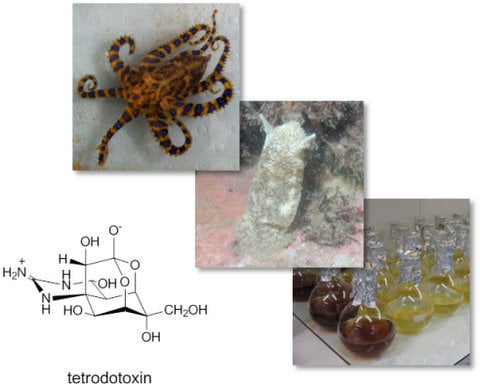
Diversity and Biosynthetic Potential of Culturable Microbes Associated with Toxic Marine Animals
Abstract
:1. Introduction

2. Results and Discussion
2.1. Bacterial Diversity in Toxic Hapalochlaena sp. and Pleurobranchaea maculata
| Strain | Closest 16S rRNA gene BLAST matcha | Identity | PCR screening | TTX | ||
|---|---|---|---|---|---|---|
| (%) | PKS | NRPS | AMT | Detectionb | ||
| HM BE02 | Alteromonadales bacterium HOT3G5 (HQ537362.1) | 99 | - | - | - | - |
| HM BE03 | Alteromonas sp. S2542 (FJ457277.1) | 99 | - | - | - | - |
| HM BE04 | Alteromonas sp. S2542 (FJ457277.1) | 99 | - | - | - | - |
| HM BE05 | Pseudoalteromonas sp. AmSamW21 (GU903211.1) | 99 | + | - | - | - |
| HM BE06 | Vibrio rotiferianus isolate AP17 (HE584775.1) | 99 | - | - | - | - |
| HM LI01 | Nautellaitalica strain R-28753 (AM944522.1) | 99 | - | - | + | NT |
| HM LI02 | Alteromonas sp. S2542 (FJ457277.1) | 99 | - | - | - | - |
| HM LI03 | Pseudoalteromonadaceae bacterium S3 (HQ164448.1) | 100 | - | - | - | NT |
| HM LI04 | Nautellaitalica strain R-28753 (AM944522.1) | 99 | - | - | - | NT |
| HM LI05 | Alteromonas sp. S2542 (FJ457277.1) | 99 | - | - | - | NT |
| HM LI06 | Pseudoalteromonas sp. AKA07-4 (AB571944.1) | 99 | - | - | - | NT |
| HM OE02 | Pseudoalteromonadaceae bacterium S3 (HQ164448.1) | 100 | - | - | + | NT |
| HM OE03 | Alteromonas sp. CF14-3 (FJ170033.1) | 99 | - | - | - | - |
| HM OE04 | Thalassomonas sp. PaD1.04 (GQ391976.1) | 98 | - | - | - | - |
| HM OE07 | Pseudoalteromonas sp. TB51 (JF273853.1) | 99 | - | - | - | - |
| HM OE08 | Colwellia sp. KMD002 (EU599214.3) | 98 | - | - | - | NT |
| HM OE09 | Pseudoalteromonadaceae bacterium S3 (HQ164448.1) | 99 | + | - | + | NT |
| HM SA02 | Pseudoalteromonas sp. AKA07-4 (AB571944.1) | 99 | + | + | + | - |
| HM SA03 | Pseudoalteromonadaceae bacterium S3 (HQ164448.1) | 99 | + | + | + | - |
| HM SA04 | Pseudoalteromonas sp. NW 4327 strain NW (FR839670.1) | 99 | + | - | - | NT |
| HM SA05 | Marinomonas communis strain LMG 2864 (DQ011528.1) | 99 | - | - | - | NT |
| HM SA06 | Alteromonas sp. KB19 (HM583350.1) | 99 | - | - | - | - |
| Strain | Closest 16S rRNA gene BLAST match a | Identity | PCR screening | TTX | ||
|---|---|---|---|---|---|---|
| (%) | PKS | NRPS | AMT | Detection b | ||
| PM DT05 | Photobacterium kishitanii strain S-27 (JF412253.1) | 99 | - | - | - | - |
| PM DT08 | Shewanella schlegeliana (AB081761.1) | 99 | - | - | - | - |
| PM DT10 | Photobacterium sp. DFC2.17 isolate DFC2.17 (FR873783.1) | 99 | - | - | - | - |
| PM DT11 | Photobacterium kishitanii strain S-27 (JF412253.1) | 99 | - | - | - | NT |
| PM DT12 | Shewanella schlegeliana (AB081761.1) | 99 | + | - | - | - |
| PM EG02 | Vibrio sp. YDO5 (GU586127.1) | 98 | - | - | - | - |
| PM EG04 | Staphylococcus warneri strain Na58 (HQ831387.1) | 99 | - | - | - | - |
| PM EG05 | Staphylococcus warneri strain Na59 (HQ831388.1) | 99 | - | - | - | - |
| PM EG08 | Pseudoalteromonas sp. 114Z-11 (GU584139.1) | 99 | - | - | - | - |
| PM EG09 | Pseudoalteromonas sp. BSs20138 (EU365489.1) | 99 | - | - | - | - |
| PM EG11 | Alteromonas sp. N98(2010) (HQ188650.1) | 99 | - | + | - | - |
| PM EG13 | Pseudoalteromonas sp. LJ1 (FJ665500.1) | 99 | - | - | - | - |
| PM EG14 | Halomonas sp. Pper-Hx-1972 (EU123940.1) | 99 | + | + | - | - |
| PM EG15 | Vibrio sp. S4639 (FJ457601.1) | 99 | - | - | - | - |
| PM EG17 | Pseudoalteromonas sp. 114Z-11 (GU584139.1) | 98 | - | - | - | - |
| PM EG18 | Pseudoalteromonas porphyrae strain HK1 (FJ205736.1) | 99 | + | + | - | - |
| PM GO01 | Shewanella schlegeliana (AB081761.1) | 99 | + | - | - | - |
| PM GO04 | Vibrio splendidus isolate PB1-10rrnM (EU091337.1) | 99 | - | - | - | - |
| PM GO05 | Photobacterium kishitanii strain calba.5.9 (AY642170.1) | 99 | - | - | - | - |
| PM GO06 | Shewanella schlegeliana (AB081761.1) | 99 | + | - | - | - |
| PM GO08 | Photobacterium kishitanii strain calba.5.9 (AY642170.1) | 99 | - | - | - | - |
| PM GO09 | Vibrio fischeri strain SI1E (AY292949.1) | 99 | - | - | - | NT |
| PM GO12 | Photobacterium kishitanii strain calba.5.9 (AY642170.1) | 99 | - | - | - | NT |
| PM GO13 | Vibrio fischeri strain SI1Ecomplete (AY292949.1) | 99 | - | - | - | NT |
| PM RT05 | Pseudoalteromonas sp. LJ1 (FJ665500.1) | 99 | - | + | - | - |
| PM RT07 | Alteromonas sp. N98(2010) (HQ188650.1) | 99 | - | + | - | - |
| PM RT10 | Vibrio parahaemolyticus strain NMGB2 (JN561593.1) | 99 | - | - | - | NT |

2.2. Overestimation of Abundance and Diversity of TTX-Producing Bacteria
| Host Organism | Tissue tested | TTX concentration (mg/kg) |
|---|---|---|
| Hapalochlaena sp. | Posterior salivary gland | BDL a |
| Beak | BDL | |
| Pleurobranchaea maculata | Eggs | 5.19 |
| Digestive tract | 4.23 | |
| Reproductive tract | 3.84 | |
| Gonads | 3.54 |
| Strain a | PCR screening | TTX detection (ELISA) | TTX detection | ||
|---|---|---|---|---|---|
| PKS | NRPS | AMT | (ng/g) b | (LC-MS) | |
| Vibrio sp. 34HU9 (EU268265) | + | + | − | 169 | BDL c |
| Marinomonas sp. 38JIA1 (EU268259) | − | + | − | 98 | BDL |
| Tenacibaculum sp. 30ORI8 (EU268261) | − | + | − | 54 | BDL |
2.3. Mining Host-Associated Bacteria for Proposed TTX Biosynthesis Genes
3. Experimental Section
3.1. Specimen Collection
3.2. Specimen Dissection
3.3. Bacterial Culturing and Genomic DNA Extraction
3.4. Identification of Bacterial Isolates
3.5. PCR Screening of Bacterial Isolates for Biosynthesis Genes
| Primer | Sequence | Tma | ATb | Reference |
|---|---|---|---|---|
| 27fl | AGAGTTTGATCCTGGCTCAG | 61 | 55 | [41] |
| 1494rc | TACGGCTACCTTGTTACGAC | 59 | 55 | [41] |
| DKF | GTGCCGGTNCC(A/G)TGNG(T/C)(T/C)TC | 67 | 55 | [42] |
| DKR | GCGATGGA(T/C)CCNCA(A/G)CA(A/G)(C/A)G | 65 | 55 | [42] |
| MTF2 | GCNGG(C/T)GG(C/T)GCNTA(C/T)GTNCC | 64 | 52 | [43] |
| MTR2 | CCNCG(A/G/T)AT(T/C)TTNAC(T/C)TG | 47 | 52 | [43] |
| ATfwd1 | GT(A/C/G)TG(T/C)CC(A/T)(A/C)G(G/C)GA(T/C)GT(A/C/G)ATG | 57 | 55 | [44] |
| ATrev1 | AT(A/G)TCCCA(A/T)(A/G)T(C/G/T)C(A/G)CA(A/G)TG | 62 | 55 | [44] |
3.6. Phylogenetic Analysis of 16S rRNA Gene Sequences
3.7. Toxin Extraction from Organ Homogenates and Bacterial Isolates
3.8. Analysis of Extracts by Liquid Chromatography-Mass Spectrometry
4. Conclusions
Acknowledgements
Conflict of Interest
References
- Pawlik, J.R. Marine invertebrate chemical defenses. Chem. Rev. 1993, 93, 1911–1922. [Google Scholar] [CrossRef]
- Ahasan, H.; Mamun, A.A.; Karim, S.R.; Bakar, M.A.; Gazi, E.A.; Bala, C.S. Paralytic complications of puffer fish (tetrodotoxin) poisoning. Singap. Med. J. 2004, 45, 73–74. [Google Scholar]
- Venkatesh, B.; Lu, S.Q.; Dandona, N.; See, S.L.; Brenner, S.; Soong, T.W. Genetic basis of tetrodotoxin resistance in pufferfishes. Curr. Biol. 2005, 15, 2069–2072. [Google Scholar] [CrossRef]
- Shiomi, K.; Yamaguchi, S.; Kikuchi, T.; Yamamori, K.; Matsui, T. Occurrence of tetrodotoxin-binding high molecular weight substances in the body fluid of shore crab (Hemigrapsus sanguineus). Toxicon 1992, 30, 1529–1537. [Google Scholar] [CrossRef]
- Chau, R.; Kalaitzis, J.A.; Neilan, B.A. On the origins and biosynthesis of tetrodotoxin. Aquat. Toxicol. 2011, 104, 61–72. [Google Scholar] [CrossRef]
- Sheumack, D.; Howden, M.; Spence, I. Occurrence of a tetrodotoxin-like compound in the eggs of the venomous blue-ringed octopus (Hapalochlaena maculosa). Toxicon 1984, 22, 811–812. [Google Scholar] [CrossRef]
- McNabb, P.; Selwood, A.I.; Munday, R.; Wood, S.A.; Taylor, D.I.; MacKenzie, L.A.; van Ginkel, R.; Rhodes, L.L.; Cornelisen, C.; Heasman, K. Detection of tetrodotoxin from the grey side-gilled sea slug—Pleurobranchaea maculata, and associated dog neurotoxicosis on beaches adjacent to the Hauraki Gulf, Auckland, New Zeala. Toxicon 2010, 56, 466–473. [Google Scholar] [CrossRef]
- Wang, X.; Yu, R.; Luo, X.; Zhou, M.; Lin, X. Toxin-screening and identification of bacteria isolated from highly toxic marine gastropod Nassarius semiplicatus. Toxicon 2008, 52, 55–61. [Google Scholar] [CrossRef]
- Noguchi, T.; Arakawa, O. Tetrodotoxin—Distribution and accumulation in aquatic organisms, and cases of Human intoxication. Mar. Drugs 2008, 6, 220–242. [Google Scholar]
- Noguchi, T.; Arakawa, O.; Takatani, T. TTX accumulation in pufferfish. Comp. Biochem. Physiol. D Genomics Proteomics 2006, 1, 145–152. [Google Scholar] [CrossRef]
- Wu, Z.; Xie, L.; Xia, G.; Zhang, J.; Nie, Y.; Hu, J.; Wang, S.; Zhang, R. A new tetrodotoxin-producing actinomycete, Nocardiopsis dassonvillei, isolated from the ovaries of puffer fish Fugu rubripes. Toxicon 2005, 45, 851–859. [Google Scholar] [CrossRef]
- Lehman, E.M.; Brodie, E.D. No evidence for an endosymbiotic bacterial origin of tetrodotoxin in the newt Taricha granulosa. Toxicon 2004, 44, 243–249. [Google Scholar] [CrossRef]
- Kotaki, Y.; Shimizu, Y. 1-Hydroxy-5,11-dideoxytetrodotoxin, the first N-hydroxy and ring-deoxy derivative of tetrodotoxin found in the newt Taricha granulosa. J. Am. Chem. Soc. 1993, 115, 827–830. [Google Scholar] [CrossRef]
- Woodward, R.; Gougoutas, J. The structure of tetrodotoxin. J. Am. Chem. Soc. 1964, 86, 5030–5030. [Google Scholar] [CrossRef]
- Yasumoto, T.; Yotsu, M.; Murata, M.; Naoki, H. New tetrodotoxin analogs from the newt Cynops ensicauda. J. Am. Chem. Soc. 1988, 110, 2344–2345. [Google Scholar] [CrossRef]
- Shimizu, Y.; Kobayashi, M. Apparent lack of tetrodotoxin biosynthesis in captured Taricha torosa and Taricha granulosa. Chem. Pharm. Bull. 1983, 31, 3625–3631. [Google Scholar] [CrossRef]
- Kellmann, R.; Mihali, T.; Jeon, Y.; Pickford, R.; Pomati, F.; Neilan, B. Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol. 2008, 74, 4044–4053. [Google Scholar] [CrossRef]
- Mihali, T.K.; Kellmann, R.; Muenchhoff, J.; Barrow, K.D.; Neilan, B.A. Characterization of the gene cluster responsible for cylindrospermopsin biosynthesis. Appl. Environ. Microbiol. 2008, 74, 716–722. [Google Scholar] [CrossRef]
- Shen, Y.; Yoon, P.; Yu, T.-W.; Floss, H.G.; Hopwood, D.; Moore, B.S. Ectopic expression of the minimal whiE polyketide synthase generates a library of aromatic polyketides of diverse sizes and shapes. Proc. Natl. Acad. Sci. USA 1999, 96, 3622–3627. [Google Scholar] [CrossRef]
- Gallacher, S.; Birkbeck, T. Effect of phosphate concentration on production of tetrodotoxin by Alteromonas tetraodonis. Appl. Environ. Microbiol. 1993, 59, 3981–3983. [Google Scholar]
- Clardy, J.; Walsh, C. Lessons from natural molecules. Nature 2004, 432, 829–837. [Google Scholar] [CrossRef]
- Pettit, R. Mixed fermentation for natural product drug discovery. Appl. Microbiol. Biotechnol. 2009, 83, 19–25. [Google Scholar] [CrossRef]
- Kurosawa, K.; Ghiviriga, I.; Sambandan, T.G.; Lessard, P.A.; Barbara, J.E.; Rha, C.; Sinskey, A.J. Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces padanus to Rhodococcus fascians. J. Am. Chem. Soc. 2008, 130, 1126–1127. [Google Scholar]
- Oh, D.-C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Nat. Prod. 2007, 70, 515–520. [Google Scholar] [CrossRef]
- Cheng, C.A.; Hwang, D.F.; Tsai, Y.H.; Chen, H.C.; Jeng, S.S.; Noguchi, T.; Ohwada, K.; Hasimoto, K. Microflora and tetrodotoxin-producing bacteria in a gastropod, Niotha clathrata. Food Chem. Toxicol. 1995, 33, 929–934. [Google Scholar] [CrossRef]
- Ruby, E.G. Symbiotic conversations are revealed under genetic interrogation. Nat. Rev. Microbiol. 2008, 6, 752–762. [Google Scholar] [CrossRef]
- Campbell, S.; Harada, R.M.; DeFelice, S.V.; Bienfang, P.K.; Li, Q.X. Bacterial production of tetrodotoxin in the pufferfish Arothron hispidus. Nat. Prod. Res. 2009, 23, 1630–1640. [Google Scholar] [CrossRef]
- Simidu, U.; Kita-Tsukamoto, K.; Yasumoto, T.; Yotsu, M. Taxonomy of four marine bacterial strains that produce tetrodotoxin. Int. J. Syst. Bacteriol. 1990, 40, 331–336. [Google Scholar] [CrossRef]
- Wang, J.; Fan, Y.; Yao, Z. Isolation of a Lysinibacillus fusiformis strain with tetrodotoxin-producing ability from puffer fish Fugu obscurus and the characterization of this strain. Toxicon 2010, 56, 640–643. [Google Scholar] [CrossRef]
- Yang, G.; Xu, J.; Liang, S.; Ren, D.; Yan, X.; Bao, B. A novel TTX-producing Aeromonas isolated from the ovary of Takifugu obscurus. Toxicon 2010, 56, 324–329. [Google Scholar] [CrossRef]
- Ritchie, K.; Nagelkerken, I.; James, S.; Smith, G. Environmental microbiology: A tetrodotoxin-producing marine pathogen. Nature 2000, 404, 354. [Google Scholar] [CrossRef]
- Wood, S.A.; Taylor, D.I.; McNabb, P.; Walker, J.; Adamson, J.; Cary, S.C. Tetrodotoxin Concentrations in Pleurobranchaea maculata: Temporal, Spatial and Individual Variability from New Zealand Populations. Mar. Drugs 2012, 10, 163–176. [Google Scholar] [CrossRef]
- Saker, M.L.; Griffiths, D.J. The effect of temperature on growth and cylindrospermopsin content of seven isolates of Cylindrospermopsis raciborskii (Nostocales, Cyanophyceae) from water bodies in northern Australia. Phycologia 2000, 39, 349–354. [Google Scholar] [CrossRef]
- Williams, B.L.; Hanifin, C.T.; Brodie, E.D.; Caldwell, R.L. Ontogeny of tetrodotoxin levels in blue-ringed octopuses: Maternal Investment and apparent independent production in offspring of Hapalochlaena lunulata. J. Chem. Ecol. 2011, 37, 10–17. [Google Scholar] [CrossRef]
- Wood, S.; Casas, M.; Taylor, D.; McNabb, P.; Salvitti, L.; Ogilvie, S.; Cary, S. Depuration of tetrodotoxin and changes in bacterial communities in Pleurobranchea maculata adults and egg masses maintained in captivity. J. Chem. Ecol. 2012, 38, 1342–1350. [Google Scholar] [CrossRef]
- Chaston, J.; Goodrich-Blair, H. Common trends in mutualism revealed by model associations between invertebrates and bacteria. FEMS Microbiol. Rev. 2010, 34, 41–58. [Google Scholar] [CrossRef]
- Hwang, D.; Arakawa, O.; Saito, T.; Noguchi, T.; Simidu, U.; Tsukamoto, K.; Shida, Y.; Hashimoto, K. Tetrodotoxin-producing bacteria from the blue-ringed octopus Octopus maculosus. Mar. Biol. 1989, 100, 327–332. [Google Scholar] [CrossRef]
- Do, H.; Kogure, K.; Simidu, U. Identification of deep-sea-sediment bacteria which produce tetrodotoxin. Appl. Environ. Microbiol. 1990, 56, 1162–1163. [Google Scholar]
- Matsumura, K. Reexamination of tetrodotoxin production by bacteria. Appl. Environ. Microbiol. 1995, 61, 3468–3470. [Google Scholar]
- Tillett, D.; Neilan, B.A. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 2000, 36, 251–258. [Google Scholar] [CrossRef]
- Neilan, B.A.; Jacobs, D.; Therese, D.D.; Blackall, L.L.; Hawkins, P.R.; Cox, P.T.; Goodman, A.E. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int. J. Syst. Evol. Microbiol. 1997, 47, 693–697. [Google Scholar]
- Moffitt, M.C.; Neilan, B.A. On the presence of peptide synthetase and polyketide synthase genes in the cyanobacterial genus Nodularia. FEMS Microbiol. Lett. 2001, 196, 207–214. [Google Scholar] [CrossRef]
- Neilan, B.A.; Dittmann, E.; Rouhiainen, L.; Bass, R.A.; Schaub, V.; Sivonen, K.; Borner, T. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J. Bacteriol. 1999, 181, 4089–4097. [Google Scholar]
- Kellmann, R. The Molecular Genetics of Cylindrospermopsis and Saxitoxin Biosynthesis. Ph.D. Dissertation, University of New South Wales, Sydney, Australia, 2005. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Los Alamos National Security: Los Alamos, NM, USA, 2005. FindModel. Available online: http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html (accessed on 18 August 2011).
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef]
- Samples Availability: Available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chau, R.; Kalaitzis, J.A.; Wood, S.A.; Neilan, B.A. Diversity and Biosynthetic Potential of Culturable Microbes Associated with Toxic Marine Animals. Mar. Drugs 2013, 11, 2695-2712. https://doi.org/10.3390/md11082695
Chau R, Kalaitzis JA, Wood SA, Neilan BA. Diversity and Biosynthetic Potential of Culturable Microbes Associated with Toxic Marine Animals. Marine Drugs. 2013; 11(8):2695-2712. https://doi.org/10.3390/md11082695
Chicago/Turabian StyleChau, Rocky, John A. Kalaitzis, Susanna A. Wood, and Brett A. Neilan. 2013. "Diversity and Biosynthetic Potential of Culturable Microbes Associated with Toxic Marine Animals" Marine Drugs 11, no. 8: 2695-2712. https://doi.org/10.3390/md11082695
APA StyleChau, R., Kalaitzis, J. A., Wood, S. A., & Neilan, B. A. (2013). Diversity and Biosynthetic Potential of Culturable Microbes Associated with Toxic Marine Animals. Marine Drugs, 11(8), 2695-2712. https://doi.org/10.3390/md11082695