Marine Invertebrate Xenobiotic-Activated Nuclear Receptors: Their Application as Sensor Elements in High-Throughput Bioassays for Marine Bioactive Compounds
Abstract
:1. Introduction

2. Bioactive Chemicals are Naturally Present in Animal Diets
3. Detoxification Pathways and Their Transcription-Level Regulation
4. Xenobiotic Receptors: Functions, Structures, and Taxonomic Distribution
4.1. Vertebrate Pregnane X Receptor

4.2. Non-Marine Invertebrate XANRs
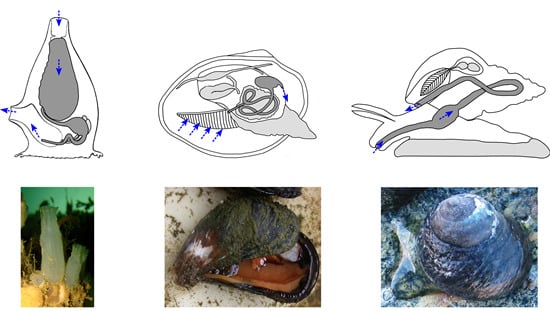
4.3. Marine Invertebrate XANRs
5. Marine Invertebrate Putative XANR LBDs are Activated by Known Marine Bioactive Compounds


6. Development of High-Throughput Bioassays Based on Marine Invertebrate XANR LBDs
6.1. XANR LBD-Based Bioassays: Technical Considerations and Challenges
6.1.1. NR LBD Bioassays Using Recombinant Yeast
6.1.2. Biosensors for High-Throughput Screening for NR LBD Ligands
6.2. XANR LBD-Based Assays: Biological, Ecological, and Evolutionary Considerations
7. Future Prospects for Marine Invertebrate XANR LBD-Based Bioassays
8. Conclusions
Abbreviations
| AD | activation domain |
| AhR | aryl hydrocarbon receptor |
| BaP | benzo [α] pyrene |
| BsVDR/PXRα | Botryllus schlosseri VDR/PXR orthologue α |
| CAR | constitutive androstane receptor |
| CiVDR/PXRα | Ciona intestinalis VDR/PXR orthologue α |
| CiVDR/PXRβ | Ciona intestinalis VDR/PXR orthologue β |
| CPRG | chlorophenol red-β-d-galactopyranoside |
| CYP | cytochrome P450 gene |
| DBD | DNA-binding domain |
| DDT | 1,1,1-trichloro-2,2-di(4-chlorophenyl)ethane |
| ER | estrogen receptor |
| GAL4 | yeast DNA-binding transcription factor |
| GST | glutathione S-transferases |
| HR96 | nuclear hormone receptor 96 |
| LBD | ligand-binding domain |
| MRP | multidrug resistance-associated protein |
| NR | nuclear receptor |
| NR1I | nuclear receptor sub-family 1, class I |
| NR1J | nuclear receptor sub-family 1, class J |
| NR1H | nuclear receptor sub-family 1, class H |
| OA | okadaic acid |
| PAHs | polycyclic aromatic hydrocarbons |
| PTX-2 | pectenotoxin-2 |
| PCN | pregnenolone 16α-carbonitrile |
| PXR | pregnane X receptor |
| RXR | retinoid X receptor |
| SRC-1 | steroid co-activator 1 |
| TCPOBOP | 1,4-Bis[2-(3,5-dichloropyridyloxy)]benzene |
| VDR | vitamin D receptor |
| VP16 | viral protein 16 |
| XANR | xenobiotic-activated nuclear receptor |
| XR | xenobiotic receptor |
Acknowledgments
Conflicts of Interest
References
- Denison, M.S.; Nagy, S.R. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 2003, 43, 309–334. [Google Scholar] [CrossRef] [PubMed]
- Kewley, R.J.; Whitelaw, M.L.; Chapman-Smith, A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004, 36, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Fujii-Kuriyama, Y.; Kawajiri, K. Molecular mechanisms of the physiological functions of the aryl hydrocarbon (dioxin) receptor, a multifunctional regulator that senses and responds to environmental stimuli. Proc. Jpn. Acad. Ser. B-Phys. Biol. Sci. 2010, 86, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.B.; Maglich, J.M.; McKee, D.D.; Wisely, B.; Willson, T.M.; Kliewer, S.A.; Lambert, M.H.; Moore, J.T. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol. Endocrinol. 2002, 16, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Krasowski, M.D.; Ni, A.; Hagey, L.R.; Ekins, S. Evolution of promiscuous nuclear hormone receptors: LXR, FXR, VDR, PXR, and CAR. Mol. Cell. Endocrinol. 2011, 334, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Reschly, E.J.; Krasowski, M.D. Evolution and function of the NR1I nuclear hormone receptor subfamily (VDR, PXR, and CAR) with respect to metabolism of xenobiotics and endogenous compounds. Curr. Drug Metab. 2006, 7, 349–365. [Google Scholar] [CrossRef] [PubMed]
- Echevarria, M.; Naar, J.P.; Tomas, C.; Pawlik, J.R. Effects of Karenia brevis on clearance rates and bioaccumulation of brevetoxins in benthic suspension feeding invertebrates. Aquat. Toxicol. 2012, 106, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Haberkorn, H.; Tran, D.; Massabuau, J.C.; Ciret, P.; Savar, V.; Soudant, P. Relationship between valve activity, microalgae concentration in the water and toxin accumulation in the digestive gland of the Pacific oyster Crassostrea gigas exposed to Alexandrium minutum. Mar. Pollut. Bull. 2011, 62, 1191–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forbey, J.S.; Dearing, M.D.; Gross, E.M.; Orians, C.M.; Sotka, E.E.; Foley, W.J. A Pharm-Ecological perspective of terrestrial and aquatic plant-herbivore interactions. J. Chem. Ecol. 2013, 39, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Raubenheimer, D.; Simpson, S.J. Nutritional PharmEcology: Doses, nutrients, toxins, and medicines. Integr. Comp. Biol. 2009, 49, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Kerppola, T.K. Regulation of Drosophila metamorphosis by xenobiotic response regulators. PLoS Genet. 2013, 9, e1003263. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Ramirez, F.A.; Vallim, M.A.; Cavalcanti, D.N.; Teixeira, V.L. Effects of the secondary metabolites from Canistrocarpus cervicornis (Dictyotales, Phaeophyceae) on fertilization and early development of the sea urchin Lytechinus variegatus. Lat. Am. J. Aquat. Res. 2013, 41, 296–304. [Google Scholar]
- Targett, N.M.; Arnold, T.M. Effects of secondary metabolites on digestion in marine herbivores. In Marine Chemical Ecology; CRC Press: Boca Raton, FL, USA, 2001; pp. 391–411. [Google Scholar]
- Hay, M.E. Marine chemical ecology: Chemical signals and cues structure marine populations, communities, and ecosystems. Annu. Rev. Mar. Sci. 2009, 1, 193–212. [Google Scholar] [CrossRef]
- Dearing, M.D.; Foley, W.J.; McLean, S. The influence of plant secondary metabolites on the nutritional ecology of herbivorous terrestrial vertebrates. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 169–189. [Google Scholar] [CrossRef]
- Marsh, K.J.; Wallis, I.R.; Andrew, R.L.; Foley, W.J. The detoxification limitation hypothesis: Where did it come from and where is it going? J. Chem. Ecol. 2006, 32, 1247–1266. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.; Avila, C.; Starmer, J.; Paul, V.J. A sequestered soft coral diterpene in the aeolid nudibranch Phyllodesmium guamensis. J. Exp. Mar. Biol. Ecol. 1998, 226, 33–49. [Google Scholar] [CrossRef]
- Glendinning, J.I. Is the bitter rejection response always adaptive? Physiol. Behav. 1994, 56, 1217–1227. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, J.; Mueller, K.L.; Hoon, M.A.; Adler, E.; Feng, L.X.; Guo, W.; Zuker, C.S.; Ryba, N.J.P. T2Rs function as bitter taste receptors. Cell 2000, 100, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Jones, G.; Zhang, S. Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol. Biol. 2009, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, J. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol. Biol. Evol. 2013, 7, 7. [Google Scholar] [CrossRef]
- Sugawara, T.; Go, Y.; Udono, T.; Morimura, N.; Tomonaga, M.; Hirai, H.; Imai, H. Diversification of bitter taste receptor gene family in western chimpanzees. Mol. Biol. Evol. 2011, 28, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.M.; Pawlik, J.R. Do coral reef fish learn to avoid unpalatable prey using visual cues? Anim. Behav. 2013, 85, 339–347. [Google Scholar] [CrossRef]
- Long, J.D.; Hay, M.E. Fishes learn aversions to a nudibranch’s chemical defense. Mar. Ecol. Prog. Ser. 2006, 307, 199–208. [Google Scholar] [CrossRef]
- Ritson-Williams, R.; Paul, V.J. Marine benthic invertebrates use multimodal cues for defense against reef fish. Mar. Ecol. Prog. Ser. 2007, 340, 29–39. [Google Scholar] [CrossRef]
- Hegaret, H.; Wikfors, G.H.; Shumway, S.E. Diverse feeding responses of five species of bivalve mollusc when exposed to three species of harmful algae. J. Shellfish Res. 2007, 26, 549–559. [Google Scholar] [CrossRef]
- Fdil, M.A.; Mouabad, A.; Outzourhit, A.; Benhra, A.; Maarouf, A.; Pihan, J.C. Valve movement response of the mussel Mytilus galloprovincialis to metals (Cu, Hg, Cd and Zn) and phosphate industry effluents from Moroccan Atlantic coast. Ecotoxicology 2006, 15, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Wildish, D.; Lassus, P.; Martin, J.; Saulnier, A.; Bardouil, M. Effect of the PSP-causing dinoflagellate, Alexandrium sp. on the initial feeding response of Crassostrea gigas. Aquat. Living Resour. 1998, 11, 35–43. [Google Scholar] [CrossRef]
- Glendinning, J.I. How do predators cope with chemically defended foods? Biol. Bull. 2007, 213, 252–266. [Google Scholar] [CrossRef] [PubMed]
- Manfrin, C.; de Moro, G.; Torboli, V.; Venier, P.; Pallavicini, A.; Gerdol, M. Physiological and molecular responses of bivalves to toxic dinoflagellates. Invertebr. Surviv. J. 2012, 9, 184–199. [Google Scholar]
- Fernandez-Reiriz, M.J.; Navarro, J.M.; Contreras, A.M.; Labarta, U. Trophic interactions between the toxic dinoflagellate Alexandrium catenella and Mytilus chilensis: Feeding and digestive behaviour to long-term exposure. Aquat. Toxicol. 2008, 87, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Sotka, E.E.; Gantz, J. Preliminary evidence that the feeding rates of generalist marine herbivores are limited by detoxification rates. Chemoecology 2013, 23, 233–240. [Google Scholar] [CrossRef]
- Sotka, E.E.; Forbey, J.; Horn, M. The emerging role of pharmacology in understanding consumer-prey interactions in marine and freshwater systems. Integr. Comp. Biol. 2009, 49, 291–313. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef] [PubMed]
- Oakley, A. Glutathione transferases: A structural perspective. Drug Metab. Rev. 2011, 43, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R.; Goldstone, J.V.; Stegeman, J.J. The cytochrome P450 genesis locus: The origin and evolution of animal cytochrome P450s. Philos. Trans. R. Soc. B-Biol. Sci. 2013, 368. [Google Scholar] [CrossRef]
- Testa, B.; Pedretti, A.; Vistoli, G. Foundation review: Reactions and enzymes in the metabolism of drugs and other xenobiotics. Drug Discov. Today 2012, 17, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Sladek, F.M. What are nuclear receptor ligands? Mol. Cell. Endocrinol. 2011, 334, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.E. Xenobiotics and the evolution of multicellular animals: Emergence and diversification of ligand-activated transcription factors. Integr. Comp. Biol. 2005, 45, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Iyanagi, T. Molecular mechanism of phase I and phase II drug-metabolizing enzymes: Implications for detoxification. In International Review of Cytology—A Survey of Cell Biology, 1st ed.; Jeon, K.W., Ed.; Elsevier Academic Press Inc: San Diego, CA, USA, 2007; Volume 260, pp. 35–112. [Google Scholar]
- Yang, Y.M.; Noh, K.; Han, C.Y.; Kim, S.G. Transactivation of genes encoding for phase II enzymes and phase III transporters by phytochemical antioxidants. Molecules 2010, 15, 6332–6348. [Google Scholar] [CrossRef] [PubMed]
- Sotka, E.E.; Whalen, K.E. Herbivore offense in the sea: The detoxification and transport of secondary metabolites. In Algal Chemical Ecology, XVIII ed.; Amsler, C.D., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 203–228. [Google Scholar]
- Court, M.H. Feline drug metabolism and disposition pharmacokinetic evidence for species differences and molecular mechanisms. Vet. Clin. N. Am. Small Anim. Pract. 2013, 43, 1039–1054. [Google Scholar] [CrossRef]
- Shrestha, B.; Reed, J.M.; Starks, P.T.; Kaufman, G.E.; Goldstone, J.V.; Roelke, M.E.; O’Brien, S.J.; Koepfli, K.-P.; Frank, L.G.; Court, M.H.; et al. Evolution of a major drug metabolizing enzyme defect in the domestic cat and other Felidae: Phylogenetic timing and the role of hypercarnivory. PLoS One 2011, 6, e18046. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.G. Idiosyncratic nutrient requirements of cats appear to be diet-induced evolutionary adaptations. Nutr. Res. Rev. 2002, 15, 153–168. [Google Scholar] [CrossRef] [PubMed]
- James, M.O.; Ambadapadi, S. Interactions of cytosolic sulfotransferases with xenobiotics. Drug Metab. Rev. 2013, 45, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Pinto, N.; Dolan, M.E. Clinically relevant genetic variations in drug metabolizing enzymes. Curr. Drug Metab. 2011, 12, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Turpeinen, M.; Klein, K.; Schwab, M. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 2008, 392, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, H. Roles of human cytochrome P450 enzymes involved in drug metabolism and toxicological studies. J. Pharm. Soc. Jpn. 2000, 120, 1347–1357. [Google Scholar]
- Pavek, P.; Dvorak, Z. Xenobiotic-induced transcriptional regulation of xenobiotic metabolizing enzymes of the cytochrome P450 superfamily in human extrahepatic tissues. Curr. Drug Metab. 2008, 9, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Thummel, K.E.; Wilkinson, G.R. In vitro and in vivo drug interactions involving human CYP3A. Annu. Rev. Pharm. Toxicol. 1998, 38, 389–430. [Google Scholar] [CrossRef]
- Dobrinas, M.; Cornuz, J.; Pedrido, L.; Eap, C.B. Influence of cytochrome P450 oxidoreductase genetic polymorphisms on CYP1A2 activity and inducibility by smoking. Pharm. Genomics 2012, 22, 143–151. [Google Scholar] [CrossRef]
- Martignoni, M.; Groothuis, G.M.M.; de Kanter, R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin. Drug Metab. Toxicol. 2006, 2, 875–894. [Google Scholar] [CrossRef] [PubMed]
- Vignati, L.A.; Bogni, A.; Grossi, P.; Monshouwer, M. A human and mouse pregnane X receptor reporter gene assay in combination with cytotoxicity measurements as a tool to evaluate species-specific CYP3A induction. Toxicology 2004, 199, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Martignoni, M.; de Kanter, R.; Grossi, P.; Mahnke, A.; Saturno, G.; Monshouwer, M. An in vivo and in vitro comparison of CYP induction in rat liver and intestine using slices and quantitative RT-PCR. Chem.-Biol. Interact. 2004, 151, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Li, A.P. Species comparison in P450 induction: Effects of dexamethasone, omeprazole, and rifampin on P450 isoforms 1A and 3A in primary cultured hepatocytes from man, Sprague-Dawley rat, minipig, and beagle dog. Chem. Biol. Interact. 2001, 134, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Kocarek, T.A.; Schuetz, E.G.; Strom, S.C.; Fisher, R.A.; Guzelian, P.S. Comparative analysis of cytochrome P4503A induction in primary cultures of rat, rabbit, and human hepatocytes. Drug Metab. Dispos. 1995, 23, 415–421. [Google Scholar] [PubMed]
- Rewitz, K.F.; Styrishave, B.; Lobner-Olesen, A.; Andersen, O. Marine invertebrate cytochrome P450: Emerging insights from vertebrate and insect analogies. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006, 143, 363–381. [Google Scholar] [PubMed]
- Schuler, M.A.; Berenbaum, M.R. Structure and function of cytochrome P450S in insect adaptation to natural and synthetic toxins: Insights gained from molecular modeling. J. Chem. Ecol. 2013, 39, 1232–1245. [Google Scholar] [CrossRef] [PubMed]
- Feyereisen, R. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim. Biophys. Acta (BBA) Proteins Proteomics 2011, 1814, 19–28. [Google Scholar] [CrossRef]
- Fisher, T.; Crane, M.; Callaghan, A. Induction of cytochrome P-450 activity in individual Chironomus riparius (Meigen) larvae exposed to xenobiotics. Ecotoxicol. Environ. Saf. 2003, 54, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kasai, S.; Scott, J.G. Expression and regulation of CYP6D3 in the house fly, Musca domestica (L.). Insect Biochem. Mol. Biol. 2001, 32, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tomita, T.; Itokawa, K.; Komagata, O.; Kasai, S. Overexpression of cytochrome P450 genes in insecticide-resistant mosquitoes. J. Pestic. Sci. 2010, 35, 562–568. [Google Scholar] [CrossRef]
- Natsuhara, K.; Shimada, K.; Tanaka, T.; Miyata, T. Phenobarbital induction of permethrin detoxification and phenobarbital metabolism in susceptible and resistant strains of the beet armyworm Spodoptera exigua (Hubner). Pestic. Biochem. Physiol. 2004, 79, 33–41. [Google Scholar] [CrossRef]
- Johnson, R.M.; Mao, W.; Pollock, H.S.; Niu, G.; Schuler, M.A.; Berenbaum, M.R. Ecologically appropriate xenobiotics induce cytochrome P450s in Apis mellifera. PLoS One 2012, 7, e31051. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Johnson, R.M.; Berenbaum, M.R. Toxicity of mycotoxins to honeybees and its amelioration by propolis. Apidologie 2011, 42, 79–87. [Google Scholar] [CrossRef]
- Morra, R.; Kuruganti, S.; Lam, V.; Lucchesi, J.C.; Ganguly, R. Functional analysis of the cis-acting elements responsible for the induction of the Cyp6a8 and Cyp6g1 genes of Drosophila melanogaster by DDT, phenobarbital and caffeine. Insect Mol. Biol. 2010, 19, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Maitra, S.; Dombrowski, S.M.; Waters, L.C.; Ganguly, R. Three second chromosome-linked clustered Cyp6 genes show differential constitutive and barbital-induced expression in DDT-resistant and susceptible strains of Drosophila melanogaster. Gene 1996, 180, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Misra, J.R.; Horner, M.A.; Lam, G.; Thummel, C.S. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 2011, 25, 1796–1806. [Google Scholar] [CrossRef]
- Danielson, P.B.; MacIntyre, R.J.; Fogleman, J.C. Molecular cloning of a family of xenobiotic-inducible drosophilid cytochrome P450s: Evidence for involvement in host-plant allelochemical resistance. PNAS 1997, 94, 10797–10802. [Google Scholar]
- Menzel, R.; Bogaert, T.; Achazi, R. A systematic gene expression screen of Caenorhabditis elegans cytochrome P450 genes reveals CYP35 as strongly xenobiotic inducible. Arch. Biochem. Biophys. 2001, 395, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Menzel, R.; Rodel, M.; Kulas, J.; Steinberg, C.E.W. CYP35: Xenobiotically induced gene expression in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2005, 438, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, P.; Mueller, M.; Krueger, A.; Steinberg, C.E.W.; Menzel, R. Cytochrome P450-dependent metabolism of PCB52 in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2009, 488, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Chakrapani, B.P.; Kumar, S.; Subramaniam, J.R. Development and evaluation of an in vivo assay in Caenorhabditis elegans for screening of compounds for their effect on cytochrome P450 expression. J. Biosci. 2008, 33, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.L.; Chen, B.; Qiu, X.Y.; Lin, K.L.; Yu, X.G. Three novel cytochrome P450 genes identified in the marine polychaete Perinereis nuntia and their transcriptional response to xenobiotics. Aquat. Toxicol. 2013, 134, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Whalen, K.E.; Starczak, V.R.; Nelson, D.R.; Goldstone, J.V.; Hahn, M.E. Cytochrome P450 diversity and induction by gorgonian allelochemicals in the marine gastropod Cyphoma gibbosum. BMC Ecol. 2010, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Bousova, I.; Skalova, L. Inhibition and induction of glutathione S-transferases by flavonoids: Possible pharmacological and toxicological consequences. Drug Metab. Rev. 2012, 44, 267–286. [Google Scholar] [CrossRef] [PubMed]
- Maglich, J.M.; Stoltz, C.M.; Goodwin, B.; Hawkins-Brown, D.; Moore, J.T.; Kliewer, S.A. Nuclear pregnane X receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol. Pharmacol. 2002, 62, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Whalen, K.E.; Morin, D.; Lin, C.Y.; Tjeerdema, R.S.; Goldstone, J.V.; Hahn, M.E. Proteomic identification, cDNA cloning and enzymatic activity of glutathione S-transferases from the generalist marine gastropod, Cyphoma gibbosum. Arch. Biochem. Biophys. 2008, 478, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Kuhajek, J.M.; Schlenk, D. Effects of the brominated phenol, lanosol, on cytochrome P-450 and glutathione transferase activities in Haliotis rufescens and Katharina tunicata. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2003, 134, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Whalen, K.E.; Lane, A.L.; Kubanek, J.; Hahn, M.E. Biochemical warfare on the reef: The role of glutathione transferases in consumer tolerance of dietary prostaglandins. PLoS One 2010, 5, 0008537. [Google Scholar] [CrossRef]
- Mao, W.; Rupasinghe, S.G.; Johnson, R.M.; Zangerl, A.R.; Schuler, M.A.; Berenbaum, M.R. Quercetin-metabolizing CYP6AS enzymes of the pollinator Apis mellifera (Hymenoptera: Apidae). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 154, 427–434. [Google Scholar] [CrossRef]
- Bainy, A.C.D.; Kubota, A.; Goldstone, J.V.; Lille-Langoy, R.; Karchner, S.I.; Celander, M.C.; Hahn, M.E.; Goksoyr, A.; Stegeman, J.J. Functional characterization of a full length pregnane X receptor, expression in vivo, and identification of PXR alleles, in Zebrafish (Danio rerio). Aquat. Toxicol. 2013, 142–143, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Chai, X.; Zeng, S.; Xie, W. Nuclear receptors PXR and CAR: Implications for drug metabolism regulation, pharmacogenomics and beyond. Expert Opin. Drug Metab. Toxicol. 2013, 9, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Chiang, J.Y.L. Nuclear receptors in drug metabolism and beyond. Drug Metab. Rev. 2013, 45, 1–2. [Google Scholar] [PubMed]
- NR1I2 nuclear receptor subfamily 1, group I, member 2. Available online: http://www.ncbi.nlm.nih.gov/gene/8856 (accessed on 26 February 2014).
- Kliewer, S.A.; Moore, J.T.; Wade, L.; Staudinger, J.L.; Watson, M.A.; Jones, S.A.; McKee, D.D.; Oliver, B.B.; Willson, T.M.; Zetterstrom, R.H.; et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 1998, 92, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Blumberg, B.; Sabbagh, W.; Juguilon, H.; Bolado, J.; van Meter, C.M.; Ono, E.S.; Evans, R.M. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998, 12, 3195–3205. [Google Scholar] [CrossRef] [PubMed]
- Bertilsson, G.; Heidrich, J.; Svensson, K.; Asman, M.; Jendeberg, L.; Sydow-Backman, M.; Ohlsson, R.; Postlind, H.; Blomquist, P.; Berkenstam, A.; et al. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. PNAS 1998, 95, 12208–12213. [Google Scholar] [CrossRef] [PubMed]
- Orans, J.; Teotico, D.G.; Redinbo, M.R. The nuclear xenobiotic receptor pregnane X receptor: Recent insights and new challenges. Mol. Endocrinol. 2005, 19, 2891–2900. [Google Scholar] [CrossRef] [PubMed]
- Li, T.G.; Chiang, J.Y.L. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 288, G74–G84. [Google Scholar] [PubMed]
- McKenna, N.J.; Lanz, R.B.; O’Malley, B.W. Nuclear receptor coregulators: Cellular and molecular biology. Endocr. Rev. 1999, 20, 321–344. [Google Scholar] [PubMed]
- Teotico, D.G.; Frazier, M.L.; Ding, F.; Dokholyan, N.V.; Temple, B.R.S.; Redinbo, M.R. Active Nuclear Receptors Exhibit Highly Correlated AF-2 Domain Motions. PLoS Comput. Biol. 2008, 4, e1000111. [Google Scholar] [CrossRef] [PubMed]
- Tolson, A.H.; Wang, H.B. Regulation of drug-metabolizing enzymes by xenobiotic receptors: PXR and CAR. Adv. Drug Deliv. Rev. 2010, 62, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Mani, S.; Redinbo, M.R.; Krasowski, M.D.; Li, H.; Ekins, S. Elucidating the “Jekyll and Hyde” nature of PXR: The case for discovering antagonists or allosteric antagonists. Pharm. Res. 2009, 26, 1807–1815. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.K.; Waxman, D.J. Synthetic drugs and natural products as modulators of constitutive androstane receptor (CAR) and pregnane X receptor (PXR). Drug Metab. Rev. 2006, 38, 51–73. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.P.; Mota, L.C.; Baldwin, W.S. Activation of CAR and PXR by Dietary, Environmental and Occupational Chemicals Alters Drug Metabolism, Intermediary Metabolism, and Cell Proliferation. Curr. Pharm. Pers. Med. 2009, 7, 81–105. [Google Scholar]
- Manez, S. A fresh insight into the interaction of natural products with pregnane X receptor. Nat. Prod. Commun. 2008, 3, 2123–2128. [Google Scholar]
- Staudinger, J.L.; Ding, X.; Lichti, K. Pregnane X receptor and natural products: Beyond drug-drug interactions. Expert Opin. Drug Metab. Toxicol. 2006, 2, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Verma, S.; Blumberg, B. The steroid and xenobiotic receptor (SXR), beyond xenobiotic metabolism. Nucl. Recept. Signal. 2009, 7, e001. [Google Scholar] [PubMed]
- Harms, M.J.; Eick, G.N.; Goswami, D.; Colucci, J.K.; Griffin, P.R.; Ortlund, E.A.; Thornton, J.W. Biophysical mechanisms for large-effect mutations in the evolution of steroid hormone receptors. PNAS 2013, 110, 11475–11480. [Google Scholar] [CrossRef] [PubMed]
- Eick, G.N.; Colucci, J.K.; Harms, M.J.; Ortlund, E.A.; Thornton, J.W. Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLoS Genet. 2012, 8, e1003072. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Li, S.; Dong, D. 3D structures and ligand specificities of nuclear xenobiotic receptors CAR, PXR and VDR. Drug Discov. Today 2013, 18, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.D.; Redinbo, M.R. Xenobiotic-sensing nuclear receptors involved in drug metabolism: A structural perspective. Drug Metab. Rev. 2013, 45, 79–100. [Google Scholar] [CrossRef] [PubMed]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schutz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839. [Google Scholar] [CrossRef] [PubMed]
- Krasowski, M.D.; Yasuda, K.; Hagey, L.R.; Schuetz, E.G. Evolution of the pregnane X receptor: Adaptation to cross-species differences in biliary bile salts. Mol. Endocrinol. 2005, 19, 1720–1739. [Google Scholar] [CrossRef]
- Krasowski, M.D.; Yasuda, K.; Hagey, L.R.; Schuetz, E.G. Evolutionary selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors). Nucl. Recept. 2005, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.D.; Burch, P.E.; Cooney, A.J.; Lanz, R.B.; Pereira, F.A.; Wu, J.Q.; Gibbs, R.A.; Weinstock, G.; Wheeler, D.A. Genomic analysis of the nuclear receptor family: New insights into structure, regulation, and evolution from the rat genome. Genome Res. 2004, 14, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Krasowski, M.D.; Ai, N.; Hagey, L.R.; Kollitz, E.M.; Kullman, S.W.; Reschly, E.J.; Ekins, S. The evolution of farnesoid X, vitamin D, and pregnane X receptors: Insights from the green-spotted pufferfish (Tetraodon nigriviridis) and other non-mammalian species. BMC Biochem. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.B.; Parks, D.J.; Jones, S.A.; Bledsoe, R.K.; Consler, T.G.; Stimmel, J.B.; Goodwin, B.; Liddle, C.; Blanchard, S.G.; Willson, T.M.; et al. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J. Biol. Chem. 2000, 275, 15122–15127. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.A.; Moore, L.B.; Shenk, J.L.; Wisely, G.B.; Hamilton, G.A.; McKee, D.D.; Tomkinson, N.C.O.; LeCluyse, E.L.; Lambert, M.H.; Willson, T.M.; et al. The pregnane X receptor: A promiscuous xenobiotic receptor that has diverged during evolution. Mol. Endocrinol. 2000, 14, 27–39. [Google Scholar] [PubMed]
- LeCluyse, E.L. Pregnane X receptor: Molecular basis for species differences in CYP3A induction by xenobiotics. Chem. Biol. Interact. 2001, 134, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J.M.; McKee, D.D.; Watson, M.A.; Willson, T.M.; Moore, J.T.; Kliewer, S.A. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J. Clin. Investig. 1998, 102, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Evans, R.M. Orphan nuclear receptors: The exotics of xenobiotics. J. Biol. Chem. 2001, 276, 37739–37742. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Barwick, J.L.; Downes, M.; Blumberg, B.; Simon, C.M.; Nelson, M.C.; Neuschwander-Tetri, B.A.; Bruntk, E.M.; Guzelian, P.S.; Evans, R.M.; et al. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature 2000, 406, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Tirona, R.G.; Leake, B.F.; Podust, L.M.; Kim, R.B. Identification of amino acids in rat pregnane X receptor that determine species-specific activation. Mol. Pharmacol. 2004, 65, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Y.C.; Kumar, S.; Hedges, S.B. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. PNAS 1999, 266, 163–171. [Google Scholar]
- Erwin, D.H.; Davidson, E.H. The last common bilaterian ancestor. Development 2002, 129, 3021–3032. [Google Scholar] [PubMed]
- Hedges, S.B.; Dudley, J.; Kumar, S. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics 2006, 22, 2971–2972. [Google Scholar] [CrossRef] [PubMed]
- King-Jones, K.; Horner, M.A.; Lam, G.; Thummel, C.S. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006, 4, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Lindblom, T.H.; Pierce, G.J.; Sluder, A.E. A C. elegans orphan nuclear receptor contributes to xenobiotic resistance. Curr. Biol. 2001, 11, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.G.H.; Kozaki, T.; Scott, J.G. Hormone receptor-like in 96 and Broad-Complex modulate phenobarbital induced transcription of cytochrome P450 CYP6D1 in Drosophila S2 cells. Insect Mol. Biol. 2011, 20, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Palanker, L.; Necakov, A.S.; Sampson, H.M.; Ni, R.; Hu, C.H.; Thummel, C.S.; Krause, H.M. Dynamic regulation of Drosophila nuclear receptor activity in vivo. Development 2006, 133, 3549–3562. [Google Scholar] [CrossRef] [PubMed]
- Sieber, M.H.; Thummel, C.S. Coordination of triacylglycerol and cholesterol homeostasis by DHR96 and the Drosophila LipA homolog magro. Cell Metab. 2012, 15, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Karimullina, E.; Li, Y.; Ginjupalli, G.K.; Baldwin, W.S. Daphnia HR96 is a promiscuous xenobiotic and endobiotic nuclear receptor. Aquat. Toxicol. 2012, 116, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tan, A.; Palli, S.R. The function of nuclear receptors in regulation of female reproduction and embryogenesis in the red flour beetle, Tribolium castaneum. J. Insect Physiol. 2010, 56, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Bonasio, R.; Zhang, G.; Ye, C.; Mutti, N.S.; Fang, X.; Qin, N.; Donahue, G.; Yang, P.; Li, Q.; Li, C.; et al. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 2010, 329, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Velarde, R.A.; Robinson, G.E.; Fahrbach, S.E. Nuclear receptors of the honey bee: Annotation and expression in the adult brain. Insect Mol. Biol. 2006, 15, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Giraudo, M.; Audant, P.; Feyereisen, R.; le Goff, G. Nuclear receptors HR96 and ultraspiracle from the fall armyworm (Spodoptera frugiperda), developmental expression and induction by xenobiotics. J. Insect Physiol. 2013, 59, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Sluder, A.E.; Mathews, S.W.; Hough, D.; Yin, V.P.; Maina, C.V. The nuclear receptor superfamily has undergone extensive proliferation and diversification in nematodes. Genome Res. 1999, 9, 103–120. [Google Scholar] [PubMed]
- Bertrand, W.; Brunet, F.G.; Escriva, H.; Parmentier, G.; Laudet, V.; Robinson-Rechavi, M. Evolutionary genomics of nuclear receptors: From twenty-five ancestral genes to derived endocrine systems. Mol. Biol. Evol. 2004, 21, 1923–1937. [Google Scholar] [CrossRef] [PubMed]
- Delsuc, F.; Brinkmann, H.; Chourrout, D.; Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 2006, 439, 965–968. [Google Scholar] [CrossRef] [PubMed]
- Delsuc, F.; Tsagkogeorga, G.; Lartillot, N.; Philippe, H. Additional molecular support for the new chordate phylogeny. Genesis 2008, 46, 592–604. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.R.; Tsagkogeorga, G.; Delsuc, F.; Blanquart, S.; Shenkar, N.; Loya, Y.; Douzery, E.J.P.; Huchon, D. Tunicate mitogenomics and phylogenetics: Peculiarities of the Herdmania momus mitochondrial genome and support for the new chordate phylogeny. BMC Genomics 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Rowe, T. Chordate phylogeny and development. In Assembling The Tree of Life, 1st ed.; Cracraft, J., Donoghue, M.J., Eds.; Oxforf University Press: Oxford, UK, 2004; pp. 384–409. [Google Scholar]
- Dehal, P.; Satou, Y.; Campbell, R.K.; Chapman, J.; Degnan, B.; de Tomaso, A.; Davidson, B.; di Gregorio, A.; Gelpke, M.; Goodstein, D.M.; et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 2002, 298, 2157–2167. [Google Scholar] [CrossRef] [PubMed]
- Voskoboynik, A.; Neff, N.F.; Sahoo, D.; Newman, A.M.; Pushkarev, D.; Koh, W.; Passarelli, B.; Fan, H.C.; Mantalas, G.L.; Palmeri, K.J.; et al. The genome sequence of the colonial chordate, Botryllus schlosseri. ELife 2013, 2. [Google Scholar] [CrossRef]
- Denoeud, F.; Henriet, S.; Mungpakdee, S.; Aury, J.-M.; da Silva, C.; Brinkmann, H.; Mikhaleva, J.; Olsen, L.C.; Jubin, C.; Canestro, C.; et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 2010, 330, 1381–1385. [Google Scholar] [CrossRef] [Green Version]
- Bracken-Grissom, H.; Collins, A.G.; Collins, T.; Crandall, K.; Distel, D.; Dunn, C.; Giribet, G.; Haddock, S.; Knowlton, N.; Martindale, M.; et al. The Global Invertebrate Genomics Alliance (GIGA): Developing community resources to study diverse invertebrate genomes. J. Hered. 2014, 105, 1–18. [Google Scholar] [PubMed]
- Dussault, I.; Forman, B.M. The nuclear receptor PXR: A master regulator of “homeland” defense. Crit. Rev. Eukaryot. Gene Expr. 2002, 12, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Kliewer, S.; Goodwin, B.; Willson, T. The nuclear pregnane X receptor: A key regulator of xenobiotic metabolism. Endocr. Rev. 2002, 23, 687–702. [Google Scholar] [CrossRef] [PubMed]
- Timsit, Y.E.; Negishi, M. CAR and PXR: The xenobiotic-sensing receptors. Steroids 2007, 72, 231–246. [Google Scholar] [CrossRef] [PubMed]
- Bridgham, J.T.; Eick, G.N.; Larroux, C.; Deshpande, K.; Harms, M.J.; Gauthier, M.E.A.; Ortlund, E.A.; Degnan, B.M.; Thornton, J.W. Protein evolution by molecular tinkering: Diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 2010, 8, e1000497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, M.; Simakov, O.; Chapman, J.; Fahey, B.; Gauthier, M.E.A.; Mitros, T.; Richards, G.S.; Conaco, C.; Dacre, M.; Hellsten, U.; et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 2010, 466, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Reitzel, A.M.; Tarrant, A.M. Nuclear receptor complement of the cnidarian Nematostella vectensis: Phylogenetic relationships and developmental expression patterns. BMC Evol. Biol. 2009, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Vogeler, S.; Galloway, T.; Lyons, B.; Bean, T. The nuclear receptor gene family in the Pacific oyster, Crassostrea gigas, contains a novel subfamily group. BMC Genomics 2014, 15, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sodergren, E.; Weinstock, G.M.; Davidson, E.H.; Cameron, R.A.; Gibbs, R.A.; Angerer, R.C.; Angerer, L.M.; Arnone, M.I.; Burgess, D.R.; Burke, R.D.; et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science 2006, 314, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Gazulha, V.; Mansur, M.C.D.; Cybis, L.F.; Azevedo, S.M.F.O. Feeding behavior of the invasive bivalve Limnoperna fortunei (Dunker, 1857) under exposure to toxic cyanobacteria Microcystis aeruginosa. Braz. J. Biol. 2012, 72, 41–49. [Google Scholar] [PubMed]
- Bridgham, J.T.; Keay, J.; Ortlund, E.A.; Thornton, J.W. Vestigialization of an allosteric switch: Genetic and structural mechanisms for the evolution of constitutive activity in a steroid hormone receptor. PLoS Genet. 2014, 10, e1004058. [Google Scholar] [CrossRef] [PubMed]
- Satou, Y.; Yamada, L.; Mochizuki, Y.; Takatori, N.; Kawashima, T.; Sasaki, A.; Hamaguchi, M.; Awazu, S.; Yagi, K.; Sasakura, Y.; et al. A cDNA resource from the basal chordate Ciona intestinalis. Genesis 2002, 33, 153–154. [Google Scholar] [CrossRef] [PubMed]
- Yagi, K.; Satou, Y.; Mazet, F.; Shimeld, S.M.; Degnan, B.; Rokhsar, D.; Levine, M.; Kohara, Y.; Satoh, N. A genomewide survey of developmentally relevant genes in Ciona intestinalis—III. Genes for Fox, ETS, nuclear receptors and NF kappa B. Dev. Genes Evol. 2003, 213, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Ekins, S.; Reschly, E.J.; Hagey, L.R.; Krasowski, M.D. Evolution of pharmacologic specificity in the pregnane X receptor. BMC Evol. Biol. 2008, 8. [Google Scholar] [CrossRef]
- Satou, Y.; Kawashima, T.; Shoguchi, E.; Nakayama, A.; Satoh, N. An integrated database of the ascidian, Ciona intestinalis: Towards functional genomics. Zool. Sci. 2005, 22, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Reschly, E.J.; Bainy, A.C.D.; Mattos, J.J.; Hagey, L.R.; Bahary, N.; Mada, S.R.; Ou, J.; Venkataramanan, R.; Krasowski, M.D. Functional evolution of the vitamin D and pregnane X receptors. BMC Evol. Biol. 2007, 7, 222. [Google Scholar] [CrossRef] [PubMed]
- Fidler, A.E.; Holland, P.T.; Reschly, E.J.; Ekins, S.; Krasowski, M.D. Activation of a tunicate (Ciona intestinalis) xenobiotic receptor orthologue by both natural toxins and synthetic toxicants. Toxicon 2012, 59, 365–372. [Google Scholar] [CrossRef] [PubMed]
- PubChem Compound Database. Available online: https://www.ncbi.nlm.nih.gov/pccompound?cmd=search (accessed on 7 September 2014).
- Pinne, M.; Raucy, J.L. Advantages of cell-based high-volume screening assays to assess nuclear receptor activation during drug discovery. Expert Opin. Drug Discov. 2014, 9, 669–686. [Google Scholar] [CrossRef] [PubMed]
- Raucy, J.L.; Lasker, J.M. Cell-based systems to assess nuclear receptor activation and their use in drug development. Drug Metab. Rev. 2013, 45, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Norcliffe, J.L.; Alvarez-Ruiz, E.; Martin-Plaza, J.J.; Steel, P.G.; Denny, P.W. The utility of yeast as a tool for cell-based, target-directed high-throughput screening. Parasitology 2013, 141, 8–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hontzeas, N.; Hafer, K.; Schiestl, R.H. Development of a microtiter plate version of the yeast DEL assay amenable to high-throughput toxicity screening of chemical libraries. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2007, 634, 228–234. [Google Scholar] [CrossRef]
- Rajasarkka, J.; Virta, M. Miniaturization of a panel of high throughput yeast-cell-based nuclear receptor assays in 384-and 1536-well microplates. Comb. Chem. High Throughput Screen. 2011, 14, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Bovee, T.F.H.; Helsdingen, R.J.R.; Hamers, A.R.M.; van Duursen, M.B.M.; Nielen, M.W.F.; Hoogenboom, R.L.A.P. A new highly specific and robust yeast androgen bioassay for the detection of agonists and antagonists. Anal. Bioanal. Chem. 2007, 389, 1549–1558. [Google Scholar] [CrossRef] [PubMed]
- An, W.F.; Tolliday, N. Cell-based assays for high-throughput screening. Mol. Biotechnol. 2010, 45, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.L.; Zhao, H.M. A highly efficient and sensitive screening method for trans-activation activity of estrogen receptors. Gene 2003, 306, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Raucy, J.L.; Lasker, J.M. Current in vitro high-throughput screening approaches to assess nuclear receptor activation. Curr. Drug Metab. 2010, 11, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.-L.; Shiizaki, K.; Kawanishi, M.; Kondo, M.; Yagi, T. Validation of a new yeast-based reporter assay consisting of human estrogen receptors α/β and coactivator SRC-1: Application for detection of estrogenic activity in environmental samples. Environ. Toxicol. 2009, 24, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Redinbo, M.R.; Venkatesh, M.; Ekins, S.; Chaudhry, A.; Bloch, N.; Negassa, A.; Mukherjee, P.; Kalpana, G.; Mani, S.; et al. Novel yeast-based strategy unveils antagonist binding regions on the nuclear xenobiotic receptor PXR. J. Biol. Chem. 2013, 288, 13655–13668. [Google Scholar] [CrossRef] [PubMed]
- McEwan, I.J. Bakers yeast rises to the challenge: Reconstitution of mammalian steroid receptor signalling in S. cerevisiae. Trends Genet. 2001, 17, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.E.; Burow, M.E.; McLachlan, J.A.; Miller, C.A. Detecting ligands and dissecting nuclear receptor-signaling pathways using recombinant strains of the yeast Saccharomyces cerevisiae. Nat. Protoc. 2008, 3, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Goldstone, J.V.; Goldstone, H.M.H.; Morrison, A.M.; Tarrant, A.; Kern, S.E.; Woodin, B.R.; Stegeman, J.J. Cytochrome P450 1 genes in early deuterostomes (tunicates and sea urchins) and vertebrates (chicken and frog): Origin and diversification of the CYP1 gene family. Mol. Biol. Evol. 2007, 24, 2619–2631. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.; Metzger, D.; Chambon, P. Role of the 2 activating domains of the estrogen receptor in the cell-type and promoter-context dependent agonistic activity of the antiestrogen 4-hydroxytamoxifen. EMBO J. 1990, 9, 2811–2818. [Google Scholar] [PubMed]
- Louvion, J.F.; Havauxcopf, B.; Picard, D. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene 1993, 131, 129–134. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, R.A.; Burgess, D.; Shema, R.; Motlekar, N.; Napper, A.D.; Diamond, S.L.; Pavitt, G.D. A Saccharomyces cerevisiae cell-based quantitative beta-galactosidase handling and assay compatible with robotic high-throughput screening. Yeast 2008, 25, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Bovee, T.F.H.; Hendriksen, P.J.M.; Portier, L.; Wang, S.; Elliott, C.T.; van Egmond, H.P.; Nielen, M.W.F.; Peijnenburg, A.; Hoogenboom, L.A.P. Tailored microarray platform for the detection of marine toxins. Environ. Sci. Technol. 2011, 45, 8965–8973. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Kumar, V.; Majumder, C.B.; Roy, P. Screening of some anti-progestin endocrine disruptors using a recombinant yeast-based in vitro bioassay. Toxicol. Vitr. 2008, 22, 788–798. [Google Scholar] [CrossRef]
- Nordeen, S.K. Luciferase reporter gene vectors for analysis of promoters and enhancers. Biotechniques 1988, 6, 454–458. [Google Scholar] [PubMed]
- Fan, F.; Wood, K.V. Bioluminescent assays for high-throughput screening. Assay Drug Dev. Technol. 2007, 5, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Hancock, M.K.; Medina, M.N.; Smith, B.M.; Orth, A.P. Microplate orbital mixing improves high-throughput cell-based reporter assay readouts. J. Biomol. Screen. 2007, 12, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Routledge, E.J.; Sumpter, J.P. Estrogenic activity of surfactants and some of their degradation products assessed using a recombinant yeast screen. Environ. Toxicol. Chem. 1996, 15, 241–248. [Google Scholar] [CrossRef]
- Miller, C.A.; Tan, X.B.; Wilson, M.; Bhattacharyya, S.; Ludwig, S. Single plasmids expressing human steroid hormone receptors and a reporter gene for use in yeast signaling assays. Plasmid 2010, 63, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Chen, J.; Sun, T.; Shen, J.H.; Shen, X.; Jiang, H.L. A yeast two-hybrid technology-based system for the discovery of PPAR gamma agonist and antagonist. Anal. Biochem. 2004, 335, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Balsiger, H.A.; de la Torre, R.; Lee, W.-Y.; Cox, M.B. A four-hour yeast bioassay for the direct measure of estrogenic activity in wastewater without sample extraction, concentration, or sterilization. Sci. Total Environ. 2010, 408, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.M.; McLachlan, J.A.; Arnold, S.F. The estrogenic and antiestrogenic activities of phytochemicals with human estrogen receptor expressed in yeast. Steroids 1997, 62, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Gaido, K.W.; Leonard, L.S.; Lovell, S.; Gould, J.C.; Babai, D.; Portier, C.J.; McDonnell, D.P. Evaluation of chemicals with endocrine modulating activity in a yeast-based steroid hormone receptor gene transcription assay. Toxicol. Appl. Pharmacol. 1997, 143, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Passos, A.L.; Pinto, P.I.; Power, D.M.; Canario, A.V. A yeast assay based on the gilthead sea bream (teleost fish) estrogen receptor beta for monitoring estrogen mimics. Ecotoxicol. Environ. Saf. 2009, 72, 1529–1537. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Chou, P.H.; Kawanishi, M.; Yagi, T. Occurrence of xenobiotic ligands for retinoid X receptors and thyroid hormone receptors in the aquatic environment of Taiwan. Mar. Pollut. Bull. 2014, 23. [Google Scholar] [CrossRef]
- Holdgate, G.A.; Anderson, M.; Edfeldt, F.; Geschwindner, S. Affinity-based, biophysical methods to detect and analyze ligand binding to recombinant proteins: Matching high information content with high throughput. J. Struct. Biol. 2010, 172, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Senveli, S.U.; Tigli, O. Biosensors in the small scale: Methods and technology trends. IET Nanobiotechnol. 2013, 7, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Fechner, P.; Gauglitz, G.; Gustafsson, J.-A. Nuclear receptors in analytics—A fruitful joint venture or a wasteful futility? Trends Anal. Chem. 2010, 29, 297–305. [Google Scholar] [CrossRef]
- Lin, W.; Liu, J.; Jeffries, C.; Yang, L.; Lu, Y.; Lee, R.E.; Chen, T. Development of BODIPY FL Vindoline as a novel and high-affinity pregnane X receptor fluorescent probe. Bioconjug. Chem. 2014, 18, 18. [Google Scholar]
- Hill, K.L.; Dutta, P.; Long, Z.; Sepaniak, M.J. Microcantilever-based nanomechanical studies of the orphan nuclear receptor pregnane X receptor-ligand interactions. J. Biomater. Nanobiotechnol. 2011, 3, 133–142. [Google Scholar] [CrossRef]
- Dagnino, S.; Bellet, V.; Grimaldi, M.; Riu, A.; Ait-Aissa, S.; Cavailles, V.; Fenet, H.; Balaguer, P. Affinity purification using recombinant PXR as a tool to characterize environmental ligands. Environ. Toxicol. 2014, 29, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Bricelj, V.M.; Connell, L.; Konoki, K.; MacQuarrie, S.P.; Scheuer, T.; Catterall, W.A.; Trainer, V.L. Sodium channel mutation leading to saxitoxin resistance in clams increases risk of PSP. Nature 2005, 434, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Bricelj, V.M.; MacQuarrie, S.P.; Doane, J.A.E.; Connell, L.B. Evidence of selection for resistance to paralytic shellfish toxins during the early life history of soft-shell clam (Mya arenaria) populations. Limnol. Oceanogr. 2010, 55, 2463–2475. [Google Scholar] [CrossRef]
- Roje-Busatto, R.; Ujević, I. PSP toxins profile in ascidian Microcosmus vulgaris (Heller, 1877) after human poisoning in Croatia (Adriatic Sea). Toxicon 2014, 79, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.L.; Settlemier, C.J.; Ballauer, J.M. An examination of the epiphytic nature of Gambierdiscus toxicus, a dinoflagellate involved in ciguatera fish poisoning. Harmful Algae 2011, 10, 598–605. [Google Scholar] [PubMed]
- Martins, A.; Vieira, H.; Gaspar, H.; Santos, S. Marketed marine natural products in the pharmaceutical and cosmeceutical industries: Tips for success. Mar. Drugs 2014, 12, 1066–1101. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.S.; Glaser, K.B.; Cuevas, C.; Jacobs, R.S.; Kem, W.; Little, R.D.; McIntosh, J.M.; Newman, D.J.; Potts, B.C.; Shuster, D.E.; et al. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol. Sci. 2010, 31, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F.; Labes, A.; Wiese, J. Bio-mining the microbial treasures of the ocean: New natural products. Biotechnol. Adv. 2011, 29, 468–482. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2013, 30, 237–323. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richter, I.; Fidler, A.E. Marine Invertebrate Xenobiotic-Activated Nuclear Receptors: Their Application as Sensor Elements in High-Throughput Bioassays for Marine Bioactive Compounds. Mar. Drugs 2014, 12, 5590-5618. https://doi.org/10.3390/md12115590
Richter I, Fidler AE. Marine Invertebrate Xenobiotic-Activated Nuclear Receptors: Their Application as Sensor Elements in High-Throughput Bioassays for Marine Bioactive Compounds. Marine Drugs. 2014; 12(11):5590-5618. https://doi.org/10.3390/md12115590
Chicago/Turabian StyleRichter, Ingrid, and Andrew E. Fidler. 2014. "Marine Invertebrate Xenobiotic-Activated Nuclear Receptors: Their Application as Sensor Elements in High-Throughput Bioassays for Marine Bioactive Compounds" Marine Drugs 12, no. 11: 5590-5618. https://doi.org/10.3390/md12115590
APA StyleRichter, I., & Fidler, A. E. (2014). Marine Invertebrate Xenobiotic-Activated Nuclear Receptors: Their Application as Sensor Elements in High-Throughput Bioassays for Marine Bioactive Compounds. Marine Drugs, 12(11), 5590-5618. https://doi.org/10.3390/md12115590