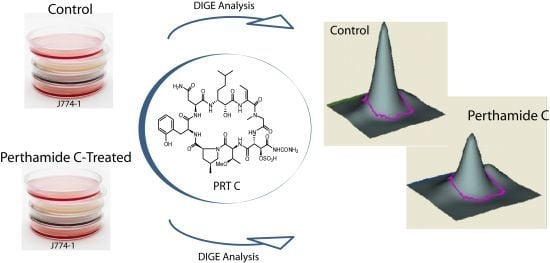
Differential in Gel Electrophoresis (DIGE) Comparative Proteomic Analysis of Macrophages Cell Cultures in Response to Perthamide C Treatment
Abstract
:1. Introduction

2. Results and Discussion

| Spot | Proteins | Ratio | LC-MSMS | |||||
|---|---|---|---|---|---|---|---|---|
| Accession No. | Mw (Da) | pI | Description | Coverage | Mascot Score | Peptide No. | ||
| 1684 | ALDOA_MOUSE | 39787 | 5.29 | Fructose-bisphosphate aldolase A | 5.13 | 9% | 120 | 6 (3) |
| 1752 | TPIS_MOUSE | 27038 | 6.05 | Triosephosphate isomerase | 2.99 | 17% | 334 | 9 (9) |
| 920 | HS90B_MOUSE | 83615 | 5.88 | Heat shock protein HSP 90-beta | 2.95 | 7% | 400 | 11 (9) |
| 1086 | TBB5_MOUSE | 50095 | 8.97 | Tubulin beta-5 chain | 2.91 | 12% | 240 | 14 (9) |
| 1754 | HPRT_MOUSE | 24783 | 5.29 | Hypoxanthine-guanine phosphoribosyl transferase | 2.76 | 14% | 152 | 8 (5) |
| 969 | MOES_MOUSE | 67839 | 4.94 | Moesin | 2.76 | 8% | 147 | 9 (4) |
| 2096 | ACTB_MOUSE | 42052 | 6.67 | Actin, cytoplasmic 1 | 2.64 | 36% | 139 | 4 (3) |
| 1965 | 1433Z_MOUSE | 27925 | 6.90 | 14-3-3 protein zeta/delta | 2.53 | 42% | 163 | 8 (4) |
| 1190 | ACTB_MOUSE | 42052 | 6.22 | Actin, cytoplasmic 1 | 2.51 | 20% | 321 | 12 (9) |
| 1698 | PGAM_MOUSE | 28928 | 6.67 | Phosphoglycerate mutase 1 | 2.27 | 20% | 188 | 12 (7) |
| 1828 | ACTB_MOUSE | 42052 | 4.73 | Actin, cytoplasmic 1 | 2.24 | 25% | 92 | 4 (3) |
| 1569 | ACTB_MOUSE | 42052 | 4.97 | Actin, cytoplasmic 1 | 2.22 | 15% | 109 | 3 (3) |
| 1533 | PDIA3_MOUSE | 57099 | 8.31 | Protein disulfide-isomerase A3 | 2.13 | 19% | 99 | 5 (2) |
| 1542 | ROA2_MOUSE | 37437 | 6.21 | Ribonucleoproteins A2/B1 | 2.08 | 44% | 93 | 2 (2) |
| 1058 | COR1A_MOUSE | 51641 | 4.78 | Coronin-1A | 2.02 | 33% | 77 | 6 (3) |
| 337 | GELS_MOUSE | 86287 | 8.02 | Gelsolin | 1.84 | 27% | 200 | 8 (6) |
| 676 | LYZ1_MOUSE | 17240 | 9.55 | Lysozyme C-1 | 2.81 | 8% | 41 | 1 (1) |
| 692 | ATPB_MOUSE | 56265 | 5.19 | ATP synthase subunit beta | 2.72 | 42% | 365 | 18 (16) |
| 477 | HNRPK_MOUSE | 51230 | 5.39 | Heterogeneous nuclear ribonucleoprotein K | 2.64 | 19% | 177 | 7 (6) |
| 476 | TCPZ_MOUSE | 58424 | 5.72 | T-complex protein 1-zeta | 2.55 | 4% | 68 | 5 (2) |
| 454 | TCPE_MOUSE | 60042 | 7.23 | T-complex protein 1-epsilon | 2.43 | 4% | 54 | 2 (1) |
| 389 | TKT_MOUSE | 68272 | 7.27 | Transketolase | 2.41 | 10% | 36 | 2 (1) |
| 541 | VIME_MOUSE | 53712 | 5.91 | Vimentin | 2.31 | 28% | 462 | 23 (16) |
| 542 | VIME_MOUSE | 53712 | 5.91 | Vimentin | 2.18 | 40% | 329 | 14 (12) |
| 441 | DPYL2_MOUSE | 62638 | 4.83 | Dihydropyrimidinase-related 2 | 2.17 | 16% | 97 | 6 (4) |
| 1117 | TALDO_MOUSE | 37534 | 6.37 | Transaldolase | 1.99 | 15% | 72 | 5 (3) |
| 561 | TCPB_MOUSE | 57783 | 6.31 | T-complex protein 1 subunit beta | 1.91 | 8% | 171 | 4 (4) |
| 1221 | RLA0_MOUSE | 34366 | 5.97 | 60S acidic ribosomal protein P0 | 1.90 | 13% | 164 | 6 (5) |
| 753 | ENOA_MOUSE | 47453 | 5.06 | Alpha-enolase | 1.86 | 29% | 110 | 6 (4) |
| 486 | UBP14_MOUSE | 56422 | 5.07 | Ubiquitin carboxyl-terminal hydrolase | 1.84 | 44% | 63 | 1 (1) |
| 1010 | RSSA_MOUSE | 32931 | 4.81 | 40S ribosomal protein SA | 1.81 | 31% | 228 | 9 (6) |
| 966 | AHSA1_MOUSE | 38321 | 6.63 | Activator of 90 kDa heat shock protein ATPase-1 | 1.81 | 10% | 29 | 2 (1) |
| 1336 | ANXA5_MOUSE | 35787 | 5.95 | Annexin A5 | 1.63 | 13% | 171 | 3 (3) |

3. Experimental Section
3.1. Cell Culture and Proteins Extraction

3.2. Minimal CyDye Labeling
3.3. Isoelectric Focusing (IEF) and SDS-PAGE
3.4. Image Scan and Data Analysis
3.5. Spot Picking and Digestion
3.6. MS Analysis and Database Searching
3.7. Immunoblotting
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Suarez-Jimenez, G.M.; Burgos-Hernandez, A.; Ezquerra-Brauer, J.M. Bioactive peptides and depsipeptides with anticancer potential: Sources from marine animals. Mar. Drugs 2012, 5, 963–986. [Google Scholar]
- Abraham, I.; El Sayed, K.; Chen, Z.S.; Guo, H. Current status on marine products with reversal effect on cancer multidrug resistance. Mar. Drugs 2012, 10, 2312–2321. [Google Scholar] [CrossRef]
- D’Orazio, N.; Gammone, M.A.; Gemello, E.; de Girolamo, M.; Cusenza, S.; Riccioni, G. Marine bioactives: Pharmacological properties and potential applications against inflammatory diseases. Mar. Drugs 2012, 4, 812–833. [Google Scholar]
- Margarucci, L.; Monti, M.C.; Tosco, A.; Riccio, R.; Casapullo, A. Chemical proteomics discloses petrosapongiolide M, an anti-inflammatory marine sesterterpene, as a proteasome inhibitor. Angew. Chem. Int. Ed. 2010, 23, 3960–3963. [Google Scholar]
- Margarucci, L.; Monti, M.C.; Fontanella, B.; Riccio, R.; Casapullo, A. Chemical proteomics reveals bolinaquinone as a clathrin-mediated endocytosis inhibitor. Mol. BioSyst. 2011, 2, 480–485. [Google Scholar]
- Cassiano, C.; Monti, M.C.; Festa, C.; Zampella, A.; Riccio, R.; Zampella, A.; Casapullo, A. Chemical proteomics reveals heat shock protein 60 to be the main cellular target of the marine bioactive sesterterpene suvanine. Chembiochem 2012, 13, 1953–1958. [Google Scholar] [CrossRef]
- Slattery, M.; Ankisetty, S.; Corrales, J.; Marsh-Hunkin, K.E.; Gochfeld, D.J.; Willett, K.L.; Rimoldi, J.M. Marine proteomics: A critical assessment of an emerging technology. J. Nat. Prod. 2012, 10, 1833–1877. [Google Scholar]
- Rabus, R. An overview of 2D DIGE analysis of marine (environmental) bacteria. Methods Mol. Biol. 2012, 854, 355–372. [Google Scholar] [CrossRef]
- Burgess, J.G. New and emerging analytical techniques for marine biotechnology. Curr. Opin. Biotechnol. 2012, 1, 29–33. [Google Scholar]
- Veldhoen, N.; Ikonomou, M.G.; Helbing, C.C. Molecular profiling of marine fauna: integration of omics with environmental assessment of the world’s oceans. Ecotoxicol. Environ. Saf. 2012, 2, 23–38. [Google Scholar] [CrossRef]
- Festa, C.; De Marino, S.; Sepe, V.; Monti, M.C.; Luciano, P.; D’Auria, M.V.; Debitus, C.; Bucci, M.; Vellecco, V.; Zampella, A. Perthamides C and D, two new potent anti-inflammatory cyclopeptides from a Solomon Lithistid sponge Theonella swinhoei. Tetrahedron 2009, 50, 10424–10429. [Google Scholar]
- Sepe, V.; D’Auria, M.V.; Bifulco, G.; Ummarino, R.; Zampella, A. Concise synthesis of AHMHA unit in perthamide C. Structural and stereochemical revision of perthamide C. Tetrahedron 2010, 66, 7520–7526. [Google Scholar] [CrossRef]
- Festa, C.; De Marino, S.; Sepe, V.; Monti, M.C.; D’Auria, M.V.; Bifulco, G.; Debitus, C.; Bucci, M.; Vellecco, V.; Zampella, A. Solomonamides A and B, new anti-inflammatory peptides from Theonella swinhoei. Org. Lett. 2011, 6, 1532–1535. [Google Scholar]
- Festa, C.; de Marino, S.; Sepe, V.; D’Auria, M.V.; Bifulco, G.; Andres, R.; Terencio, M.C.; Paya, M.; Debitus, C.; Zampella, A. Perthamides C–F, potent human antipsoriatic cyclopeptides. Tetrahedron 2011, 67, 7780–7786. [Google Scholar]
- Bucci, M.R.; Cantalupo, A.; Vellecco, V.; Panza, E.; Monti, M.C.; Zampella, A.; Ianaro, A.; Cirino, G. Perthamide C inhibits eNOS and iNOS expression and has immunomodulating activity in vivo. PLos One 2013, 8, e57801. [Google Scholar] [CrossRef]
- Margarucci, L.; Monti, M.C.; Mencarelli, A.; Cassiano, C.; Fiorucci, S.; Riccio, R.; Zampella, A.; Casapullo, A. Heat shock proteins as key biological targets of the marine natural cyclopeptide perthamide C. Mol. BioSyst. 2012, 5, 1412–1417. [Google Scholar]
- Huaiyu, M.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef]
- McLaughlin, M.; Vandenbroeck, K. The endoplasmic reticulum protein folding factory and its chaperones: New targets for drug discovery? Br. J. Pharmacol. 2011, 2, 328–334. [Google Scholar] [CrossRef]
- Myung, S.J.; Yoon, J.H.; Kim, B.H.; Lee, J.H.; Jung, E.U.; Lee, H.S. Heat shock protein 90 inhibitor induces apoptosis and attenuates activation of hepatic stellate cells. J. Pharmacol. Exp. Ther. 2009, 1, 276–282. [Google Scholar]
- Chen, S.; Brown, I.R. Translocation of constitutively expressed heat shock protein Hsc70 to synapse-enriched areas of the cerebral cortex after hyperthermic stress. J. Neurosci. Res. 2007, 85, 402–409. [Google Scholar] [CrossRef]
- Macha, M.A.; Matta, A.; Chauhan, S.; Siu, K.M.; Ralhan, R. 14-3-3 zeta is a molecular target in guggulsterone induced apoptosis in head and neck cancer cells. BMC Cancer 2010, 30, 655–659. [Google Scholar]
- Lin, Y.F.; Tsai, P.; Liu, H.G.; Liang, P.H. Intracellular beta-tubulin/chaperonin containing TCP1 beta complex serves as a novel chemotherapeutic target against drug-resistant tumors. Cancer Res. 2009, 17, 6879–6888. [Google Scholar]
- Macleod, K.; Mullen, P.; Sewell, J.; Rabiasz, G.; Lawrie, S.; Miller, E.; Smyth, J.F.; Langdon, S.P. Altered ErbB receptor signaling and gene expression in cisplatin-resistant ovarian cancer. Cancer Res. 2005, 15, 6789–6800. [Google Scholar]
- Brackley, K.I.; Grantham, J. Activities of the chaperonin containing TCP-1 (CCT): Implications for cell cycle progression and cytoskeletal organisation. Cell Stress Chaperones 2009, 14, 23–31. [Google Scholar] [CrossRef]
- Yokota, S.I.; Yanagi, H.; Yura, T.; Kubota, H. Upregulation of cytosolic chaperonin CCT subunits during recovery from chemical stress that causes accumulation of unfolded proteins. Eur. J. Biochem. 2000, 6, 1658–1664. [Google Scholar]
- Hawkins, T.E.; Das, D.; Young, B.; Moss, S.E. DT40 cells lacking the Ca2+-binding protein annexin 5 are resistant to Ca2+-dependent apoptosis. Proc. Natl. Acad. Sci. USA 2002, 12, 8054–8059. [Google Scholar] [CrossRef]
- Ohtsu, M.; Sakai, N.; Fujita, H.; Kashiwagi, M.; Gasa, S.; Shimizu, S.; Eguchi, Y.; Tsujimoto, Y.; Sakiyama, Y.; Kobayashi, K.; et al. Inhibition of apoptosis by the actin-regulatory protein gelsolin. EMBO J. 1997, 15, 4650–4656. [Google Scholar]
- Castro-Rivera, E.; Ran, S.; Thorpe, P.; Minna, J.D. Semaphorin 3B (SEMA3B) induces apoptosis in lung and breast cancer, whereas VEGF165 antagonizes this effect. Proc. Natl. Acad. Sci. USA 2004, 31, 11432–11437. [Google Scholar] [CrossRef]
- Walerych, D.; Gutkowska, M.; Klejman, M.P.; Wawrzynow, P.; Tracz, Z.; Wiech, M.; Zylicz, M.; Zylicz, A. ATP binding to Hsp90 is sufficient for effective chaperoning of p53 protein. J. Biol. Chem. 2010, 42, 32020–32028. [Google Scholar]
- Al-Fageeh, M.B.; Smales, C.M. Control and regulation of the cellular responses to cold shock: The responses in yeast and mammalian systems. Biochem. J. 2006, 397, 247–259. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Vilasi, A.; Monti, M.C.; Tosco, A.; Marino, S.D.; Margarucci, L.; Riccio, R.; Casapullo, A. Differential in Gel Electrophoresis (DIGE) Comparative Proteomic Analysis of Macrophages Cell Cultures in Response to Perthamide C Treatment. Mar. Drugs 2013, 11, 1288-1299. https://doi.org/10.3390/md11041288
Vilasi A, Monti MC, Tosco A, Marino SD, Margarucci L, Riccio R, Casapullo A. Differential in Gel Electrophoresis (DIGE) Comparative Proteomic Analysis of Macrophages Cell Cultures in Response to Perthamide C Treatment. Marine Drugs. 2013; 11(4):1288-1299. https://doi.org/10.3390/md11041288
Chicago/Turabian StyleVilasi, Annalisa, Maria Chiara Monti, Alessandra Tosco, Simona De Marino, Luigi Margarucci, Raffaele Riccio, and Agostino Casapullo. 2013. "Differential in Gel Electrophoresis (DIGE) Comparative Proteomic Analysis of Macrophages Cell Cultures in Response to Perthamide C Treatment" Marine Drugs 11, no. 4: 1288-1299. https://doi.org/10.3390/md11041288
APA StyleVilasi, A., Monti, M. C., Tosco, A., Marino, S. D., Margarucci, L., Riccio, R., & Casapullo, A. (2013). Differential in Gel Electrophoresis (DIGE) Comparative Proteomic Analysis of Macrophages Cell Cultures in Response to Perthamide C Treatment. Marine Drugs, 11(4), 1288-1299. https://doi.org/10.3390/md11041288