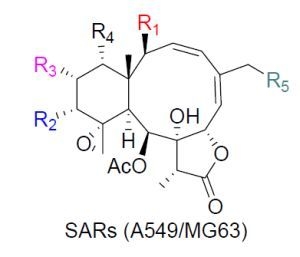
2. Results and Discussion
Freshly collected specimens of
D. gemmacea were immediately frozen to −20 °C and stored at this temperature before extraction. The workup for the extraction and isolation of cembrane diterpenoids was basically performed as previously reported [
10,
11,
12]. This common procedure yielded twenty-one pure compounds (
1–
21).
Gemmacolide AA (
1), a white amorphous powder, had the molecular formula of C
31H
40O
16 based on its HRESI-MS. The IR spectrum showed absorption bands of hydroxyl (3470 cm
−1), a γ-lactone (1775 cm
−1), and ester (1741 cm
−1) functionalities. This observation was in agreement with the signals in the
13C NMR and DEPT spectra (
Table 1) for ten
sp2 carbon atoms (6 × OC = O, CH = CH, CH = C) at lower field and twenty one
sp3 carbon atoms at higher field (1 × C, 2 × CH, 6 × CH
3, 2 × OC, 6 × OCH, 3 × OCH
2, 1 × OCH
3), accounting for eight double bond equivalents (
Table 1,
Table 2). The remaining double bond equivalents were due to the presence of four rings in the molecule.
Table 1.
13C NMR data for gemmacolides AA–AI (1–9) a.
Table 1.
13C NMR data for gemmacolides AA–AI (1–9) a.
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|
| 1 | 46.6, C | 46.6, C | 46.5, C | 46.5, C | 47.3, C | 46.4, C | 46.4, C | 46.4, C | 46.5, C |
| 2 | 75.4, CH | 75.4, CH | 74.1, CH | 74.1, CH | 76.1, CH | 74.2, CH | 74.2, CH | 75.5, CH | 75.5, CH |
| 3 | 130.6, CH | 130.6, CH | 131.3, CH | 131.3, CH | 131.0, CH | 132.1, CH | 132.1, CH | 131.3, CH | 131.5, CH |
| 4 | 129.5, CH | 129.5, CH | 128.7, CH | 128.6,CH | 128.9, CH | 127.8, CH | 127.8, CH | 128.5, CH | 128.3, CH |
| 5 | 141.3, C | 141.3, C | 140.3, C | 141.6, C | 141.3, C | 139.8, C | 139.8, C | 139.7, C | 138.7, C |
| 6 | 123.1, CH | 123.2, CH | 122.8, CH | 122.7, CH | 122.9, CH | 122.5, CH | 122.4, CH | 122.5, CH | 122.2, CH |
| 7 | 78.9, CH | 78.9, CH | 78.9, CH | 79.0, CH | 79.0, CH | 78.7, CH | 78.7, CH | 78.7, CH | 78.3, CH |
| 8 | 81.1, C | 81.0, C | 81.0, C | 80.9, C | 80.9, C | 81.0, C | 81.1, C | 81.0, C | 81.0, C |
| 9 | 63.8, CH | 63.9, CH | 63.9, CH | 63.9, CH | 64.7, CH | 63.9, CH | 63.8, CH | 63.8, CH | 63.9, CH |
| 10 | 32.7, CH | 32.8, CH | 32.8, CH | 32.7, CH | 37.8, CH | 32.8, CH | 32.7, CH | 32.7, CH | 31.4, CH |
| 11 | 58.1, C | 58.2, C | 58.3, C | 58.2, C | 60.0, C | 58.4, C | 58.4, C | 58.3, C | 60.4, C |
| 12 | 73.2, CH | 72.7, CH | 72.9, CH | 72.9, CH | 29.2, CH2 | 72.8, CH | 73.3, CH | 72.8, CH | 75.2, CH |
| 13 | 66.5, CH | 66.4, CH | 66.5, CH | 66.5, CH | 25.1, CH2 | 66.5, CH | 66.7, CH | 66.4, CH | 67.4, CH |
| 14 | 73.9, CH | 73.9, CH | 73.8, CH | 73.4, CH | 74.6, CH | 73.6, CH | 73.7, CH | 73.6, CH | 75.9, CH |
| 15 | 14.3, CH3 | 14.4, CH3 | 14.5, CH3 | 14.5, CH3 | 14.4, CH3 | 14.5, CH3 | 14.5, CH3 | 14.5, CH3 | 14.6, CH3 |
| 16 | 72.2, CH2 | 72.2, CH2 | 72.2, CH2 | 72.1, CH2 | 72.1, CH2 | 62.8, CH2 | 62.8, CH2 | 62.7, CH2 | 62.7, CH2 |
| 17 | 44.2, CH | 44.2, CH | 44.2, CH | 44.2, CH | 44.1, CH | 44.1, CH | 44.1, CH | 44.1, CH | 44.0, CH |
| 18 | 6.3, CH3 | 6.3, CH3 | 6.3, CH3 | 6.3, CH3 | 6.4, CH3 | 6.3, CH3 | 6.3, CH3 | 6.3, CH3 | 6.3, CH3 |
| 19 | 175.2, C | 175.3, C | 175.3, C | 175.3, C | 175.7, C | 175.3, C | 175.2, C | 175.2, C | 175.3, C |
| 20 | 48.9, CH2 | 49.1, CH2 | 49.1, CH2 | 49.2, CH2 | 50.4, CH2 | 49.1, CH2 | 48.8, CH2 | 49.0, CH2 | 48.1, CH2 |
| 9-OAc | 170.2, C | 170.2, C | 170.2, C | 170.2, C | 170.3, C | 170.2, C | 170.2, C | 170.2, C | 170.2, C |
| | 21.5, CH3 | 21.5, CH3 | 21.5, CH3 | 21.6, CH3 | 21.6, CH3 | 21.5, CH3 | 21.5, CH3 | 21.5, CH3 | 21.5, CH3 |
| R1 | 171.9, C | 171.9, C | 169.7, C | 169.7, C | 166.5, C | 169.6, C | 169.5, C | 166.6, C | 166.8, C |
| | 61.1, CH2 | 61.1, CH2 | 21.2, CH3 | 20.6, CH3 | 60.6, CH2 | 21.3, CH3 | 20.5, CH3 | 60.9, CH2 | 60.9, CH2 |
| | | | | | 172.2, C | | | 172.4, C | 172.4, C |
| | | | | | 42.8, CH2 | | | 44.5, CH2 | 42.7, CH2 |
| | | | | | 25.7, CH | | | 25.6, CH | 25.7, CH |
| | | | | | 22.4, 2 × CH3 | | | 22.3, 2 × CH3 | 22.4, 2 × CH3 |
| R2 | 169.7, C | 171.8, C | 171.8, C | 171.9, C | | 171.9, C | 169.8, C | 171.9, C | |
| | 20.9, CH3 | 43.5, CH2 | 43.5, CH2 | 43.5, CH2 | | 43.4, CH2 | 20.6, CH3 | 43.3, CH2 | |
| | | 25.7, CH | 25.7, CH | 24.9, CH | | 25.7, CH | | 25.6, CH | |
| | | 22.3, CH3 | 22.3, CH3 | 22.5, 2 × CH3 | | 22.4, 2 × CH3 | | 22.3, 2 × CH3 | |
| | | 22.4, CH3 | 22.4, CH3 | | | | | | |
| R3 | 169.7, C | 169.7, C | 169.7, C | 169.7, C | | 169.8, C | 170.5, C | 169.7, C | 169.9, C |
| | 20.5, CH3 | 20.5, CH3 | 20.9, CH3 | 21.3, CH3 | | 20.5, CH3 | 20.7, CH3 | 20.5, CH3 | 20.6, CH3 |
| R4 | 170.6, C | 170.6, C | 170.1, C | 172.3, C | 170.1, C | 170.5, C | 170.5, C | 170.2, C | 170.2, C |
| | 20.9, CH3 | 21.0, CH3 | 21.4, CH3 | 43.5, CH2 | 21.2, CH3 | 20.9, CH3 | 20.9, CH3 | 20.8, CH3 | 20.8, CH3 |
| | | | | 25.1, CH | | | | | |
| | | | | 22.3, 2 × CH3 | | | | | |
| R5 | 58.5, CH3 | 58.5, CH3 | 58.5, CH3 | 58.5, CH3 | 58.5, CH3 | 172.1, C | 172.0, C | 172.0, C | 172.6, C |
| | | | | | | 43.3, CH2 | 43.3, CH2 | 43.5, CH2 | 43.2, CH2 |
| | | | | | | 25.7, CH | 25.7, CH | 25.7, CH | 25.8, CH |
| | | | | | | 22.5, 2 × CH3 | 22.4, 2 × CH3 | 22.5, 2 × CH3 | 22.4, 2 × CH3 |
Table 2.
1H NMR data for gemmacolides AA–AF (1–6) a.
Table 2.
1H NMR data for gemmacolides AA–AF (1–6) a.
| Position | 1 | 2 | 3 | 4 | 5 | 6 |
|---|
| 2 | 5.70, d (9.5) | 5.70, d (9.5) | 5.57, ov | 5.52, ov | 5.68, d (9.6) | 5.61, ov |
| 3 | 5.60, dd (10.4, 9.7) | 5.60, dd (10.2, 9.5) | 5.57, ov | 5.53, ov | 5.56, dd (10.6, 9.6) | 5.61, ov |
| 4 | 6.33, d (10.4) | 6.33, d (10.2) | 6.29, d (10.7) | 6.28, d (10.1) | 6.30, d (10.6) | 6.29, ov |
| 6 | 5.91, d (8.7) | 5.90, d (8.8) | 5.89, d (8.8) | 5.88, d (8.6) | 5.88, d (8.7) | 5.71, d (8.7) |
| 7 | 4.99, d (8.7) | 4.98, d (8.8) | 4.99, d (8.8) | 5.01, d (8.6) | 4.97, d (8.7) | 4.97, d (8.7) |
| 9 | 4.76, d (4.5) | 4.76, d (4.5) | 4.75, d (4.6) | 4.74, d (4.6) | 4.75, d (5.0) | 4.74, d (4.7) |
| 10 | 3.63, d (4.5) | 3.62, d (4.5) | 3.61, d (4.6) | 3.61, ov | 3.14, d (5.0) | 3.61, ov |
| 12 | 4.88, d (3.2) | 4.92, d (3.3) | 4.91, d (3.0) | 4.92, d (3.1) | 2.19, m | 4.91, d (3.3) |
| | | | | | 1.11, m | |
| 13β | 5.06, dd (3.2, 3.2) | 5.08, dd (3.3, 3.5) | 5.09, dd (3.0, 3.0) | 5.1, dd (3.1, 3.2) | 1.95, m | 5.08, dd (3.3, 3.2) |
| 13α | | | | | 1.74, m | |
| 14 | 5.17, d (3.2) | 5.17, d (3.5) | 5.21, d (3.0) | 5.26, d (3.2) | 4.86, br s | 5.22, d (3.2) |
| 15 | 1.14, s | 1.14, s | 1.13, s | 1.13, s | 1.13, s | 1.14, s |
| 16a | 4.51, d (15.1) | 4.51, d (14.8) | 4.49, d (14.7) | 4.5, d (14.4) | 4.47, d (14.9) | 5.42, d (15.7) |
| 16b | 4.23, d (15.1) | 4.23, d (14.8) | 4.23, d (14.7) | 4.23, d (14.4) | 4.16, d (14.9) | 4.64, d (15.7) |
| 17 | 2.31, q (7.1) | 2.30, ov | 2.30, q (7.1) | 2.31, q (6.9) | 2.27, q (7.1) | 2.30, q (7.0) |
| 18 | 1.16, d (7.1) | 1.13, ov | 1.14, d (7.1) | 1.14, d (6.9) | 1.14, d (7.1) | 1.13, d (7.0) |
| 20a | 3.60, d (2.4) | 3.60, d (2.0) | 3.61, ov | 3.60, ov | 3.49, br s | 3.61, ov |
| 20b | 2.92, d (2.4) | 2.92, d (2.0) | 2.92, d (2.0) | 2.94, br s | 2.64, d (2.3) | 2.92, br s |
| 8-OH | | 2.72, s | | | | |
| 9-OAc | 2.19, s | 2.19, s | 2.19, s | 2.19, s | 2.16, s | 2.19, s |
| R1 | 4.13, d (16.9) | 4.13, d (16.8) | 1.94, s | 1.95, s | 4.53, d (15.7) | 1.95, s |
| | 4.00, d (16.9) | 4.00, d (16.8) | | | 4.48, d (15.7) | |
| | | | | | 2.32, ov | |
| | | | | | 2.29, ov | |
| | | | | | 2.13, m | |
| | | | | | 0.98, d (6.5) (×2) | |
| R2 | 2.16, s | 2.22, ov | 2.32, ov | 2.27, ov | | 2.30, ov (×2) |
| | | 2.32, ov | 2.23, ov | 2.15, ov | | |
| | | 2.17, m | 2.15, m | 2.04, m | | 2.16, m |
| | | 1.01, d (6.5) | 1.01, d (6.6) | 0.99, d (6.5) (×2) | | 0.99, d (6.4) (×2) |
| | | 0.99, d (6.5) | 0.99, d (6.6) | | | |
| R3 | 1.94, s | 1.93, s | 1.95, s | 1.95, s | | 1.94, s |
| R4 | 2.08, s | 2.08, s | 2.06, s | 2.32, ov (×2) | 2.07, s | 2.10, s |
| | | | | 2.15, m | | |
| | | | | 0.99, d (6.6) (×2) | | |
| R5 | 3.46, s | 3.45, s | 3.45, s | 3.45, s | 3.42, s | 2.28, ov (×2) |
| | | | | | | 2.13, m |
| | | | | | | 0.99, d (6.4) (×2) |
Analysis of the
1H and
13C NMR spectra of
1 (
Table 1,
Table 2) revealed a great similarity to those of gemmacolide N (
22) [
11]. An additional glycolyl group was observed. The location of the glycolyl group at C-2 was indicated by the distinct HMBC correlations of both H-2 and H-2′ with C-1′. The four acetyl groups were assigned at C-9, C-12, C-13 and C-14 due to the obvious HMBC correlations from the secondary alcohol protons to the respective ester carbonyl groups. The established planar structure of
1 was further supported by the COSY and HMBC spectra as shown in
Figure 1. The relative configuration of
1 at the chiral centers was proved the same as that of juncenolide D (
23) by a NOESY experiment (
Figure 2), showing a β configuration of H-7, H-12, H-13, H-14, Me-15, H-17, and CH
2-20, and an α configuration of H-2, H-9, H-10, and Me-18. The geometry of the Δ
3 double bond was assigned as
Z based on the proton coupling constant between H-3 and H-4 (
J = 10.4 Hz) while that of Δ
5 was determined as
E due to the NOESY correlation between H-6 and H
2-16. The relative configuration of
1 was thus determined as (1
S*,2
S*,7
S*,8
S*,9
S*,10
S*,11
R*,12
R*,13
R*,14
R*,17
R*).
Figure 1.
Key HMBC (arrow) and COSY (bond) correlations for compound 1.
Figure 1.
Key HMBC (arrow) and COSY (bond) correlations for compound 1.
Figure 2.
Key NOESY correlations for compound 1.
Figure 2.
Key NOESY correlations for compound 1.
As gemmacolide AA (
1) contained the same lactone and diene chromophores as gemmacolide N (
22) and they differed only in the nature of the ester group at C-2 and the R
3, the ECD spectrum of gemmacolide N could therefore be used as an ECD reference for the configurational assignment of gemmacolide AA (
1) and analogues. Since the absolute configuration of gemmacolide N had been unambiguously determined by a TDDFT calculation of its solution ECD spectrum [
11], the absolute configuration of
1 was then suggested as (1
S,2
S,7
S,8
S,9
S,10
S,11
R,12
R,13
R,14
R,17
R) due to the congruent ECD curves for
1 and that of gemmacolide N. The assignment of the absolute configuration was in agreement with that of dichotellide T, an analogue recently isolated from the same species of animals with its absolute stereochemistry being determined by X-ray single crystal diffraction analysis [
5].
Gemmacolide AB (
2) was obtained as a white amorphous powder with the molecular formula of C
34H
46O
16 being established by HRESI-MS. The structure of
2 differed from that of
1 by the presence of an isovaleryl group instead of an acetyl group at C-12 (
Table 1,
Table 2). The assignment of the isovalerate ester at C-12 was indicated by the HMBC correlations from both H-12 and H-2″ to the isovaleryl carbonyl carbon. The structure of
2 was thus determined. Its absolute configuration was proved the same as that of
1 on the basis of their similar ECD spectrum.
Gemmacolide AC (
3) was isolated as a white amorphous powder, had a molecular formula of C
34H
46O
15 as deduced from its HRESI-MS. Its
1H and
13C NMR spectra data (
Table 1,
Table 2) were similar to those of
2 with the only difference of the glycolyl group at C-2 in
2 being replaced by an acetyl group in
3. The location of the acetyl group at C-2 was confirmed by the HMBC correlations of H-2 with C-1′. Its absolute configuration was proved the same as that of
2 based on their similar ECD spectrum.
Gemmacolide AD (
4) was isolated as a white amorphous powder. Its molecular formula was established as C
37H
52O
15 by HRESI-MS.
1H and
13C NMR spectra data of
4 (
Table 1,
Table 2) greatly resembled to those of
3 except that the acetyl group at C-14 in
3 was replaced by an isovaleryl group in
4. The location of the two isovalery groups at C-12 and C-14 were indicated by the HMBC correlations from both H-12 and H-14 to the isovaleryl carbonyl carbon. The established structure of
4 was further supported by detailed analysis of its 2D NMR data. Its absolute configuration was proven the same as that of
3 based on their similar ECD spectrum.
Gemmacolide AE (
5) was isolated as a white amorphous powder. The molecular formula C
32H
44O
13 was established by the HRESI-MS. Comparison of overall
1H and
13C NMR spectra data (
Table 1,
Table 2) of
5 with those of
19 revealed great similarity. However, signals for two acetyl groups in
19 were disappeared in
5. The de-acetyl groups were found to be CH
2-12 and CH
2-13 based on the proton sequence from H
2-12 to H-14 as deduced from the
1H-
1H COSY experiment. The relative configuration for other chiral centers remained intact due to the NOESY experiment. The ECD experiment suggested (−)-(1
R,2
S,7
S,8
S,9
S,10
S,11
R,14
S,17
R) absolute configuration for compound
5.
Gemmacolide AF (
6) was isolated as a white amorphous powder. The molecular formula C
38H
52O
16 was established by the HRESI-MS.
1H and
13C NMR spectra of
6 (
Table 1,
Table 2) were similar to those of compound
3 except that oxygenated methyl group in
3 was replaced by an isovaleryl group in
6. Two isovaleryl groups were attached at C-12 and C-16 due to the HMBC correlations. The relative and absolute configuration of
6 was proved the same as that of
3 by the analysis of NOESY and ECD spectra.
Gemmacolide AG (
7), a white amorphous powder, showed a molecular formula of C
35H
46O
16 in the HRESI-MS.
1H and
13C NMR spectroscopic data of
7 were almost identical to those of
6 (
Table 1,
Table 3) except for the replacement of the 12-isovaleryl group in
6 by an acetyl group in
7. The absolute structure of
7 was proved the same as that of
6 by the analysis of NOESY and ECD spectra.
Table 3.
1H NMR data for gemmacolides AG-AL (7–12) a.
Table 3.
1H NMR data for gemmacolides AG-AL (7–12) a.
| Position | 7 | 8 | 9 | 10 | 11 | 12 |
|---|
| 2 | 5.61, ov | 5.67, ov | 5.78, d (9.9) | 5.58, d (9.9) | 5.7, d (9.9) | 5.64, d (9.7) |
| 3 | 5.61, ov | 5.62, dd (10.0, 9.9) | 5.61, dd (9.9, 10.3) | 5.60, dd (10.1, 9.9) | 5.6, dd (9.9, 10.3) | 5.60, dd (9.7, 9.8) |
| 4 | 6.29, br s | 6.34, d (10.4) | 6.32, d (10.3) | 6.40, d (10.1) | 6.40, d (10.3) | 6.33, d (9.8) |
| 6 | 5.71, d (8.6) | 5.69, ov | 5.65, ov | 6.06, d (8.4) | 5.9, d (8.6) | 5.89, d (8.7) |
| 7 | 4.97, d (8.6) | 4.96, d (9.0) | 4.96, ov | 4.95, d (8.4) | 4.99, d (8.6) | 4.98, d (8.7) |
| 9 | 4.74, d (4.4) | 4.74, d (4.7) | 4.74, d (4.8) | 4.74, d (4.7) | 4.74, d (4.8) | 4.75, d (4.5) |
| 10 | 3.61, ov | 3.61, d (4.7) | 3.6, ov | 3.59, ov | 3.62, d (4.8) | 3.62, d (4.5) |
| 12 | 4.86, br s | 4.91, d (3.0) | 3.45, br s | 4.91, d (3.2) | 4.9, d (3.4) | 4.89, d (3.1) |
| 13β | 5.08, dd (3.0, 2.8) | 5.08, dd (3.0, 2.8) | 4.97, ov | 5.08, dd (3.3, 3.2) | 5.08, dd (3.4, 3.5) | 5.09, dd (3.1, 3.0) |
| 14 | 5.22, br s | 5.20, d (2.8) | 5.30, br s | 5.18, d (3.3) | 5.16, d (3.5) | 5.17, d (3.0) |
| 15 | 1.14, s | 1.13, s | 1.13, s | 1.13, s | 1.13, s | 1.13, s |
| 16a | 5.45, d (15.6) | 5.42, d (15.6) | 5.46, d (15.6) | 4.68, d (16.2) | 4.5, d (14.7) | 4.53, d (15.3) |
| 16b | 4.64, d (15.6) | 4.59, d (15.6) | 4.56, d (15.6) | 4.45, ov | 4.25, d (14.7) | 4.11, d (15.3) |
| 17 | 2.31, q (7.0) | 2.30, q (6.9) | 2.28, q (6.9) | 2.30, q (7.0) | 2.31, q (6.9) | 2.29, q (6.9) |
| 18 | 1.15, d (7.0) | 1.13, d (6.9) | 1.13, d (6.9) | 1.14, d (7.0) | 1.15, d (6.9) | 1.15, d (6.9) |
| 20a | 3.61, ov | 3.60, ov | 3.60, br s | 3.59, ov | 3.60, d (2.6) | 3.60, d (2.1) |
| 20b | 2.92, br s | 2.93, d (2.2) | 2.76, d (2.1) | 2.93, d (1.4) | 2.97, d (2.6) | 2.92, d (2.1) |
| 9-OAc | 2.19, s | 2.19, s | 2.19, s | 2.18, s | 2.19, s | 2.18, s |
| R1 | 1.95, s | 4.52, d (15.6) | 4.52, d (15.6) | 4.54, d (15.7) | 4.15,d (16.9) | 4.53, d (15.7) |
| | | 4.43, d (15.6) | 4.42, d (15.6) | 4.44, d (15.7) | 4.0, d (16.9) | 4.43, d (15.7) |
| | | 2.28, ov (×2) | 2.28, ov (×2) | 2.29, ov (×2) | | 2.28, ov (×2) |
| | | 2.16, m | 2.13, m | 2.13, m | | 2.15, ov |
| | | 0.98, ov (×2) | 0.99, d (6.5) (×2) | 0.98, ov (×2) | | 0.98, ov (×2) |
| R2 | 2.16, s | 2.35, ov (×2) | | 2.32, ov | 2.16, s | 2.14, s |
| | | | | 2.23, ov | | |
| | | 2.16, m | | 2.17, m | | |
| | | 0.98, ov (×2) | | 1.00, ov (×2) | | |
| R3 | 1.95, s | 1.94, s | 2.03, s | 1.94, s | 2.2, ov | 2.09, ov (×2) |
| | | | | | 2.08, ov | 1.99, ov |
| | | | | | 1.97, m | 0.92, d (6.5) |
| | | | | | 0.9, d (6.5) (×2) | 0.91, d (6.5) |
| R4 | 2.10, s | 2.10, s | 2.13, s | 2.07, s | 2.06, s | 2.05, s |
| R5 | 2.3, ov (×2) | 2.26, ov (×2) | 2.28, ov (×2) | | 3.45, s | 3.44, s |
| | 2.05, m | 2.16, m | 2.13, m | | | |
| | 0.99, d (6.6) (×2) | 0.98, ov (×2) | 0.99, d (6.6) (×2) | | | |
Gemmacolide AH (
8) was isolated as a white amorphous powder with the molecular formula of C
43H
60O
18 being established by HRESI-MS. Its
1H and
13C NMR spectra data (
Table 1,
Table 3) were similar to those of
6 with the only difference of the acetyl group at C-2 in
6 being replaced by an isovaleric acetyl group in
8. This assignment was clearly indicated by the long range correlation from both H
2-4′ and H
2-2′ to C-3′, and from both H-2 and H
2-2′ to C-1′. The relative configuration of all the chiral centers remained intact, which was supported by a NOESY experiment. The absolute configuration of
8 was obtained based on the ECD experiment.
Gemmacolide AI (
9), a white amorphous powder, had a molecular formula of C
38H
52O
17 as deduced from its HRESI-MS.
1H and
13C NMR spectra of
9 (
Table 1,
Table 3) showed similarity to those of compound
8. The isovaleryl group in at C-12 in
8 was replaced by a hydroxy group in
9. This conclusion was supported by extensive 2D NMR analysis. Its ECD spectrum indicated the same absolute configuration as that of
8.
Gemmacolide AJ (
10) had a molecular of C
38H
51O
16Cl as established by HRESI-MS. An isotopic ratio of 3:1 observed in the molecular ion peak at
m/
z 821/823 ([M + Na]
+) confirmed the appearance of a chlorine atom in the molecule.
1H and
13C NMR spectra of
10 closely resembled those of
8 (
Table 3,
Table 4), except for the absence of the signals of one isovaleryl group. This fact together with the up-field shifted signal of C-16 in the
13C NMR spectra (δ
C 72.0,
t in
4 and 44.2,
t in
10) led to the location of chlorine atom at C-16 [
14]. The structure of
10 was thus determined. Its relative and absolute stereochemistry was proved the same as that of
8 by NOESY and ECD measurements.
Table 4.
13C NMR data for gemmacolides AJ–AR (10–18) a.
Table 4.
13C NMR data for gemmacolides AJ–AR (10–18) a.
| Position | 10 b | 11 b | 12 b | 13 b | 14 c | 15 b | 16 d | 17 b | 18 b |
|---|
| 1 | 46.5, C | 46.6, C | 46.5, C | 46.5, C | 46.5, C | 46.5, C | 46.5, C | 46.5, C | 46.5, C |
| 2 | 75.3, CH | 75.4, CH | 75.4, CH | 75.2, CH | 75.2, CH | 75.5, CH | 75.2, CH | 75.5, CH | 74.1, CH |
| 3 | 131.1, CH | 130.6, CH | 129.4, CH | 131, CH | 131.1, CH | 131.5, CH | 131.1, CH | 131.3, CH | 132.1, CH |
| 4 | 129.0, CH | 129.5, CH | 130.5, CH | 129, CH | 129.1, CH | 128.5, CH | 129.0, CH | 129.5, CH | 127.8, CH |
| 5 | 139.9, C | 141.3, C | 141.6, C | 139.9, C | 139.7, C | 139.5, C | 144.3, C | 144.7, C | 139.9, C |
| 6 | 126.0, CH | 123.2, CH | 122.6, CH | 125, CH | 126.3,CH | 122.8, CH | 126.2, CH | 123.4, CH | 122.8, CH |
| 7 | 78.5, CH | 78.9, CH | 79.0, CH | 78.5, CH | 78.5, CH | 78.7, CH | 78.5, CH | 78.7, CH | 78.7, CH |
| 8 | 80.9, C | 81.0, C | 81.1, C | 81.1, C | 81.0, C | 81.0, C | 80.0, C | 81.1, C | 80.2, C |
| 9 | 63.7, CH | 63.8, CH | 63.9, CH | 63.9, CH | 63.6, CH | 63.8, CH | 63.5, CH | 63.8, CH | 63.8, CH |
| 10 | 32.7, CH | 32.7, CH | 32.7, CH | 32.1, CH | 32.6, CH | 32.7, CH | 31.6, CH | 32.7, CH | 32.6, CH |
| 11 | 58.3, C | 58.2, C | 58.2, C | 58.2, C | 58.1, C | 58.3, C | 58.0, C | 58.4, C | 58.5, C |
| 12 | 72.7, CH | 73.2, CH | 73.2, CH | 73.2, CH | 73.0, CH | 72.7, CH | 73.1, CH | 73.3, CH | 73.3, CH |
| 13 | 66.3, CH | 66.2, CH | 66.3, CH | 66.3, CH | 66.4, CH | 66.2, CH | 66.3, CH | 66.5, CH | 66.6, CH |
| 14 | 73.6, CH | 74.0, CH | 73.9, CH | 73.9, CH | 73.8, CH | 73.8, CH | 73.3, CH | 73.4, CH | 73.3, CH |
| 15 | 14.4, CH3 | 14.4, CH3 | 14.4, CH3 | 14.4, CH3 | 14.3, CH3 | 14.4, CH3 | 14.7, CH3 | 14.5, CH3 | 14.5, CH3 |
| 16 | 44.2, CH2 | 72.2, CH2 | 72.0, CH2 | 44.2, CH2 | 44.5, CH2 | 62.8, CH2 | 44.4, CH2 | 63.2, CH2 | 63.2, CH2 |
| 17 | 44.0, CH | 44.2, CH | 44.2, CH | 44.0, CH | 44.0, CH | 44.1, CH | 43.2, CH | 44.1, CH | 44.1, CH |
| 18 | 6.3, CH3 | 6.3, CH3 | 6.3, CH3 | 6.9, CH3 | 6.3, CH3 | 6.3, CH3 | 6.3, CH3 | 6.3, CH3 | 6.3, CH3 |
| 19 | 175.0, C | 175.6, C | 175.2, C | 175.2, C | 174.9, C | 175.2, C | 175.1, C | 175.3, C | 175.2, C |
| 20 | 49.0, CH2 | 48.9, CH2 | 48.9, CH2 | 48.9, CH2 | 49.8, CH2 | 49.0, CH2 | 49.0, CH2 | 49.1, CH2 | 49.0, CH2 |
| 9-OAc | 170.1, C | 170.3, C | 170.2, C | 170.2, C | 170.1, C | 170.1, C | 170.2, C | 170.3, C | 170.2, C |
| | 21.5, CH3 | 21.5, CH3 | 21.5, CH3 | 21.5, CH3 | 21.5, CH3 | 21.6, CH3 | 21.5, CH3 | 21.6, CH3 | 21.5, CH3 |
| R1 | 167.0, C | 171.9, C | 166.6, C | 166.6, C | 172.2, C | 172.0, C | 172.8, C | 170.7, C | 169.4, C |
| | 60.8, CH2 | 61.2, CH2 | 60.9, CH2 | 60.9, CH2 | 61.1, CH2 | 61.2, CH2 | 61.1, CH2 | 21.5, CH3 | 21.3, CH3 |
| | 172.4, C | | 172.4, C | 172.4, C | | | | | |
| | 42.7, CH2 | | 42.8, CH2 | 42.7, CH2 | | | | | |
| | 25.6, CH | | 25.7, CH | 25.7, CH | | | | | |
| | 22.3, 2 × CH3 | | 22.4, 2 × CH3 | 22.4, 2 × CH3 | | | | | |
| R2 | 171.8, C | 169.6, C | 169.6, C | 169.6, C | 169.7, C | 171.8, C | 169.8, C | 169.9, C | 169.9, C |
| | 43.6, CH2 | 21.5, CH3 | 20.9, CH3 | 20.9, CH3 | 20.9, CH3 | 43.4, CH2 | 21.3, CH3 | 21.1, CH3 | 20.8, CH3 |
| | 25.7, CH | | | | | 25.7, CH | | | |
| | 22.4, 2 × CH3 | | | | | 22.4, 2 × CH3 | | | |
| R3 | 169.7, C | 171.8, C | 171.7, C | 171.7, CH | 171.4, C | 171.7, C | 169.7, C | 169.9, C | 169.8, C |
| | 20.5, CH3 | 42.6, CH2 | 42.6, CH2 | 42.6, CH2 | 44.5, CH2 | 42.6, CH2 | 20.5, CH3 | 21.1, CH3 | 20.6, CH3 |
| | | 25.0, CH | 25.0, CH | 25.0, CH | 25.7, CH | 25.0, CH | | | |
| | | 22.3, 2 × CH3 | 22.5, CH3 | 22.4, 2 × CH3 | 22.4, CH3 | 22.3, 2 × CH3 | | | |
| | | | 22.4, CH3 | | 22.3, CH3 | | | | |
| R4 | 170.3, C | 170.4, C | 170.0, C | 170.0, C | 170.6, C | 170.9, C | 172.4, C | 172.4, C | 172.5, C |
| | 20.8, CH3 | 21.0, CH3 | 20.8, CH3 | 20.8, CH3 | 20.8, CH3 | 20.9, CH3 | 42.6, CH2 | 43.1, CH2 | 43.1, CH2 |
| | | | | | | | 25.0, CH | 25.1, CH | 25.1, CH |
| | | | | | | | 22.3, 2 × CH3 | 22.5, CH3 | 22.5, CH3 |
| | | | | | | | | 22.4, CH3 | 22.4, CH3 |
| R5 | | 58.5, CH3 | 58.5, CH3 | | | 172.2, C | | | 170.0, C |
| | | | | | | 43.3, CH2 | | | 21.0, CH3 |
| | | | | | | 25.6, CH | | | |
| | | | | | | 22.5, 2 × CH3 | | | |
Gemmacolide AK (
11), a white amorphous powder, displayed the molecular formula of C
34H
46O
16 in the HRESI-MS. Its
1H and
13C NMR spectra data (
Table 3,
Table 4) showed great similarity to those of
2. However, the substitutions of isovaleric acetyl group at C-12 and acetyl group at C-13 in
2 had to be interchanged in
11 based on the HMBC experiment. The relative configuration for all chiral centers remained intact due to the NOEY experiment. Its absolute configuration was proved the same as that of
2 due to their similarity in ECD spectrum.
Gemmacolide AL (
12) was isolated as a white amorphous powder, had a molecular of C
39H
54O
17 as established by HRESI-MS.
1H and
13C NMR spectroscopic data of
12 were almost identical to those of
11 (
Table 3,
Table 4) except for the replacement of glycolyl group by the isovaleric acetyl. Detailed analysis of
1H-
1H COSY and HMBC spectra clarified the isovaleric acetyl at C-2. The structure of
12 was thus determined, showing the same relative and absolute configuration as that of
11, as further confirmed by the NOESY and ECD experiments.
Gemmacolide AM (
13) was isolated as a white amorphous powder. The molecular formula C
38H
51ClO
16 was established by the HRESI-MS. Its
1H and
13C NMR spectra data (
Table 4,
Table 5) closely resembled to those of
10. The substitutions of isovaleric acetyl group at C-12 and acetyl group at C-13 in
10 had to be interchanged in
13 due to the detailed analysis on the HMBC spectra. NOESY and ECD experiments led to the same absolute configuration for both compounds.
Table 5.
1H NMR data for gemmacolides AM–AR (13–18) a.
Table 5.
1H NMR data for gemmacolides AM–AR (13–18) a.
| Position | 13 b | 14 c | 15 b | 16 d | 17 b | 18 b |
|---|
| 2 | 5.57, d (9.6) | 5.64, ov | 5.73, d (9.8) | 5.6, d (9.6) | 5.61, ov | 5.54, d (9.7) |
| 3 | 5.65, dd (9.6, 10.3) | 5.65, ov | 5.63, dd (10.3, 9.8) | 5.65, dd (9.6, 10.3) | 5.60, ov | 5.60, dd (9.7, 10.3) |
| 4 | 6.40, d (10.3) | 6.41, d (8) | 6.33, d (10.3) | 6.40, d (10.3) | 6.35, d (8.5) | 6.29, d (10.3) |
| 6 | 6.06, d (8.6) | 6.07, d (8.6) | 5.73, ov | 6.07, d (8.6) | 5.81, d (8.5) | 5.75, d (8.5) |
| 7 | 4.95, d (8.6) | 4.94, d (8.6) | 4.96, d (8.5) | 4.93, d (8.6) | 4.96, d (8.5) | 4.97, d (8.5) |
| 9 | 4.74, d (4.7) | 4.75, d (4.5) | 4.75, d (4.5) | 4.74, d (4.7) | 4.74, d (4.8) | 4.74, d (4.8) |
| 10 | 3.61, ov | 3.61, ov | 3.62, d (4.5) | 3.61, ov | 3.60, d (4.8) | 3.61, d (4.8) |
| 12 | 4.96, d (3.0) | 4.92, d (3.4) | 4.92, d (3.3) | 4.88, br s | 4.89, d (3.3) | 4.88, d (3.2) |
| 13β | 5.08, dd (3.0, 3.1) | 5.23, dd (3.4, 3.5) | 5.09, dd (3.2, 3.3) | 5.08, dd (3.2, 3.3) | 5.10, dd (3.3, 3.3) | 5.09, dd (3.2, 3.0) |
| 14 | 5.16, d (3.1) | 5.18, d (3.5) | 5.19, d (3.2) | 5.21, br s | 5.28, d (3.3) | 5.26, d (3.0) |
| 15 | 1.13, s | 1.14, s | 1.15, s | 1.13, s | 1.14, s | 1.14, s |
| 16a | 4.67, d (13.6) | 4.67, d (13.5) | 5.46, d (15.6) | 4.65, d (13.8) | 4.49, br s | 5.31, d (16.1) |
| 16b | 4.47, d (13.6) | 4.54, d (13.5) | 4.62, d (15.6) | 4.57, d (13.8) | 4.49, br s | 4.72, d (16.1) |
| 17 | 2.31, ov | 2.3, ov | 2.30, ov | 2.31, ov | 2.30, ov | 2.31, q (7.1) |
| 18 | 1.13, d (6.9) | 1.14, d (6.9) | 1.14, ov | 1.14, d (7.2) | 1.15, ov | 1.15, ov |
| 20a | 3.60, ov | 3.60, ov | 3.60, ov | 3.60, ov | 3.63, br s | 3.60, br s |
| 20b | 2.94, d (2.2) | 2.94, d (2.0) | 2.94, br s | 2.95, br s | 2.93, br s | 2.93, br s |
| 9-OAc | 2.19, s | 2.19, s | 2.19, s | 2.19, s | 2.20, s | 2.19, s |
| R1 | 4.54, d (15.6) | 4.17, d (16.8) | 4.12, d (15.2) | 4.15, d (16.8) | 1.98, s | 1.94, s |
| | 4.44, d (15.6) | 4.02, d (16.8) | 3.99, d (15.2) | 4.06, d (16.8) | | |
| | 2.28, ov (×2) | | | | | |
| | 2.10, ov | | | | | |
| | 0.99, d (6.5) (×2) | | | | | |
| R2 | 2.16, s | 2.16, s | 2.31, ov (×2) | 2.16, s | 2.17, s | 2.13, s |
| | | | 2.16, ov | | | |
| | | | 0.99, ov (×2) | | | |
| R3 | 2.32, ov | 2.08, ov (×2) | 2.06, ov (×2) | 1.95, s | 1.95, s | 1.95, s |
| | 2.25, ov | | | | | |
| | 1.99, ov | 1.99, ov | 1.99, ov | | | |
| | 0.90, d (6.0) | 0.91, d (6.5) | 0.99, d (6.7) | | | |
| | 0.93, d (6.0) | 0.90, d (6.5) | 0.99, d (6.7) | | | |
| R4 | 2.07, s | 2.09, s | 2.12, s | 2.3, ov (×2) | 2.31, ov | 2.29, ov |
| | | | | | 2.18, ov | 2.21, ov |
| | | | | 2.1, ov | 2.11, ov | 2.09, ov |
| | | | | 0.99, d (6.6) | 1.00, d (6.8) | 0.99, d (6.8) |
| | | | | 0.99, d (6.6) | 0.98, d (6.8) | 0.97, d (6.8) |
| R5 | | | 2.29, ov (×2) | | | 2.17, s |
| | | | 2.18, ov | | | |
| | | | 0.99, ov (×2) | | | |
Gemmacolide AN (
14) was obtained as a white amorphous powder with the molecular formula of C
33H
43ClO
15 being established by HRESI-MS. The structure of
14 was similar to those of
13 with the only difference of the isovaleric acetyl at C-2 in
13 being replaced by a glycolyl group in
14 (
Table 4,
Table 5). The location of the glycolyl group at C-2 was confirmed by the HMBC correlations of both H-2′ and H-2 with C-1′. The structure of
14 was thus determined. Its absolute configuration was proved the same as that of
13 based on their similar ECD spectra data.
Gemmacolide AO (
15) was obtained as a white amorphous powder and exhibited a molecular formula of C
41H
58O
17 as deduced from its HRESI-MS.
1H and
13C NMR spectra of
15 were similar to those of
6. However, two of the acetyl groups in
6 were replaced by a glycolyl at C-2 and an isovaleryl group at C-13 in
15 (
Table 4,
Table 5). The planar and relative structure of
15 was confirmed by extensive 2D NMR analysis and correlations with co-isolated analogues. Its ECD spectrum suggested the same absolute configuration as that of
6.
Gemmacolide AP (
16) was isolated as a white amorphous powder, had a molecular of C
33H
43ClO
15 as established by HRESI-MS.
1H and
13C NMR spectroscopic data of
16 were almost identical to those of
14 (
Table 4,
Table 5). Detailed analysis of
1H-
1H COSY and HMBC spectra indicated an interchangeable acetyl group and isovaleryl group in
16 with respected to those in
14. Their absolute configuration was proven the same based on NOESY and ECD experiment.
Gemmacolide AQ (
17) was isolated as a white amorphous powder, showed a molecular formula of C
33H
44O
15 as deduced from its HRESI-MS.
1H and
13C NMR spectroscopic data of
17 were almost identical to those of
3 (
Table 4,
Table 5), showing a similar substituted functionalities with the exception of the disappearance of the signals for the oxygenated methyl group. The isovaleryl group, however, was proven to be attached to C-14 instead of C-12 based on the analysis of
1H-
1H COSY and HMBC spectra. The structure of
17 was then determined, having the same relative and absolute stereochemistry as that of
3 due to the NOESY and ECD measurements.
Gemmacolide AR (
18) was found to be a white amorphous powder, having the molecular formula of C
35H
46O
16 based on the HRESI-MS.
1H and
13C NMR spectra of
18 resembled to those of compound
17 except for the appearance of an additional acetyl group (
Table 4,
Table 5). The five acetyl groups were thus assigned to C-2, C-9, C-12, C-13, and C-16, which was supported by the
1H-
1H COSY and HMBC experiments. The replacement of the hydroxyl group by an acetoxyl group at C-16 was further supported by the remarkable downfield proton signal of H
2-16 from δ
H 4.47 (2H, br s) in
17 to in
18 δ
H 5.31, 4.72 (each d,
J = 16.0 Hz). The relative and absolute configuration of
18 was also proved the same as those of
17 by the NOESY and ECD experiments.
All the compounds were evaluated for their tumor cell growth inhibition activity towards tumor cell lines A549 and MG63 [
18]. In the
in vitro bioassays, compounds
1–
3,
5,
6,
8–
12, and
14–
19 exhibited different level of growth inhibition against tested tumor cells whereas compounds
4,
7,
13,
20, and
21 were not active (
Table 6). Compound
8 showed potent growth inhibition towards both tumor cell lines, being similar as that of positive control adriamycin. This observation, when comparing with the activity of
9 and
20, showed a positive contribution of the 12-
O-isovalerate to the activity as described previously [
10,
11]. The increased activity of
6 with respect to that of
7 further supported the above conclusion. The replacement of an acetyl group by an isovaleryl group at C-13 will marked decrease the activity as observed in
12 and
19, and
1 and
11 as well. The observation was in good agreement with the remarkably decreasing activity of
2 and
10 with respect to their 12,13-interchangeble analogues
11 and
13, respectively. Similar situation was also suggested for C-14 by comparing the activity of
3 and
21 with those
4 and
17, respectively. These facts suggested that 13- or 14-
O-isovalerate may decrease the activity. As for the isovaleric acetyl substitution at C-2,
8 and
12 showed a marked increasing activity comparing with their 2-OAc or glycolyl analogues whereas
13 showed a marked decreasing activity comparing with its 2-glycolyl analogue. This observation led to somewhat confliction for the contribution of isovaleric acetyl or glycoly to the activity. This confliction was also observed for 16-substitutio when 16-OMe briaranes
11 and
12 compared with their 16-Cl analogues
14 and
13, respectively.
Table 6.
Cytotoxic assay for compounds 1–21 (IC50 μM).
Table 6.
Cytotoxic assay for compounds 1–21 (IC50 μM).
| Compound | A549 | MG63 | Compound | A549 | MG63 |
|---|
| 1 | 14.7 | 28.7 | 12 | >37.8 | 37.8 |
| 2 | 19.4 | 22.8 | 13 | - | - |
| 3 | 17.9 | 42.7 | 14 | 13.4 | 12.1 |
| 4 | - | - | 15 | 78.5 | 25.8 |
| 5 | 20.1 | 41.3 | 16 | 10.1 | 17.7 |
| 6 | 27.4 | 33.0 | 17 | 28.7 | >100.0 |
| 7 | - | - | 18 | 16.8 | - |
| 8 | 5.0 | 5.0 | 19 | 9.7 | 14.9 |
| 9 | 27.7 | 37.5 | 20 | - | - |
| 10 | 39.9 | 9.1 | 21 | - | - |
| 11 | - | 39.0 | Adriamycin | 2.8 | 3.2 |
3. Experimental Section
3.1. General Experimental Procedures
Commercial silica gel (Yantai, China, 200–300; 400–500 mesh) was used for column chromatography. Precoated silica gel plates (Yantai, China, HSGF-254) were used for analytical Thin Layer Chromatography (TLC). Spots were detected on TLC under UV or by heating after spraying with anisaldehyde-sulphuric acid reagent. The NMR spectra were recorded at 300 K on Bruker DRX 400 and Avance 600 spectrometers. Chemical shifts are reported in parts per million (δ), with use of the residual CDCl3 signal (δH = 7.27 ppm) as an internal standard for 1H NMR and CDCl3 (δC = 77.02 ppm) for 13C NMR; Coupling constants (J) in Hz. 1H NMR and 13C NMR assignments were supported by 1H-1H COSY, HSQC, HMBC and NOESY experiments. The following abbreviations are used to describe spin multiplicity: s = singlet, d = doublet, t = triplet, m = multiplet, br s = broad singlet, br d = broad doublet, dd = doublet of doublets, ov = overlapped signals. Optical rotations were measured in CHCl3 on an Autopol IV polarimeter at the sodium D line (590 nm). Infrared spectra were recorded in thin polymer films on a Nexus 470 FT-IR spectrophotometer (Nicolet); peaks are reported in cm−1. UV absorption spectra were recorded on a Varian Cary 100 UV-Vis spectrophotometer; peaks wavelengths are reported in nm. Circular dichroism (CD) spectra were recorded on a JASCO J-715 CD Spectropolarimeter. The mass spectra and high resolution mass spectra were performed on a Q-TOF Micro mass spectrometer, resolution 5000. An isopropyl alcohol solution of sodium iodide (2 mg/mL) was used as a reference compound. Semi-preparative RP-HPLC was performed on an Agilent 1100 system equipped with a refractive index detector using an YMC Pack ODS-A column (particle size 5 μm, 250 × 10 mm).
3.2. Animal Material
The South China Sea gorgonian coral Dichotella gemmacea (ZS-3, 3.5 kg, wet weight and ZH-1, 10.0 kg, wet weight) were collected from the South China Sea, in August 2007 and December 2011, and identified by Dr. Xiu-Bao Li, South China Sea Institute of Oceanology, Chinese Academy of Sciences. The voucher specimens (ZS-3, ZH-1) were deposited in the Second Military Medical University.
3.3. Extraction and Isolation
The frozen animals of ZS-3 (3.5 kg, wet weight) were extracted ultrasonically for three times with acetone and MeOH, respectively. The combined residue was partitioned between H2O and EtOAc to afford 16.1 g of EtOAc extract. The EtOAc extract was further partitioned between MeOH and hexane, affording 11.2 g of MeOH soluble residue. The MeOH extract was subjected to column chromatography (CC) on silica to give 16 fractions, using hexane/acetone (from 100:0 to 0:100) as eluent. Fraction 4 was further fractionated by RP-silical gel column chromatography (gradient elution from MeOH/H2O, 3:7 to MeOH, in 5% increments) and purified by HPLC (MeOH/H2O, 80:20, 1.5 mL/min) to yield 8 (2.5 mg, 49.2 min). Fraction 5 was subjected to repeated CCs on normal phase silica gel, Sephadex LH-20, and RP-silical gel, to give two subfractions 5-A and 5-B. Subfraction 5-A was purified by HPLC (MeOH/H2O, 75:25, 1.5 mL/min) to yield 2 (3.1 mg, 22.6 min) while subfraction 5-B was split by HPLC (MeOH/H2O, 77:23, 1.5 mL/min) into 12 (0.8 mg, 38.2 min), 10 (4.5 mg, 47.7 min) and 15 (1.8 mg, 50.8 min). Fraction 6 was chromomatographied over Sephadex LH-20 (CHCl3/MeOH, 1:1) to give four sub-fractions (A–D). HPLC purification on sub-fractions A (MeOH/H2O, 70:30, 1.5 mL/min) and B (MeOH/H2O, 70:30, 1.5 mL/min) gave 5 (2.7 mg, 25.7 min) and 6 (1.7 mg, 61.7 min), respectively. Fraction 7 was subjected to CCs on normal phase silica gel and Sephadex LH-20 to give two subfraction 7-A and 7-B. Further purification on both subfrations yielded 19 (4.3 mg, 51.7 min) from 7-A (MeOH/H2O, 70:30, 1.5 mL/min) and 3 (1.6 mg, 66.7 min) from7-B (eluent MeOH/H2O, 65:35, 1.5 mL/min). Fraction 9 was purified by HPLC (MeOH/H2O, 67:33, 1.5 mL/min) to yield 1 (1.6 mg, 24.5 min). Fraction 10 was chromatographied over normal phase silica gel and Sephadex LH-20 and then purified by HPLC (eluent MeOH/H2O, 70:30, 1.5 mL/min) yielding 18 (3.8 mg, 28.7 min). Fraction 11 was further fractionated by RP-silical gel CC (MeOH/H2O, 27:73 to 76:24) to give five subfractions (11A–11E). Subfraction 11C was purified by HPLC (eluent MeOH/H2O, 63:37, 1.5 mL/min) to yield 11 (1.1 mg, 26.8 min) and 14 (1.0 mg, 27.9 min). Subfraction 11D was purified by HPLC (MeOH/H2O, 72:28, 1.5 mL/min), yielding 4 (0.9 mg, 27.7 min) and 16 (1.0 mg, 28.1 min). Fraction 12 was further fractionated by normal phase silica gel CC (n-hexane/acetone, 4:1 to 1:1, in 5 increments) to give three subfractions (12A–12C). 13 (1.1 mg, 29.5 min) was obtained from subfraction 12B by HPLC (MeOH/H2O, 73:27, 1.5 mL/min). Fraction 13 was fractionated by RP-silical gel column chromatography (gradient elution from MeOH/H2O, 2:7 to 2:1, in 5% increments) to give 6 sub-fractions (13A–13F), a HPLC purification (eluent MeOH/H2O, 65:35, 1.5 mL/min) on 13E yielded 17 (4.6 mg, 32.6 min).
The frozen animals of ZH-1 (10.0 kg, wet weight) were extracted partitioned using the above procedure to afford 20.0 g of Et2O extract. The residue was subjected to Sephadex LH-20 (CHCl3/MeOH, 1:1) give 8 fractions. Fraction 2 was subjected to reversed-phasesilica gel (gradient MeOH/H2O, from 1:9 to 4:1), followed by HPLC (MeOH/H2O, 65:35, 1.5 mL/min) to yield 9 (2.5 mg, 52.5 min), 20 (5.2 mg, 67.5 min). Fraction 3 was chromatographed on a silica gel column (gradient n-hexane/acetone, from 5:1 to 1:2) and HPLC (MeOH/H2O, 50:50, 1.5 mL/min) to yield 7 (5.5 mg, 69.2 min), 21 (3.5 mg, 16.1 min).
Gemmacolide AA (
1): White amorphous powder; [α]
24D = −44 (
c 0.24, CHCl
3); UV (MeOH) λ
max (log ε) 206(1.85) nm; CD (CH
3CN,
c 2.0 × 10
−4) λ
max (Δε) positive below 190 nm, 201.5(−7.26) nm; IR (film) ν
max 3470, 1775, 1741 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 1,
Table 2; ESI-MS
m/
z 691 [M + Na]
+; HRESI-MS
m/
z 691.2150 [M + Na]
+ (calcd. for C
31H
40O
16Na, 691.2214).
Gemmacolide AB (
2): White amorphous powder; [α]
24D = −52 (
c 0.105, CHCl
3); UV (MeOH) λ
max (log ε) 205(1.78) nm; CD (CH
3CN,
c 1.8 × 10
−4) λ
max (Δε) positive below 190 nm, 216(−5.61) nm; IR (film) ν
max 3485, 1778, 1744 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 1,
Table 2; ESI-MS
m/
z 733 [M + Na]
+; HRESI-MS
m/
z 733.2680 [M + Na]
+ (calcd. for C
34H
46O
16Na, 733.2680).
Gemmacolide AC (
3): White amorphous powder; [α]
24D = −28 (
c 0.04, CHCl
3); UV (MeOH) λ
max (log ε) 205(1.71) nm; CD (CH
3CN,
c 2.6 × 10
−4) λ
max (Δε) positive below 190 nm, 215.5(−6.41) nm; IR (film) ν
max 3467, 1774, 1742 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 1,
Table 2; ESI-MS
m/
z 717 [M + Na]
+; HRESI-MS
m/
z 717.2737 [M + Na]
+ (calcd. for C
34H
46O
15Na, 717.2734).
Gemmacolide AD (
4): White amorphous powder; [α]
24D = −11 (
c 0.09, CHCl
3); UV (MeOH) λ
max (log ε) 213(1.73) nm; CD (CH
3CN,
c 2.3 × 10
−4) λ
max (Δε) positive below 197 nm, 212.5(−6.01) nm; IR (film) ν
max 3477, 1778, 1743 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 1,
Table 2; ESI-MS
m/
z 759 [M + Na]
+; HRESI-MS
m/
z 759.3207 [M + Na]
+ (calcd. for C
37H
52O
15Na, 759.3204).
Gemmacolide AE (
5): White amorphous powder; [α]
24D = −5 (
c 0.085, CHCl
3); UV (MeOH) λ
max (log ε) 204(1.41) nm; CD (CH
3CN,
c 3.0 × 10
−4) λ
max (Δε) positive below 190 nm, 209(−6.01) nm; IR (film) ν
max 3483, 1777, 1742 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 1,
Table 2; ESI-MS
m/
z 659 [M + Na]
+; HRESI-MS
m/
z 659.2676 [M + Na]
+ (calcd. for C
32H
44O
13Na, 659.2680).
Gemmacolide AF (
6): White amorphous powder; [α]
24D = −40 (
c 0.085, CHCl
3); UV (MeOH) λ
max (log ε) 204(1.36) nm; CD (CH
3CN,
c 1.6 × 10
−4) λ
max (Δε) positive below 190 nm, 200(−10.53) nm; IR (film) ν
max 3462, 1776, 1743 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 1,
Table 3; ESI-MS
m/
z 787 [M + Na]
+; HRESI-MS
m/
z 787.3150 [M + Na]
+ (calcd. for C
38H
52O
16Na, 787.3153).
Gemmacolide AG (
7): White amorphous powder; [α]
24D = −26.9 (
c 0.17, CHCl
3); UV (MeOH) λ
max (log ε) 209(1.65) nm; CD (CH
3CN,
c 2.3 × 10
−4) λ
max (Δε) positive below 190 nm, 202.0(−10.05) nm; IR (film) ν
max 3473, 1778, 1738 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 1,
Table 3; ESI-MS
m/
z 745 [M + Na]
+; HRESI-MS
m/
z 745.2686 [M + Na]
+ (calcd. for C
35H
46O
16Na, 745.2684).
Gemmacolide AH (
8): White amorphous powder; [α]
24D = −11 (
c 0.035, CHCl
3); UV (MeOH) λ
max (log ε) 206(1.88) nm; CD (CH
3CN,
c 2.8 × 10
−4) λ
max (Δε) positive below 190 nm, 199.0(−11.96) nm; IR (film) ν
max 3469, 1775, 1744 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 1,
Table 3; ESI-MS
m/
z 887 [M + Na]
+; HRESI-MS
m/
z 887.3673 [M + Na]
+ (calcd. for C
43H
60O
18Na, 887.3677).
Gemmacolide AI (
9): White amorphous powder; [α]
24D = −21 (
c 0.25, CHCl
3); UV (MeOH) λ
max (log ε) 212(1.72) nm; CD (CH
3CN,
c 2.5 × 10
−4) λ
max (Δε) positive below 190 nm, 204.5(−12.5) nm; IR (film) ν
max 3469, 1778, 1741 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 1,
Table 3; ESI-MS
m/
z 815 [M + Cl]
−; HRESI-MS
m/
z 815.2888 [M + Cl]
− (calcd. for C
38H
52ClO
17, 815.2893).
Gemmacolide AJ (
10): White amorphous powder; [α]
24D = −49 (
c 0.10, CHCl
3); UV (MeOH) λ
max (log ε) 204(1.49) nm; CD (CH
3CN,
c 3.1 × 10
−4) λ
max (Δε) positive below 190 nm, 203(−13.32) nm; IR (film) ν
max 3474, 1778, 1744 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 3,
Table 4; ESI-MS
m/
z 821 [M + Na]
+; HRESI-MS
m/
z 821.2767 [M + Na]
+ (calcd. for C
38H
51O
16NaCl, 821.2763).
Gemmacolide AK (
11): White amorphous powde; [α]
24D = −11.5 (
c 0.11, CHCl
3); UV (MeOH) λ
max (log ε) 201(1.16) nm; CD (CH
3CN,
c 3.4 × 10
−4) λ
max (Δε) positive below 198 nm, 210.5(−6.89) nm; IR (film) ν
max 3478, 1778, 1744 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 3,
Table 4; ESI-MS
m/
z 733 [M + Na]
+; HRESI-MS
m/
z 733.2690 [M + Na]
+ (calcd. for C
34H
46O
16Na, 733.2684).
Gemmacolide AL (
12): White amorphous powder; [α]
24D = −36 (
c 0.10, CHCl
3); UV (MeOH) λ
max (log ε) 205 (1.78) nm; CD (CH
3CN,
c 3.3 × 10
−4) λ
max (Δε) positive below 190 nm, 205(−8.58) nm; IR (film) ν
max 3475, 1776, 1745 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 4,
Table 5; ESI-MS
m/
z 817 [M + Na]
+; HRESI-MS
m/
z 817.3252 [M + Na]
+ (calcd. for C
39H
54O
17Na, 817.3259).
Gemmacolide AM (
13): White amorphous powder; [α]
24D = −11.3 (
c 0.11, CHCl
3); UV (MeOH) λ
max (log ε) 206(1.55) nm; CD (CH
3CN,
c 3.2 × 10
−4) λ
max (Δε) positive below 190 nm, 199(−4.05) nm; IR (film) ν
max 3468, 1779, 1731 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 4,
Table 5; ESI-MS
m/
z 833 [M + Cl]
−, HRESI-MS
m/
z 833.2549 [M + Cl]
− (calcd. for C
38H
51Cl
2O
16 833.2554).
Gemmacolide AN (
14): White amorphous powder; [α]
24D = −14.8 (
c 0.10, CHCl
3); UV (MeOH) λ
max (log ε) 205(1.58) nm; CD (CH
3CN,
c 3.3 × 10
−4) λ
max (Δε) positive below 190 nm, 203.5(−4.02) nm; IR (film) ν
max 3465, 1778, 1738 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 4,
Table 5; ESI-MS
m/
z 737 [M + Na]
+; HRESI-MS
m/
z 737.2182 [M + Na]
+ (calcd. for C
33H
43O
15ClNa, 737.2188).
Gemmacolide AO (
15): White amorphous powder; [α]
24D = −39 (
c 0.08, CHCl
3); UV (MeOH) λ
max (log ε) 204(1.52) nm; CD (CH
3CN,
c 1.5 × 10
−4) λ
max (Δε) positive below 190 nm, 204.5(−13.93) nm; IR (film) ν
max 3468, 1778, 1743 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 4,
Table 5; ESI-MS
m/
z 845 [M + Na]
+; HRESI-MS
m/
z 845.3575 [M + Na]
+ (calcd. for C
41H
58O
17Na, 845.3572).
Gemmacolide AP (
16): White amorphous powder; [α]
24D = −51.3 (
c 0.10, CHCl
3); UV (MeOH) λ
max (log ε) 214(1.89) nm; CD (CH
3CN,
c 2.0 × 10
−4) λ
max (Δε) positive below 195 nm, 203.5(−2.25) nm; IR (film) ν
max 3478, 1778, 1743 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 4,
Table 5; ESI-MS
m/
z 749 [M + Cl]
−; HRESI-MS
m/
z 749.1981 [M + Cl]
− (calcd. for C
33H
43Cl
2O
15, 749.1979).
Gemmacolide AQ (
17): White amorphous powder; [α]
24D = −36 (
c 0.175, CHCl
3); UV (MeOH) λ
max (log ε) 204(1.41) nm; CD (CH
3CN,
c 2.1 × 10
−4) λ
max (Δε) positive below 190 nm, 200(−4.85) nm; IR (film) ν
max 3479, 1770, 1743 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 4,
Table 5; ESI-MS
m/
z 703 [M + Na]
+; HRESI-MS
m/
z 703.2572 [M + Na]
+ (calcd. for C
33H
44O
15Na, 703.2578).
Gemmacolide AR (
18): White amorphous powder; [α]
24D = −38 (
c 0.08, CHCl
3); UV (MeOH) λ
max (log ε) 210(2.54) nm; CD (CH
3CN,
c 8.8 × 10
−4) λ
max (Δε) positive below 190 nm, 198(−4.11) nm; IR (film) ν
max 3469, 1777, 1742 cm
−1;
1H and
13C NMR spectroscopic data, see
Table 4,
Table 5; ESI-MS
m/
z 803 [M + Na]
+; HRESI-MS
m/
z 745.2689 [M + Na]
+ (calcd. for C
35H
46O
16Na, 745.2684).
3.4. Cytotoxicity Assay
Cytotoxicity was tested against human lung adenocarcinoma (A549) and human osteosarcoma cell (MG63), using a modification of the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) colorimetric method [
18]. Adriamycin was used as positive control, IC
50 = 2.8 μM for A549 cells and 3.2 μM for MG63 cells.