Matrix Production, Pigment Synthesis, and Sporulation in a Marine Isolated Strain of Bacillus pumilus
Abstract
:1. Introduction

2. Results and Discussion
2.1. Matrix Genes in B. pumilus ATCC 7061T and SF214

| Genes | Putative Encoded Protein | Protein Identity (%) | |
|---|---|---|---|
| B. subtilis 168 vs. B. pumilus ATCC 7061T | B. pumilus ATCC 7061T vs. B. pumilus SF214 | ||
| Eps Operon | |||
| epsO | pyruvil transferase | - | - |
| epsN | pyridoxal phosphate-dependent aminotransferase | 68 | 97 |
| epsM | acetyl transferase | 50 | 94 |
| epsL | sugar transferase | 65 | 97 |
| epsK | membrane protein | 50 | 96 |
| epsJ | glycosyl transferase | - | - |
| epsI | pyruvil transferase | 60 | 95 |
| epsH | glycosyl transferase | 46 | 96 |
| epsG | membrane protein | 72 | 96 |
| epsF | glycosyl transferase | 54 | 95 |
| epsE | glycosyl transferase | 61 | 96 |
| epsD | glycosyl transferase | 54 | 94 |
| epsC | polysaccharide biosynthesis protein | 62 | 99 |
| epsB | tyrosine-protein kinase | 63 | 99 |
| epsA | capsular biosynthesis protein | 43 | 100 |
| TasA Operon | |||
| tapA | lipoprotein for formation | 42 | 93 |
| sipW | signal peptidase I | 52 | 98 |
| tasA | spore coat protein N | 62 | 97 |
2.2. Physiological and Chemical Characterization of the Matrix of B. pumilus SF214


| Monosaccharide | Relative Abundance a |
|---|---|
| Galactose | 1.00 |
| Mannose | 0.75 |
| Glucosamine | 0.54 |
| Glucose | 0.52 |
| Galacturonic acid | 0.21 |
| Glucuronic acid | 0.15 |
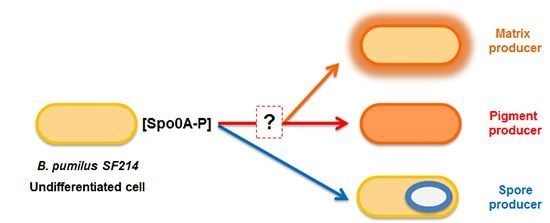
2.3. Mutants with Altered Pigmentation Are Also Altered in Matrix Formation and Sporulation




2.4. In Search of the Mutations
| Genes | Putative Encoded Protein | Protein Identity (%) | |
|---|---|---|---|
| B. subtilis 168 vs. B. pumilus ATCC 7061T | B. pumilus ATCC 7061T vs. B. pumilus SF214 | ||
| sinI | sinR antagonist | 45 | 96 |
| sinR | transcriptional regulator | 93 | 98 |
| spo0A | sporulation protein A | 88 | 100 |
| spo0B | phosphotransferase B | 58 | 98 |
| spo0F | phosphotransferase F | 92 | 100 |
3. Experimental Section
3.1. Bacterial Strains and Growth Conditions
3.2. Fluorescence Microscopy
3.3. Biofilm Production Assay
3.4. EPS Extraction
3.5. Purification of Exopolysaccharides
3.6. Carbohydrate Compositional Analysis
3.7. DNA Extraction and PCR Procedure
3.8. Bioinformatic Analysis
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- McKenney, P.T.; Driks, A.; Eichemberger, P. The Bacillus subtilis endospore: Assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 2013, 11, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, W.J.; Munakata, N.; Horneck, G.; Melosh, H.J.; Setlow, P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 2000, 64, 548–572. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.A.; To, E.; Fakhry, S.; Baccigalupi, L.; Ricca, E.; Cutting, S.M. Defining the natural habitat of Bacillus sporeformers. Res. Microbiol. 2009, 160, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Dworkin, J.; Shah, I.M. Exit from dormancy in microbial organisms. Nat. Rev. Microbiol. 2010, 8, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Lopez, D.; Vlamakis, H.; Kolter, R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 2009, 33, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Vlamakis, H.; Aguilar, C.; Losick, R.; Kolter, R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008, 22, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Norman, T.; Kolter, R.; Losick, R. Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillus subtilis. EMBO J. 2011, 30, 1402–1413. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Chu, F.; Kolter, R.; Losick, R. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 2008, 67, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Veening, J.W.; Smits, W.K.; Kuipers, O.P. Bistability, epigenetics, and bet-hedging in bacteria. Ann. Rev. Microbiol. 2008, 62, 193–210. [Google Scholar] [CrossRef] [PubMed]
- Vlamakis, H.; Chai, Y.; Beauregard, P.; Losick, R.; Kolter, R. Sticking together: Building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 2013, 11, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Mielich-Suss, B.; Lopez, D. Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ. Microbiol. 2015, 17, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Leiman, S.A.; Arboleda, L.C.; Spina, J.S.; McLoon, A.L. SinR is a mutational target for fine-tuning biofilm formation in laboratory-evolved strains of Bacillus subtilis. BMC Microbiol. 2014, 14, 301. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; Gonzalez-Pastor, J.E.; Ben-Yehuda, S.; Losick, R.; Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2001, 98, 11621–11626. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Foulston, L.; Chai, Y.; Wang, Q.; Losick, R. Alternative modes of biofilm formation by plant-associated Bacillus cereus. Microbiologyopen 2015, 4, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Hayrapetyan, H.; Muller, L.; Tempelaars, M.; Abee, T.; Groot, M.N. Comparative analysis of biofilm formation by Bacillus cereus reference strains and undomesticated food isolates and the effect of free iron. Int. J. Food Microbiol. 2015, 200, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Verplaetse, E.; Slamti, L.; Gohar, M.; Lereclus, D. Cell differentiation in a Bacillus thuringiensis population during planktonic growth, biofilm formation and host infection. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Khaneja, R.; Perez-Fons, L.; Fakhry, S.; Baccigalupi, L.; Steiger, S.; To, E.; Sandmann, G.; Dong, T.C.; Ricca, E.; Fraser, P.D.; Cutting, S.M. Carotenoids found in Bacillus. J. Appl. Microbiol. 2010, 108, 1889–1902. [Google Scholar] [PubMed]
- Manzo, N.; di Luccia, B.; Isticato, R.; D’Apuzzo, E.; de Felice, M.; Ricca, E. Pigmentation and sporulation are alternative cell fates in Bacillus pumilus SF214. PLoS ONE 2013, 8, e62093. [Google Scholar] [CrossRef] [PubMed]
- Nagorska, K.; Ostrowski, A.; Hinc, K.; Holland, I.B.; Obuchowski, M. Importance of eps genes from Bacillus subtilis in biofilm formation and swarming. J. Appl. Genet. 2010, 51, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; Vlamakis, H.; Losick, R.; Kolter, R. Functional analysis of the accessory protein TapA in Bacillus subtilis amyloid fiber assembly. J. Bacteriol. 2014, 196, 1505–1513. [Google Scholar] [CrossRef] [PubMed]
- McColl, D.J.M.; Silva, A.; Foley, M.; Kun, J.F.; Favaloro, J.M.; Thompson, J.K.; Marshall, V.M.; Coppel, R.L.; Kemp, D.J.; Anders, R.F. Molecular variation in a novel polymorphic antigen associated with Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 1994, 68, 53–67. [Google Scholar] [CrossRef]
- Mulhern, T.D.; Howlett, G.J.; Reid, G.E.; Simpson, R.J.; McColl, D.J.; Anders, R.F.; Norton, R.S. Solution structure of a polypeptide containing four heptad repeat units from a merozoite surface antigen of Plasmodium falciparum. Biochemistry 1995, 34, 3479–3491. [Google Scholar] [CrossRef] [PubMed]
- Lazarevic, V.; Soldo, B.; Médico, N.; Pooley, H.; Bron, S.; Karamata, D. Bacillus subtiis alpha-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl. Environ. Microbiol. 2005, 71, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Cozy, L.M.; Phillips, A.M.; Calvo, R.A.; Bate, A.R.; Hsueh, Y.H.; Bonneau, R.; Eichenberger, P.; Kearns, D.B. SlrA/SinR/SlrR inhibits motility gene expression upstream of a hypersensitive and hysteretic switch at the level of σD in Bacillus subtilis. Mol. Microbiol. 2012, 83, 1210–1228. [Google Scholar] [CrossRef] [PubMed]
- Laffey, S.F.; Butler, G. Phenotype switching affects biofilm formation by Candida parapsilosis. Microbiology 2005, 151, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.Y.; Lee, K-M.; Park, Y.; Yoon, S.S. Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS ONE 2011, 6, e16105. [Google Scholar] [CrossRef] [PubMed]
- Winkelman, J.T.; Blair, K.M.; Kearns, D.B. RemA (YlzA) and RemB (YaaB) regulate extracellular matrix operon expression and biofilm formation in Bacillus subtilis. J. Bacteriol. 2009, 191, 3981–3991. [Google Scholar] [CrossRef] [PubMed]
- Winkelman, J.T.; Bree, A.C.; Bate, A.R.; Eichenberger, P.; Gourse, R.L.; Kearns, D.B. RemA is a DNA-binding protein that activates biofilm matrix gene expression in Bacillus subtilis. Mol. Microbiol. 2013, 88, 984–997. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Oshima, T.; Ogasawara, N.; Ishikawa, S. Functional analysis of the protein Veg, which stimulates biofilm formation in Bacillus subtilis. J. Bacteriol. 2013, 195, 1697–1705. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K. SlrR/SlrA controls the initiation of biofilm formation in Bacillus subtilis. Mol. Microbiol. 2008, 69, 1399–1410. [Google Scholar] [PubMed]
- Zhao, X.; Wang, Y.; Shang, Q.; Li, Y.; Hao, H.; Zhang, Y.; Guo, Z.; Yang, G.; Xie, Z.; Wang, R. Collagen-like proteins (ClpA, ClpB, ClpC, and ClpD) are required for biofilm formation and adhesion to plant roots by Bacillus amyloliquefaciens FZB42. PLoS ONE 2015, 10, e0117414. [Google Scholar] [CrossRef] [PubMed]
- Bendori, O.; Pollak, S.; Hizi, D.; Eldar, A. The RapP-PhrP quorum-sensing system of Bacillus subtilis strain NCIB3610 affects biofilm formation through multiple targets, due to an atypical signal-insensitive allele of RapP. J. Bacteriol. 2015, 197, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Takada, H.; Morita, M.; Shiwa, Y.; Sugimoto, R.; Suzuki, S.; Kawamura, F.; Yoshikawa, H. Cell motility and biofilm formation in Bacillus subtilis are affected by the ribosomal proteins, S11 and S21. Biosci. Biotechnol. Biochem. 2014, 78, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Chagneau, C.; Saier, M.H., Jr. Biofilm-defective mutants of Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 2004, 8, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Youngman, P.; Perkins, J.B.; Losick, R. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertion. Mol. Gen. Genet. 1984, 195, 424–433. [Google Scholar] [CrossRef] [PubMed]
- De Castro, C.; Parrilli, M.; Holst, O.; Molinaro, A. Microbe-associated molecular patterns in innate immunity: Extraction and chemical analysis of gram-negative bacterial lipopolysaccharides. Meth. Enzymol. 2010, 480, 89–115. [Google Scholar] [PubMed]
- Cutting, S.; Vander Horn, P.B. Genetic analysis. In Molecular Biological Methods for Bacillus; Harwood, C., Cutting, S.M., Eds.; John Wiley and Sons: Chichester, UK, 1990; pp. 27–74. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Luccia, B.; Riccio, A.; Vanacore, A.; Baccigalupi, L.; Molinaro, A.; Ricca, E. Matrix Production, Pigment Synthesis, and Sporulation in a Marine Isolated Strain of Bacillus pumilus. Mar. Drugs 2015, 13, 6472-6488. https://doi.org/10.3390/md13106472
Di Luccia B, Riccio A, Vanacore A, Baccigalupi L, Molinaro A, Ricca E. Matrix Production, Pigment Synthesis, and Sporulation in a Marine Isolated Strain of Bacillus pumilus. Marine Drugs. 2015; 13(10):6472-6488. https://doi.org/10.3390/md13106472
Chicago/Turabian StyleDi Luccia, Blanda, Antonio Riccio, Adele Vanacore, Loredana Baccigalupi, Antonio Molinaro, and Ezio Ricca. 2015. "Matrix Production, Pigment Synthesis, and Sporulation in a Marine Isolated Strain of Bacillus pumilus" Marine Drugs 13, no. 10: 6472-6488. https://doi.org/10.3390/md13106472
APA StyleDi Luccia, B., Riccio, A., Vanacore, A., Baccigalupi, L., Molinaro, A., & Ricca, E. (2015). Matrix Production, Pigment Synthesis, and Sporulation in a Marine Isolated Strain of Bacillus pumilus. Marine Drugs, 13(10), 6472-6488. https://doi.org/10.3390/md13106472