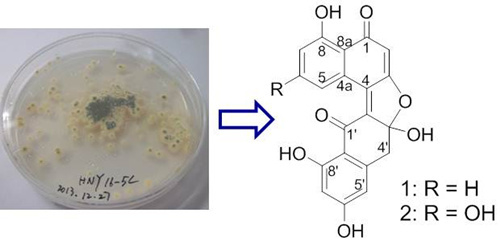
Asperlones A and B, Dinaphthalenone Derivatives from a Mangrove Endophytic Fungus Aspergillus sp. 16-5C
Abstract
:1. Introduction

2. Results and Discussion
| Position | 1 | 2 | ||
|---|---|---|---|---|
| δC, mult. | δH (J in Hz) | δC, mult. | δH (J in Hz) | |
| 1 | 191.8, qC | 190.5, qC | ||
| 2 | 102.1, CH | 6.17 s | 101.6, CH | 6.03 s |
| 3 | 171.6, qC | 170.6, qC | ||
| 4 | 129.8, qC | 130.2, qC | ||
| 4a | 128.4, qC | 129.8, qC | ||
| 5 | 121.8, CH | 8.80 d (7.8) | 111.2, CH | 8.34 d (2.2) |
| 6 | 135.3, CH | 7.68 m | 163.4, qC | |
| 7 | 122.5, CH | 7.20 d (8.3) | 106.7, CH | 6.47 d (2.2) |
| 8 | 162.1, qC | 164.4, qC | ||
| 8a | 113.8, qC | 107.2, qC | ||
| 1′ | 186.7, qC | 186.5, qC | ||
| 2′ | 142.7, qC | 142.2, qC | ||
| 3′ | 113.5, qC | 113.2, qC | ||
| 4′ | 41.0, CH2 | 3.41 d (15.7); 3.58 d (15.7) | 41.1, CH2 | 3.39 d (15.8); 3.55 d (15.8) |
| 4a′ | 144.0, qC | 144.0, qC | ||
| 5′ | 110.4, CH | 6.40 d (1.6) | 110.4, CH | 6.40 d (1.6) |
| 6′ | 167.1, qC | 167.1, qC | ||
| 7′ | 101.6, CH | 6.25 d (1.6) | 101.6, CH | 6.25 d (1.6) |
| 8′ | 167.3, qC | 167.0, qC | ||
| 8a′ | 112.0, qC | 112.0, qC | ||
| 6-OH | 10.83 brs | |||
| 8-OH | 13.58 s | 13.76 s | ||
| 3′-OH | 8.23 brs | 8.14 brs | ||
| 6′-OH | 11.30 brs | 11.17 brs | ||
| 8′-OH | 12.84 s | 12.93 s | ||


| Position | 3 | 4 | ||
|---|---|---|---|---|
| δC, mult. | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 155.5, CH | 8.32 s | 69.8, CH2 | 3.96 d (12.4); 4.68 d (12.4) |
| 3 | 153.3, qC | 156.7, qC | ||
| 4 | 116.2, CH | 7.14 s | 109.1, CH | 6.46 s |
| 4a | 142.4, qC | 148.1, qC | ||
| 5 | 109.2, CH | 5.73 s | 120.2, CH | 6.21 s |
| 6 | 192.2, qC | 192.2, qC | ||
| 7 | 85.5, qC | 84.1, qC | ||
| 7-CH3 | 22.3, CH3 | 1.56 s | 23.9, CH2 | 1.82 s |
| 8 | 192.6, qC | 200.0, qC | ||
| 8a | 114.7, qC | 66.2, qC | ||
| 1′ | 134.0, CH | 7.28 d (15.7) | 136.7, CH | 7.13 d (15.6) |
| 2′ | 125.0, CH | 6.43 d (15.7) | 124.2, CH | 6.34 d (15.6) |
| 3′ | 166.6, qC | 167.0, qC | ||
| 1″ | 168.7, qC | 168.2, qC | ||
| 2″ | 105.0, qC | 105.1, qC | ||
| 3″ | 155.2, qC | 163.1, qC | ||
| 4″ | 101.0, CH | 6.24 s | 101.0, CH | 6.16 d (2.0) |
| 5″ | 152.2, qC | 162.8, qC | ||
| 6″ | 137.3, qC | 111.6, CH | 6.24 d (2.0) | |
| 7″ | 126.2, qC | 142.8, qC | ||
| 7″-CH3 | 14.6, CH2 | 2.36 s | 23.1, CH2 | 2.44 s |
| 8a-OH | 7.26 s | |||
| 3′-OH | 12.86 brs | 12.82 brs | ||
| 3″-OH | 9.68 s | 10.26 s | ||
| 5″-OH | 10.28 brs | 10.32 brs | ||
| 6″-OH | 8.00 brs | |||



| Compound | (±)-1 | (±)-2 | 3 | 5 | 6 | Positive Control |
|---|---|---|---|---|---|---|
| IC50 (μM) | 4.24 ± 0.41 | 4.32 ± 0.60 | >50 | >50 | 3.99 ± 0.34 | 0.05 |
3. Experimental Section
3.1. General
3.2. Fungal Material
3.3. Extraction and Isolation
3.4. Materials and Methods for mPTPB Assay
3.4.1. Cloning, Expression and Purification of MptpB
3.4.2. MptpB Inhibition Assay
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2014. Available online: http://www.who.int/tb/publications/global_report/en/ (assessed on 17 July 2014).
- Butler, D. New fronts in an old war. Nature 2000, 406, 670–672. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Rao, V.; Shakila, H.; Gupta, R.; Khera, A.; Dhar, N.; Singh, A.; Koul, A.; Singh, Y.; Naseema, M.; et al. Disruption of mptpB impairs the ability of Mycobacterium tuberculosis to survive in guinea pigs. Mol. Microbiol. 2003, 50, 751–762. [Google Scholar] [CrossRef]
- Bialy, L.; Waldmann, H. Inhibitors of Protein Tyrosine Phosphatases: Next-Generation Drugs? Angew. Chem. Int. Ed. 2005, 44, 3814–3839. [Google Scholar] [CrossRef]
- Singh, R.; Singh, A.; Tyagi, A.K. Deciphering the genes involved in pathogenesis of Mycobacterium tuberculosis. Tuberculosis 2005, 85, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Castandet, J.; Prost, J.F.; Peyron, P.; Astarie-Dequeker, C.; Anes, E.; Cozzone, A.J.; Griffiths, G.; Maridonneau-Parini, I. Tyrosine phosphatase MptpA of Mycobacterium tuberculosis inhibits phagocytosis and increases actin polymerization in macrophages. Res. Microbiol. 2005, 156, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Beresford, N.; Patel, S.; Armstrong, J.; Szöor, B.; Fordham-Skelton, A.P.; Tabernero, L. MptpB, a virulence factor from Mycobacterium tuberculosis, exhibits triple-specificity phosphatase activity. Biochem. J. 2007, 406, 13–18. [Google Scholar] [CrossRef]
- Nören-Müller, A.; Wilk, W.; Saxena, K.; Schwalbe, H.; Kaiser, M.; Waldmann, H. Discovery of a New Class of Inhibitors of Mycobacterium tuberculosis Protein Tyrosine Phosphatase B by Biology-Oriented Synthesis. Angew. Chem. Int. Ed. 2008, 47, 5973–5977. [Google Scholar] [CrossRef]
- Beresford, N.J.; Mulhearn, D.; Szczepankiewicz, B.; Liu, G.; Johnson, M.E.; Fordham-Skelton, A.; Adab-Zapatero, C.; Cavet, J.S.; Tabernero, L.J. Inhibition of MptpB phosphatase from Mycobacterium tuberculosis impairs mycobacterial survival in macrophages. Antimicrob. Chemother. 2009, 63, 928–936. [Google Scholar] [CrossRef]
- Vintonyak, V.V.; Warburg, K.; Kruse, H.; Grimme, S.; Hubel, K.; Rauh, D.; Waldmann, H. Identification of Thiazolidinones Spiro-Fused to Indolin-2-ones as Potent and Selective Inhibitors of the Mycobacterium tuberculosis Protein Tyrosine Phosphatase B. Angew. Chem. Int. Ed. 2010, 49, 5902–5905. [Google Scholar] [CrossRef]
- Ecco, G.; Vernal, J.; Razzera, G.; Martins, P.A.; Matiollo, C.; Terenzi, H. Mycobacterium tuberculosis tyrosine phosphatase A (PtpA) activity is modulated by S-nitrosylation. Chem. Commun. 2010, 46, 7501–7503. [Google Scholar] [CrossRef]
- Silva, A.P.G.; Tabernero, L. New strategies in fighting TB: Targeting Mycobacterium tuberculosis-secreted phosphatases MptpA & MptpB. Future Med. Chem. 2010, 2, 1325–1337. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; He, Y.; Zhang, X.; Xu, J.; Luo, Y.; Wang, Y.; Franzblau, S.G.; Yang, Z.; Chan, R.J.; Liu, Y.; et al. Targeting Mycobacterium protein tyrosine phosphatase B for antituberculosis agents. Proc. Natl. Acad. Sci. USA 2010, 107, 4573–4578. [Google Scholar] [CrossRef]
- Wen, L.; Cai, X.; Xu, F.; She, Z.; Chan, W.; Vrijmoed, L.L.P.; Jones, E.B.G.; Lin, Y. Three Metabolites from the Mangrove Endophytic Fungus Sporothrix sp. (#4335) from the South China Sea. J. Org. Chem. 2009, 74, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Feng, X.; Xiao, Z.; Liu, L.; Li, H.; Ma, L.; Lu, Y.; Ju, J.; She, Z.; Lin, Y. Azaphilones and p-Terphenyls from the Mangrove Endophytic Fungus Penicillium chermesinum (ZH4-E2) Isolated from the South China Sea. J. Nat. Prod. 2011, 74, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huang, H.; Shao, C.; Huang, H.; Jiang, J.; Zhu, X.; Liu, Y.; Liu, L.; Lu, Y.; Li, M.; et al. Cytotoxic Norsesquiterpene Peroxides from the Endophytic Fungus Talaromyces flavus Isolated from the Mangrove Plant Sonneratia apetala. J. Nat. Prod. 2011, 74, 1230–1235. [Google Scholar] [CrossRef]
- Wang, C.; Wang, J.; Huang, Y.H.; Chen, H.; Li, Y.; Zhong, L.L.; Chen, Y.; Chen, S.P.; Wang, J.; Kang, J.L.; et al. Anti-mycobacterial activity of marine fungus-derived 4-deoxybostrycin and nigrosporin. Molecules 2013, 18, 1728–1740. [Google Scholar] [CrossRef]
- Huang, X.S.; Huang, H.B.; Li, H.X.; Sun, X.F.; Huang, H.R.; Lu, Y.J.; Lin, Y.C.; Long, Y.H.; She, Z.G. Asperterpenoid A, a new sesterterpenoid as an inhibitor of Mycobacterium tuberculosis protein tyrosine phosphatase B from the culture of Aspergillus sp. 16-5C. Org. Lett. 2013, 15, 721–723. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Huang, H.; Shao, C.; Xia, X.; Ma, L.; Huang, X.; Lu, Y.; Lin, Y.; Long, Y.; She, Z. Asperterpenols A and B, New Sesterterpenoids Isolated from a Mangrove Endophytic Fungus Aspergillus sp. 085242. Org. Lett. 2013, 15, 2522–2525. [Google Scholar] [CrossRef] [PubMed]
- Li, H.X.; Jiang, J.Y.; Liu, Z.M.; Lin, S.E.; Xia, G.P.; Xia, X.K.; Ding, B.; He, L.; Lu, Y.J.; She, Z.G. Peniphenones A–D from the mangrove fungus Penicillium dipodomyicola HN4–3A as inhibitors of Mycobacterium tuberculosis Phosphatase MptpB. J. Nat. Prod. 2014, 77, 800–806. [Google Scholar] [CrossRef]
- Miller, K.A.; Tsukamoto, S.; Williams, R.M. Asymmetric total syntheses of (+)- and (−)-versicolamide B and biosynthetic implications. Nat. Chem. 2009, 1, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Finefield, J.M.; Sherman, D.H.; Kreitman, M.; Williams, R.M. Enantiomeric Natural Products: Occurrence and Biogenesis. Angew. Chem. Int. Ed. 2012, 51, 4802–4836. [Google Scholar] [CrossRef]
- Lopes, T.I.B.; Coelho, R.G.; Yoshida, N.C.; Honda, N.K. Radical-Scavenging Activity of Orsellinates. Chem. Pharm. Bull. 2008, 56, 1551–1554. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.L.; Porco, J.A. Asymmetric Syntheses of (−)-Mitorubrin and Related Azaphilone Natural Products. Org. Lett. 2006, 8, 5169–5171. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, Y.; Wang, W.; Fu, P.; Liu, P.P.; Zhu, W.M. Anti-influenza Virus Polyketides from the Acid-Tolerant Fungus Penicillium purpurogenum JS03-21. J. Nat. Prod. 2011, 74, 2014–2018. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.A.; Wheeler, M.H. Biosynthesis and function of fungal melanins. Ann. Rev. Phytopathol. 1986, 24, 411–415. [Google Scholar] [CrossRef]
- Bode, H.B.; Zeeck, A. Sphaerolone and dihydrosphaerolone, two bisnaphthyl-pigments from the fungus Sphaeropsidales sp. F-24′707. Phytochemistry 2000, 54, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Zhang, J.; Jiang, N.; Lu, Y.H.; Wang, L.; Xu, S.H.; Wang, W.; Zhang, G.F.; Xu, Q.; Ge, H.M.; et al. Immunosuppressive Polyketides from Mantis-Associated Daldinia eschscholzii. J. Am. Chem. Soc. 2011, 133, 5931–5940. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Ge, H.M.; Zhao, W.; Dong, H.; Xu, Q.; Li, S.H.; Li, J.; Zhang, J.; Song, Y.C.; Tan, R.X. Unprecedented Immunosuppressive Polyketides from Daldinia eschscholzii, a Mantis-Associated Fungus. Angew. Chem. Int. Ed. 2008, 47, 5823–5826. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Z.; Lin, S.; Tan, C.; Lu, Y.; He, L.; Huang, X.; She, Z. Asperlones A and B, Dinaphthalenone Derivatives from a Mangrove Endophytic Fungus Aspergillus sp. 16-5C. Mar. Drugs 2015, 13, 366-378. https://doi.org/10.3390/md13010366
Xiao Z, Lin S, Tan C, Lu Y, He L, Huang X, She Z. Asperlones A and B, Dinaphthalenone Derivatives from a Mangrove Endophytic Fungus Aspergillus sp. 16-5C. Marine Drugs. 2015; 13(1):366-378. https://doi.org/10.3390/md13010366
Chicago/Turabian StyleXiao, Ze'en, Shao'e Lin, Chunbing Tan, Yongjun Lu, Lei He, Xishan Huang, and Zhigang She. 2015. "Asperlones A and B, Dinaphthalenone Derivatives from a Mangrove Endophytic Fungus Aspergillus sp. 16-5C" Marine Drugs 13, no. 1: 366-378. https://doi.org/10.3390/md13010366
APA StyleXiao, Z., Lin, S., Tan, C., Lu, Y., He, L., Huang, X., & She, Z. (2015). Asperlones A and B, Dinaphthalenone Derivatives from a Mangrove Endophytic Fungus Aspergillus sp. 16-5C. Marine Drugs, 13(1), 366-378. https://doi.org/10.3390/md13010366