha-naphthoflavone interferes with UDP-glucuronosyltransferase (UGT) activity in intact but not in permeabilized hepatic microsomes from 3-methylcholanthrene-treated rats: Possible involvement of UGT-P450 interactions. Biol. Pharm. Bull. 2004, 27, 56–60. [Google Scholar] [CrossRef]
- Fremont, J.J.; Wang, R.W.; King, C.D. Coimmunoprecipitation of UDP-glucuronosyltransferase isoforms and cytochrome P450 3A4. Mol. Pharmacol. 2005, 67, 260–262. [Google Scholar] [CrossRef]
- Takeda, S.; Ishii, Y.; Iwanaga, M.; Mackenzie, P.I.; Nagata, K.; Yamazoe, Y.; Oguri, K.; Yamada, H. Modulation of UDP-glucuronosyltransferase function by cytochrome P450: Evidence for the alteration of UGT2B7-catalyzed glucuronidation of morphine by CYP3A4. Mol. Pharmacol. 2005, 67, 665–672. [Google Scholar] [CrossRef]
- Takeda, S.; Ishii, Y.; Iwanaga, M.; Nurrochmad, A.; Ito, Y.; Mackenzie, P.I.; Nagata, K.; Yamazoe, Y.; Oguri, K.; Yamada, H. Interaction of cytochrome P450 3A4 and UDP-glucuronosyltransferase 2B7: Evidence for protein-protein association and possible involvement of CYP3A4 J-helix in the interaction. Mol. Pharmacol. 2009, 75, 956–964. [Google Scholar] [CrossRef]
- Ishii, Y.; Koba, H.; Kinoshita, K.; Oizaki, T.; Iwamoto, Y.; Takeda, S.; Miyauchi, Y.; Nishimura, Y.; Egoshi, N.; Taura, F.; et al. Alteration of the function of the UDP-glucuronosyltransferase 1A subfamily by cytochrome P450 3A4: Different susceptibility for UGT isoforms and UGT1A1/7 variants. Drug Metab. Dispos. 2014, 42, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, Y.; Nagata, K.; Yamazoe, Y.; Mackenzie, P.I.; Yamada, H.; Ishii, Y. Suppression of Cytochrome P450 3A4 Function by UDP-Glucuronosyltransferase 2B7 through a Protein-Protein Interaction: Cooperative Roles of the Cytosolic Carboxyl-Terminal Domain and the Luminal Anchoring Region. Mol. Pharmacol. 2015, 88, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, Y.; Tanaka, Y.; Nagata, K.; Yamazoe, Y.; Mackenzie, P.I.; Yamada, H.; Ishii, Y. UDP-Glucuronosyltransferase (UGT)-mediated attenuations of cytochrome P450 3A4 activity: UGT isoform-dependent mechanism of suppression. Br. J. Pharmacol. 2020, 177, 1077–1089. [Google Scholar] [CrossRef]
- Dragan, C.A.; Buchheit, D.; Bischoff, D.; Ebner, T.; Bureik, M. Glucuronide Production by Whole-Cell Biotransformation Using Genetically Engineered Fission Yeast Schizosaccharomyces pombe. Drug Metab. Dispos. 2010, 38, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Machalz, D.; Zollner, A.; Sorensen, E.J.; Wolber, G.; Bureik, M. Efficient substrate screening and inhibitor testing of human CYP4Z1 using permeabilized recombinant fission yeast. Biochem. Pharmacol. 2017, 146, 174–187. [Google Scholar] [CrossRef]
- Sharma, S.S.; Sharma, S.; Zhao, J.; Bureik, M. Mutual Influence of Human Cytochrome P450 Enzymes and UDP-Glucuronosyltransferases on Their Respective Activities in Recombinant Fission Yeast. Biomedicines 2023, 11, 281. [Google Scholar] [CrossRef]
- Bureik, M.; Schiffler, B.; Hiraoka, Y.; Vogel, F.; Bernhardt, R. Functional Expression of Human Mitochondrial CYP11B2 in Fission Yeast and Identification of a New Internal Electron Transfer Protein, etp1. Biochemistry 2002, 41, 2311–2321. [Google Scholar] [CrossRef]
- Ikushiro, S.-i.; Sahara, M.; Emi, Y.; Yabusaki, Y.; Iyanagi, T. Functional co-expression of xenobiotic metabolizing enzymes, rat cytochrome P450 1A1 and UDP-glucuronosyltransferase 1A6, in yeast microsomes. Biochim. Et. Biophys. Acta (BBA)-Gen. Subj. 2004, 1672, 86–92. [Google Scholar] [CrossRef]
- Routledge, P.A.; Shand, D.G. Clinical pharmacokinetics of propranolol. Clin. Pharmacokinet. 1979, 4, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Brosen, K. Drug interactions and the cytochrome P450 system. The role of cytochrome P450 1A2. Clin. Pharmacokinet. 1995, 29 (Suppl. S1), 20–25. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, K.; Echizen, H.; Chiba, K.; Tani, M.; Ishizaki, T. Identification of human CYP isoforms involved in the metabolism of propranolol enantiomers--N-desisopropylation is mediated mainly by CYP1A2. Br. J. Clin. Pharmacol. 1995, 39, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yao, T.W.; Zeng, S. Chiral reversed phase high-performance liquid chromatography for determining propranolol enantiomers in transgenic Chinese hamster CHL cell lines expressing human cytochrome P450. J. Biochem. Biophys. Methods 2002, 54, 369–376. [Google Scholar] [CrossRef]
- Bichara, N.; Ching, M.S.; Blake, C.L.; Ghabrial, H.; Smallwood, R.A. Propranolol hydroxylation and N-desisopropylation by cytochrome P4502D6: Studies using the yeast-expressed enzyme and NADPH/O2 and cumene hydroperoxide-supported reactions. Drug Metab. Dispos. 1996, 24, 112–118. [Google Scholar] [PubMed]
- Masubuchi, Y.; Hosokawa, S.; Horie, T.; Suzuki, T.; Ohmori, S.; Kitada, M.; Narimatsu, S. Cytochrome P450 isozymes involved in propranolol metabolism in human liver microsomes. The role of CYP2D6 as ring-hydroxylase and CYP1A2 as N-desisopropylase. Drug Metab. Dispos. 1994, 22, 909–915. [Google Scholar]
- Thompson, J.A.; Hull, J.E.; Norris, K.J. Glucuronidation of propranolol and 4′-hydroxypropranolol. Substrate specificity and stereoselectivity of rat liver microsomal glucuronyltransferases. Drug Metab. Dispos. 1981, 9, 466–471. [Google Scholar] [PubMed]
- Johnson, J.A.; Herring, V.L.; Wolfe, M.S.; Relling, M.V. CYP1A2 and CYP2D6 4-Hydroxylate Propranolol and Both Reactions Exhibit Racial Differences. J. Pharmacol. Exp. Ther. 2000, 294, 1099–1105. [Google Scholar]
- Yang, F.; Liu, S.; Wolber, G.; Bureik, M.; Parr, M.K. Complete Reaction Phenotyping of Propranolol and 4-Hydroxypropranolol with the 19 Enzymes of the Human UGT1 and UGT2 Families. Int. J. Mol. Sci. 2022, 23, 7476. [Google Scholar] [CrossRef]
- Harps, L.C.; Schipperges, S.; Bredendiek, F.; Wuest, B.; Borowiak, A.; Parr, M.K. Two dimensional chromatography mass spectrometry: Quantitation of chiral shifts in metabolism of propranolol in bioanalysis. J. Chromatogr. A 2020, 1617, 460828. [Google Scholar] [CrossRef]
- Oatis Jr, J.E.; Russell, M.P.; Knapp, D.R.; Walle, T. Ring-Hydroxylated Propranolol: Synthesis and beta-Receptor Antagonist and Vasodilating Activities of the Seven Isomers. J. Med. Chem. 1981, 24, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Durairaj, P.; Bureik, M. Rapid and convenient biotransformation procedure for human drug metabolizing enzymes using permeabilized fission yeast cells. Anal. Biochem. 2020, 607, 113704. [Google Scholar] [CrossRef] [PubMed]
- Alfa, C.; Fantes, P.; Hyams, J.; McLeod, M.; Warbrick, E. Experiments with Fission Yeast. A Laboratory Course Manual; Cold Spring Harbor Press: Cold Spring Harbor, NY, USA, 1993. [Google Scholar]
- Yamashita, T.; Nishimura, I.; Nakamura, T.; Fukami, T. A system for LogD screening of new drug candidates using a water-plug injection method and automated liquid handler. JALA J. Assoc. Lab. Autom. 2009, 14, 76–81. [Google Scholar] [CrossRef]
- Taylor, C.; Crosby, I.; Yip, V.; Maguire, P.; Pirmohamed, M.; Turner, R.M. A Review of the Important Role of CYP2D6 in Pharmacogenomics. Genes. 2020, 11, 1295. [Google Scholar] [CrossRef]
- Walle, T.; Oatis, J.E.; Walle, U.K.; Knapp, D.R. New ring-hydroxylated metabolites of propranolol: Species differences and stereospecific 7-hydroxylation. Drug Metab. Dispos. 1982, 10, 122–127. [Google Scholar]
- Walle, T.; Walle, U.K.; Olanoff, L.S. Quantitative account of propranolol metabolism in urine of normal man. Drug Metab. Dispos. Biol. Fate Chem. 1985, 13, 204–209. [Google Scholar]
- Distlerath, L.M.; Reilly, P.E.; Martin, M.V.; Davis, G.G.; Wilkinson, G.R.; Guengerich, F.P. Purification and characterization of the human liver cytochromes P-450 involved in debrisoquine 4-hydroxylation and phenacetin O-deethylation, two prototypes for genetic polymorphism in oxidative drug metabolism. J. Biol. Chem. 1985, 260, 9057–9067. [Google Scholar] [CrossRef]
- Herring, V.L.; Johnson, J.A. Direct high-performance liquid chromatographic determination in urine of the enantiomers of propranolol and its major basic metabolite 4-hydroxypropranolol. J. Chromatogr. B Biomed. Sci. Appl. 1993, 612, 215–221. [Google Scholar] [CrossRef]
- Fitzgerald, J.; O’DONNELL, S.R. Pharmacology of 4-hydroxypropranolol, a metabolite of propranolol. Br. J. Pharmacol. 1971, 43, 222–235. [Google Scholar] [CrossRef]
- Pritchard, J.F.; Schneck, D.W.; Hayes Jr, A.H. Determination of propranolol and six metabolites in human urine by high-pressure liquid chromatography. J. Chromatogr. B Biomed. Sci. Appl. 1979, 162, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Ciotti, M.; Marrone, A.; Potter, C.; Owens, I.S. Genetic polymorphism in the human UGT1A6 (planar phenol) UDP-glucuronosyltransferase: Pharmacological implications. Pharmacogenetics 1997, 7, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Krishnaswamy, S.; Hao, Q.; Al-Rohaimi, A.; Hesse, L.M.; von Moltke, L.L.; Greenblatt, D.J.; Court, M.H. UDP glucuronosyltransferase (UGT) 1A6 pharmacogenetics: II. Functional impact of the three most common nonsynonymous UGT1A6 polymorphisms (S7A, T181A, and R184S). J. Pharmacol. Exp. Ther. 2005, 313, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Villeneuve, L.; Girard, H.; Fortier, L.C.; Gagne, J.F.; Guillemette, C. Novel functional polymorphisms in the UGT1A7 and UGT1A9 glucuronidating enzymes in Caucasian and African-American subjects and their impact on the metabolism of 7-ethyl-10-hydroxycamptothecin and flavopiridol anticancer drugs. J. Pharmacol. Exp. Ther. 2003, 307, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Girard, H.; Court, M.H.; Bernard, O.; Fortier, L.C.; Villeneuve, L.; Hao, Q.; Greenblatt, D.J.; von Moltke, L.L.; Perussed, L.; Guillemette, C. Identification of common polymorphisms in the promoter of the UGT1A9 gene: Evidence that UGT1A9 protein and activity levels are strongly genetically controlled in the liver. Pharmacogenetics 2004, 14, 501–515. [Google Scholar] [CrossRef]
- Bai, S.A.; Walle, T. Isolation, purification, and structure identification of glucuronic acid conjugates of propranolol and alprenolol and their ring-hydroxylated metabolites. Drug Metab. Dispos. 1984, 12, 749–754. [Google Scholar]
- Walle, T.; Conradi, E.C.; Walle, U.K.; Fagan, T.C.; Gaffney, T.E. 4-Hydroxypropranolol and its glucuronide after single and long-term doses of propranolol. Clin. Pharmacol. Ther. 1980, 27, 22–31. [Google Scholar] [CrossRef]







| Gene | Forward Primer | Reverse Primer | Product Size |
|---|---|---|---|
| act1 | 5′-GTTATGTCTGGTGGTACCACT-3′ | 5′-GATCCACCAATCCAGACAGA-3′ | 140 bp |
| CYP2D6 | 5′-AGGTCCATTGCCACTTCCAG-3′ | 5′-CCGAACAGCTGCTAGACCAT-3′ | 160 bp |
| Strain | Expressed Protein | Genotype | Reference |
|---|---|---|---|
| DB24 | UGT1A7 | h- ura4-D18 leu1::pCAD1UGT1A7 | [17] |
| DB25 | UGT1A8 | h- ura4-D18 leu1::pCAD1UGT1A8 | [17] |
| CAD200 | UGT1A9 | h- ura4-D18 leu1::pCAD1UGT1A9 | [17] |
| DB3 | UGT2A1 | h- ura4-D18 leu1::pCAD1UGT2A1 | [17] |
| SAN300 | hCPR, CYP2D6 | h+/h− ade6-M210/ade6-M216 ura4-D18/ura4-D18 his3.Δ1/his3.Δ1 leu1::pCAD1-CPR/leu1::pCAD1/pREP1-CYP2D6 | [19] |
| SAN306 | hCPR, CYP2D6, UGT1A7 | h+/h− ade6-M210/ade6-M216 ura4-D18/ura4-D18 his3.Δ1/his3.Δ1 leu1::pCAD1-CPR/leu1::pCAD1-UGT1A7/pREP1-CYP2D6 | [19] |
| SAN307 | hCPR, CYP2D6, UGT1A8 | h+/h− ade6-M210/ade6-M216 ura4-D18/ura4-D18 his3.Δ1/his3.Δ1 leu1::pCAD1-CPR/leu1::pCAD1-UGT1A8/pREP1-CYP2D6 | [19] |
| SAN308 | hCPR, CYP2D6, UGT1A9 | h+/h− ade6-M210/ade6-M216 ura4-D18/ura4-D18 his3.Δ1/his3.Δ1 leu1::pCAD1-CPR/leu1::pCAD1-UGT1A9/pREP1-CYP2D6 | [19] |
| SAN310 | hCPR, CYP2D6, UGT2A1 | h+/h− ade6-M210/ade6-M216 ura4-D18/ura4-D18 his3.Δ1/his3.Δ1 leu1::pCAD1-CPR/leu1::pCAD1-UGT2A1/pREP1-CYP2D6 | [19] |
| Analytes | Precursor Ions (m/z) | Product Ions (m/z) | CE (V) | ESI |
|---|---|---|---|---|
| Propranolol glucuronide | 436 | 258 | 12 | + |
| 436 | 183 | 16 | + | |
| 436 | 116 | 28 | + | |
| 4-/5-Hydroxypropranolol glucuronide | 452 | 276 | 12 | + |
| 452 | 116 | 28 | + | |
| 452 | 72 | 44 | + | |
| 4-/5-Hydroxypropranolol | 276 | 116 | 12 | + |
| 276 | 72 | 16 | + | |
| 276 | 58 | 44 | + | |
| 4-Methoxypropranolol | 290 | 187 | 12 | + |
| 290 | 116 | 16 | + | |
| 290 | 72 | 44 | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Sharma, S.S.; Bureik, M.; Parr, M.K. Mutual Modulation of the Activities of Human CYP2D6 and Four UGTs during the Metabolism of Propranolol. Curr. Issues Mol. Biol. 2023, 45, 7130-7146. https://doi.org/10.3390/cimb45090451
Yang F, Sharma SS, Bureik M, Parr MK. Mutual Modulation of the Activities of Human CYP2D6 and Four UGTs during the Metabolism of Propranolol. Current Issues in Molecular Biology. 2023; 45(9):7130-7146. https://doi.org/10.3390/cimb45090451
Chicago/Turabian StyleYang, Fan, Sangeeta Shrestha Sharma, Matthias Bureik, and Maria Kristina Parr. 2023. "Mutual Modulation of the Activities of Human CYP2D6 and Four UGTs during the Metabolism of Propranolol" Current Issues in Molecular Biology 45, no. 9: 7130-7146. https://doi.org/10.3390/cimb45090451
APA StyleYang, F., Sharma, S. S., Bureik, M., & Parr, M. K. (2023). Mutual Modulation of the Activities of Human CYP2D6 and Four UGTs during the Metabolism of Propranolol. Current Issues in Molecular Biology, 45(9), 7130-7146. https://doi.org/10.3390/cimb45090451